Genome-Wide Identification, Characterization and Expression Profile of F-Box Protein Family Genes Shed Light on Lateral Branch Development in Cultivated Peanut (Arachis hypogaea L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome–Wide Identification and Chromosome Localization
2.2. Physicochemical Properties, Phylogenetic Tree and Gene Structure Analysis
2.3. Prediction of Cis–Acting Elements in the Promoter Region
2.4. Collinearity and Estimation of Ka/Ks Ratios Analysis
2.5. In Silico Expression Analysis of AhFBX Genes in Different Tissues
2.6. Plant Materials
2.7. Transcription Expression Analysis in Lateral Branch Development
2.8. RNA Extracted and qRT–PCR Analysis
3. Results
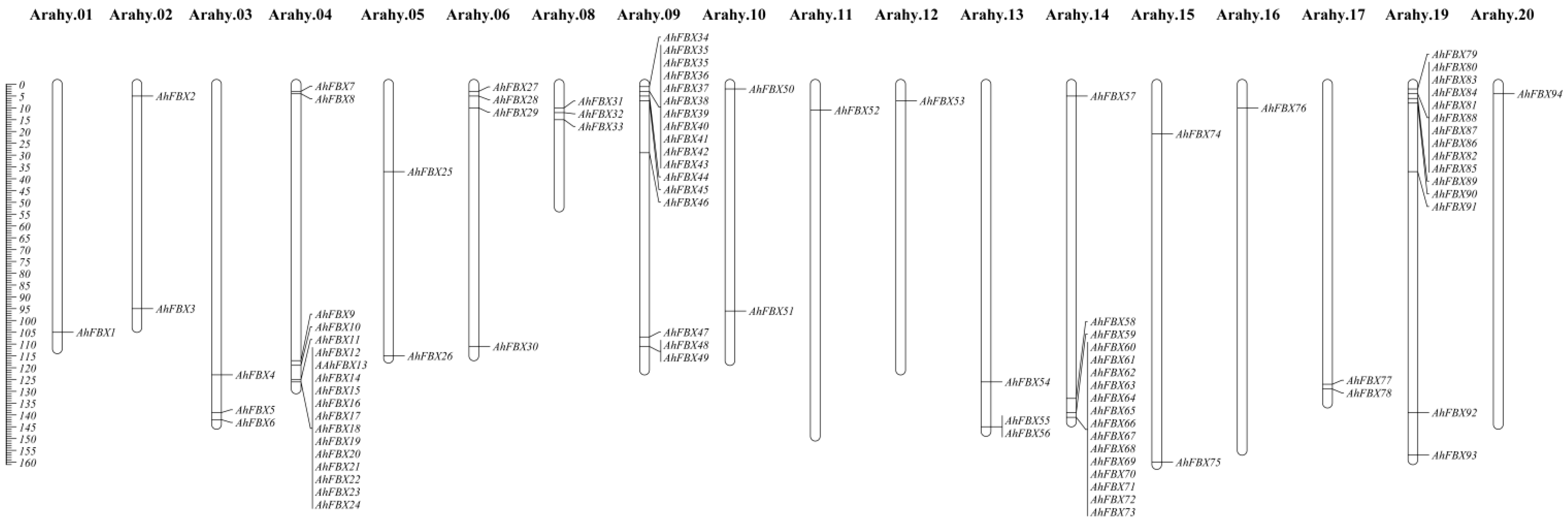
3.1. Identification and Chromosome Location of AhFBXs
3.2. Physicochemical Properties and Subcellular Localization Prediction of AhFBXs
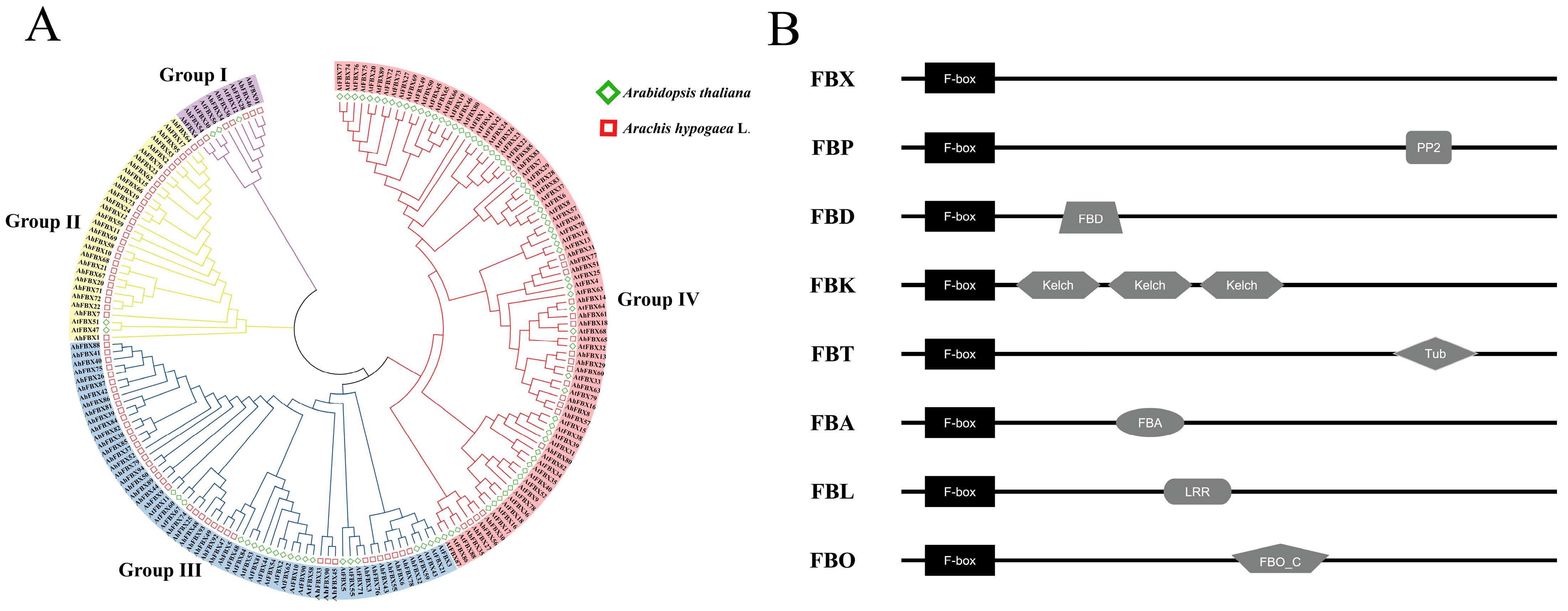
3.3. Classification and Phylogenetic Tree of AhFBXs
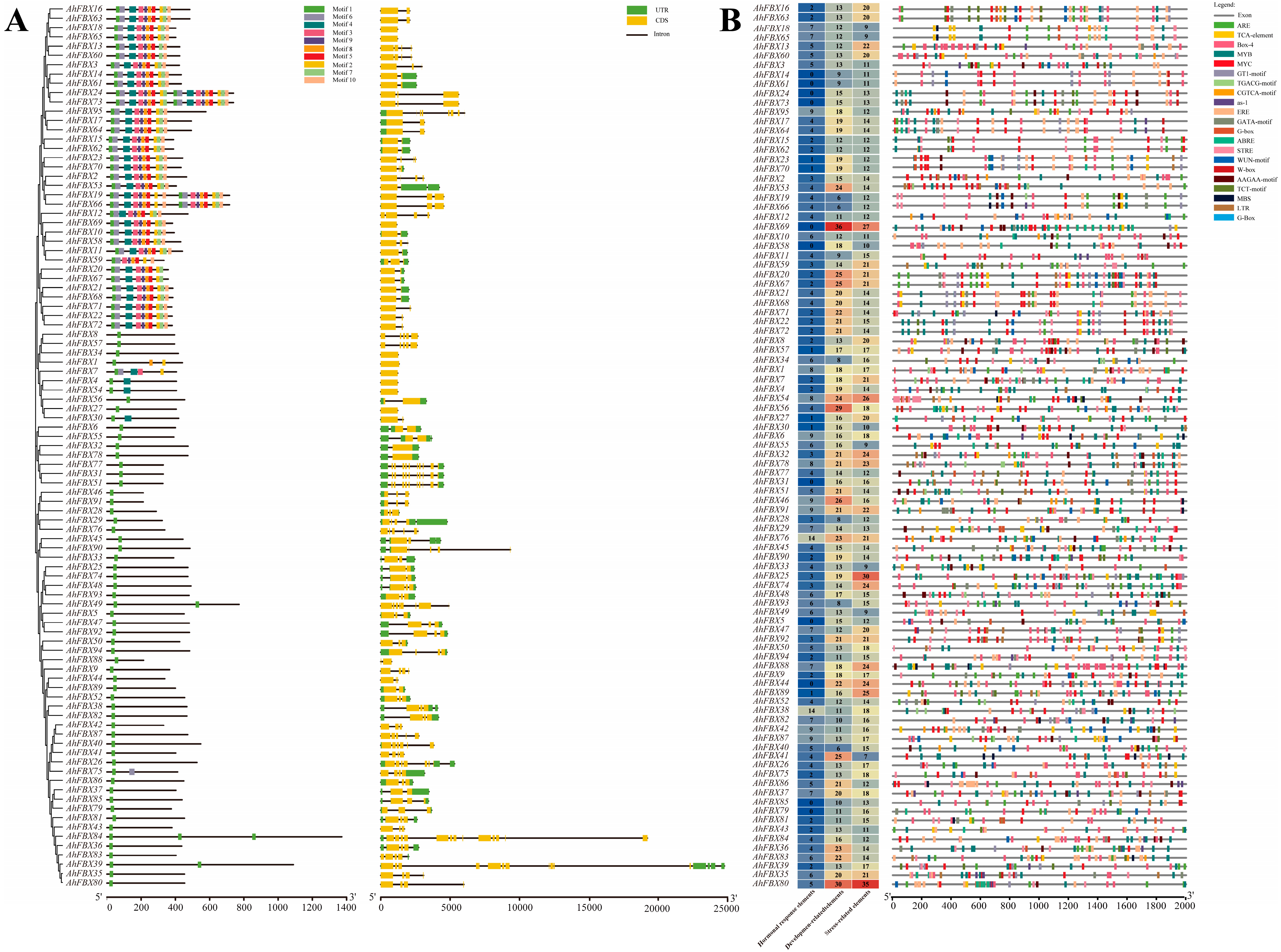
3.4. Conserved Motifs and Gene Structural of AhFBXs
3.5. Cis–Acting Elements in the Promoter Region of AhFBXs
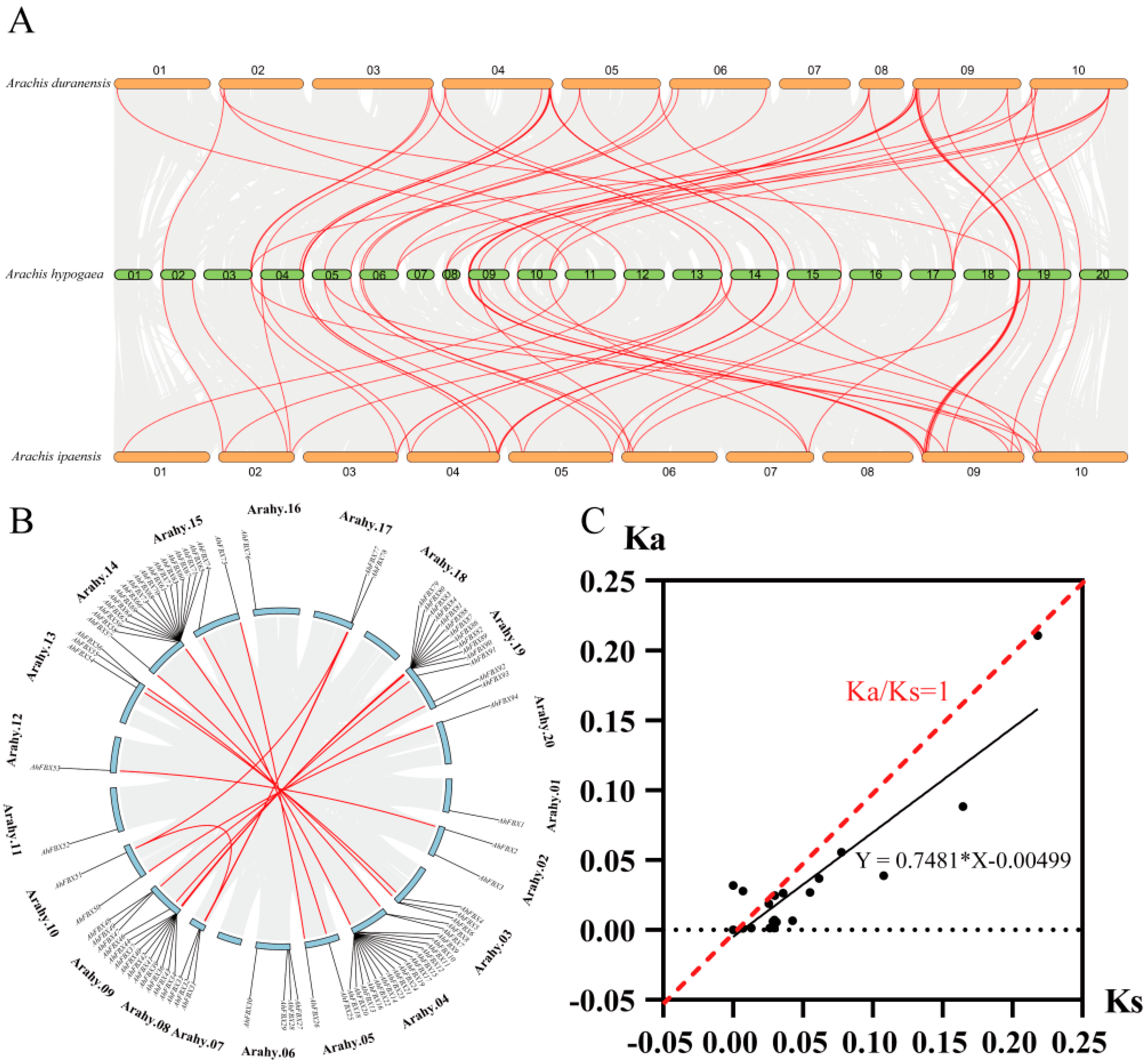
3.6. Collinearity and Estimation of Ka/Ks Ratios of AhFBXs
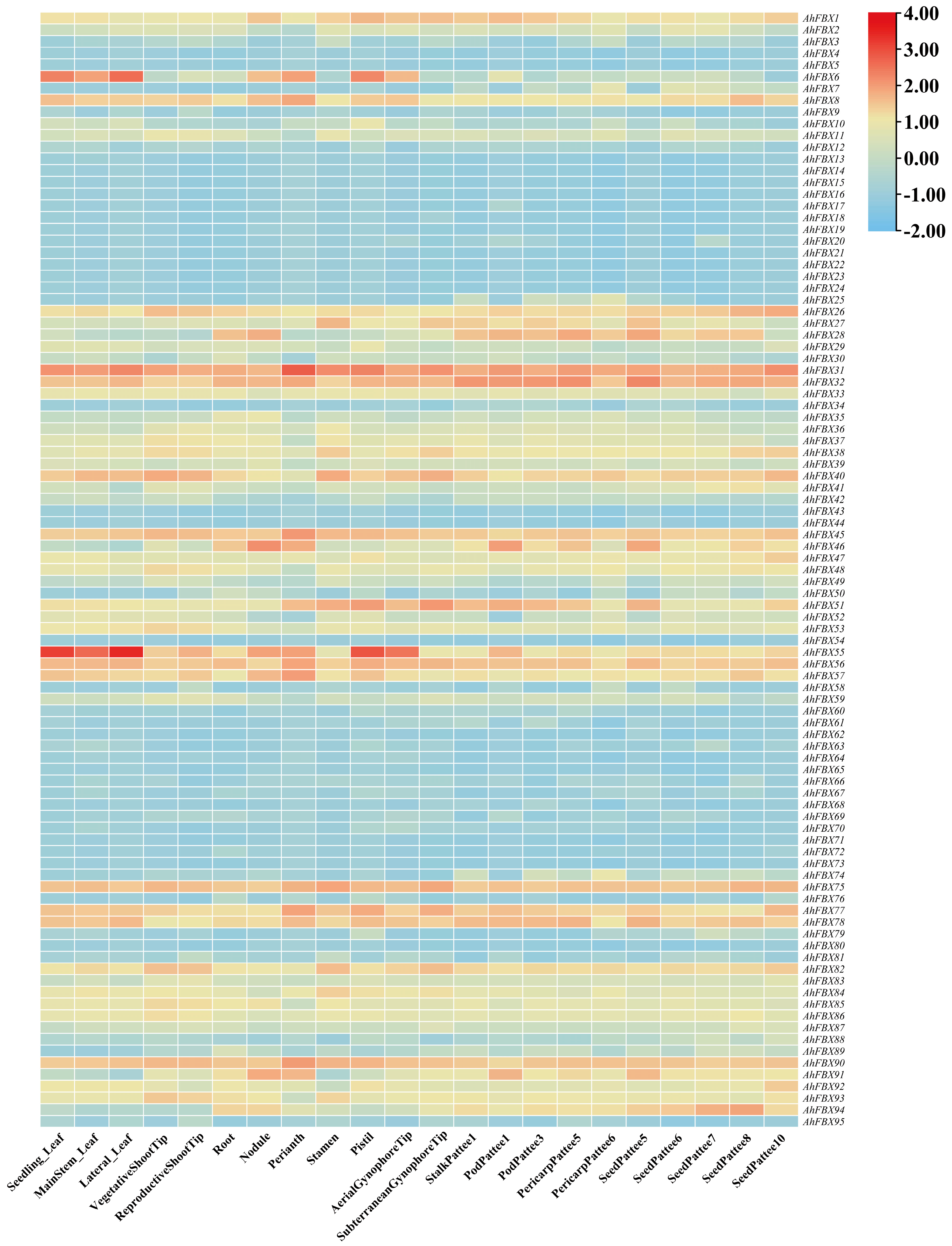
3.7. In Silico Expression Patterns of AhFBXs in Different Tissues
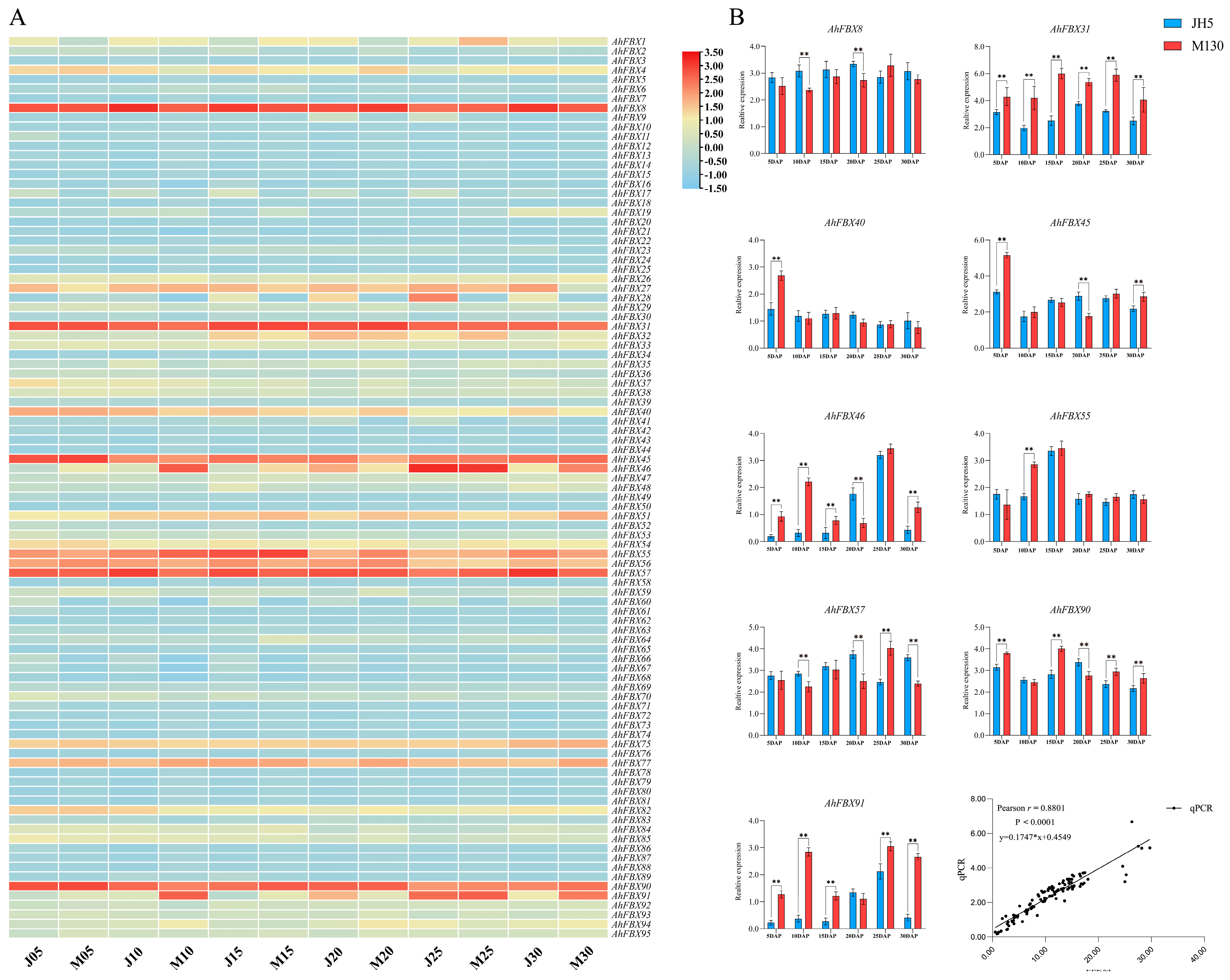
3.8. Expression of AhFBX Genes during the Lateral Branch Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sadanandom, A.; Bailey, M.; Ewan, R.; Lee, J.; Nelis, S. The ubiquitin-proteasome system: Central modifier of plant signalling. New Phytol. 2012, 196, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hellmann, H. Plant E3 Ligases: Flexible Enzymes in a Sessile World. Mol. Plant 2013, 6, 1388–1404. [Google Scholar] [CrossRef] [PubMed]
- Somers, D.E.; Fujiwara, S. Thinking outside the F-box: Novel ligands for novel receptors. Trends Plant Sci. 2009, 14, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Lechner, E.; Achard, P.; Vansiri, A.; Potuschak, T.; Genschik, P. F-box proteins everywhere. Curr. Opin. Plant Biol. 2006, 9, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Kipreos, E.T.; Pagano, M. The F-box protein family. Genome Biol. 2000, 1, s3001–s3002. [Google Scholar] [CrossRef]
- Xu, G.; Ma, H.; Nei, M.; Kong, H. Evolution of F-box genes in plants: Different modes of sequence divergence and their relationships with functional diversification. Proc. Natl. Acad. Sci. USA 2009, 106, 835–840. [Google Scholar] [CrossRef]
- Gagne, J.M.; Downes, B.P.; Shiu, S.; Durski, A.M.; Vierstra, R.D. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 11519–11524. [Google Scholar] [CrossRef]
- Jain, M.; Nijhawan, A.; Arora, R.; Agarwal, P.; Ray, S.; Sharma, P.; Kapoor, S.; Tyagi, A.K.; Khurana, J.P. F-box proteins in rice. genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 2007, 143, 1467–1483. [Google Scholar] [CrossRef]
- Jia, F.; Wu, B.; Li, H.; Huang, J.; Zheng, C. Genome-wide identification and characterisation of F-box family in maize. Mol. Genet. Genom. 2013, 288, 559–577. [Google Scholar] [CrossRef] [PubMed]
- Song, J.B.; Wang, Y.X.; Li, H.B.; Li, B.W.; Zhou, Z.S.; Gao, S.; Yang, Z.M. The F-box family genes as key elements in response to salt, heavy mental, and drought stresses in Medicago truncatula. Funct. Integr. Genom. 2015, 15, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.; Xiao, Z.X.; Wong, F.L.; Sun, S.; Liang, K.J.; Lam, H.M. Genome-wide analyses of the soybean F-box gene family in response to salt stress. Int. J. Mol. Sci. 2017, 18, 818. [Google Scholar] [CrossRef]
- Mo, F.; Zhang, N.; Qiu, Y.; Meng, L.; Cheng, M.; Liu, J.; Yao, L.; Lv, R.; Liu, Y.; Zhang, Y.; et al. Molecular characterization, gene evolution and expression analysis of the F-box gene family in Tomato (Solanum lycopersicum). Genes 2021, 12, 417. [Google Scholar] [CrossRef]
- Wang, Y.; Li, C.; Yan, S.; Yu, B.; Gan, Y.; Liu, R.; Qiu, Z.; Cao, B. Genome-wide analysis and characterization of eggplant F-box gene superfamily: Gene evolution and expression analysis under stress. Int. J. Mol. Sci. 2022, 23, 16049. [Google Scholar] [CrossRef]
- Feng, C.H.; Niu, M.X.; Liu, X.; Bao, Y.; Liu, S.; Liu, M.; He, F.; Han, S.; Liu, C.; Wang, H.L.; et al. Genome-wide analysis of the FBA Subfamily of the poplar F-box gene family and its role under drought stress. Int. J. Mol. Sci. 2023, 24, 4823. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Luong, P.; Huq, E. The F-box Protein MAX2 Functions as a Positive Regulator of Photomorphogenesis in Arabidopsis. Plant Physiol. 2007, 145, 1471–1483. [Google Scholar] [CrossRef] [PubMed]
- Song, J.B.; Huang, S.Q.; Dalmay, T.; Yang, Z.M. Regulation of leaf morphology by microRNA394 and its target LEAF CURLING RESPONSIVENESS. Plant Cell Physiol. 2012, 53, 1283–1294. [Google Scholar] [CrossRef] [PubMed]
- Marrocco, K.; Zhou, Y.; Bury, E.; Dieterle, M.; Funk, M.; Genschik, P.; Krenz, M.; Stolpe, T.; Kretsch, T. Functional analysis of EID1, an F-box protein involved in phytochrome A-dependent light signal transduction. Plant J. 2006, 45, 423–438. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Tian, S.; Zhang, W.; Dong, X.; Ma, C.; Wang, Y.; Yan, J.; Yue, B. Q(Dtbn1), an F-box gene affecting maize tassel branch number by a dominant model. Plant Biotechnol. J. 2021, 19, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.C.M.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lu, Q.; Liu, H.; Zhang, J.; Hong, Y.; Lan, H.; Li, H.; Wang, J.; Liu, H.; Li, S.; et al. Sequencing of cultivated peanut, Arachis hypogaea, yields insights into genome evolution and oil improvement. Mol. Plant. 2019, 12, 920–934. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Peng, Z.; Liu, Y.; Li, G.; Wan, S. Genome-wide analysis of the SNARE family in cultivated peanut (Arachis hypogaea L.) reveals that some members are involved in stress responses. Int. J. Mol. Sci. 2023, 24, 7103. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xiao, L.; Chi, J.; Li, R.; Han, Y.; Cui, F.; Peng, Z.; Wan, S.; Li, G. Genome-wide identification and expression of SAUR gene family in peanut (Arachis hypogaea L.) and functional identification of AhSAUR3 in drought tolerance. BMC Plant Biol. 2022, 22, 178. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, Y.; Huai, D.; Chen, Y.; Jiang, Y.; Ding, Y.; Kang, Y.; Wang, Z.; Yan, L.; Jiang, H.; et al. Genome-wide identification of peanut PIF family genes and their potential roles in early pod development. Gene 2021, 781, 145539. [Google Scholar] [CrossRef]
- Han, Y.; Li, R.; Liu, Y.; Fan, S.; Wan, S.; Zhang, X.; Li, G. The major intrinsic protein family and their function under salt-stress in peanut. Front. Genet. 2021, 12, 639585. [Google Scholar] [CrossRef]
- Gallavotti, A. The role of auxin in shaping shoot architecture. J. Exp. Bot. 2013, 64, 2593–2608. [Google Scholar] [CrossRef]
- Pittman, R.N. United States Peanut Descriptors; Agricultural Research Service of USDA: Washington, DC, USA, 1995; pp. 2–3. [Google Scholar]
- Kayam, G.; Brand, Y.; Faigenboim-Doron, A.; Patil, A.; Hedvat, I.; Hovav, R. Fine-mapping the branching habit trait in cultivated peanut by combining bulked segregant analysis and high-throughput sequencing. Front. Plant Sci. 2017, 8, 467. [Google Scholar] [CrossRef][Green Version]
- Donald, C.M. The breeding of crop ideotypes. Euphytica 1968, 17, 385–403. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, H.; Ma, B.; Liu, G.; Wang, J.; Wang, J.; Gao, R.; Li, J.; Liu, J.; Xu, J.; et al. A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat. Commun. 2017, 8, 14789. [Google Scholar] [CrossRef]
- Tian, J.; Wang, C.; Xia, J.; Wu, L.; Xu, G.; Wu, W.; Li, D.; Qin, W.; Han, X.; Chen, Q.; et al. Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 2016, 365, 658–664. [Google Scholar] [CrossRef]
- Li, L.; Cui, S.; Dang, P.; Yang, X.; Wei, X.; Chen, K.; Liu, L.; Chen, C.Y. GWAS and bulked segregant analysis reveal the Loci controlling growth habit-related traits in cultivated Peanut (Arachis hypogaea L.). BMC Genom. 2022, 23, 403. [Google Scholar] [CrossRef]
- Zhao, N.; He, M.; Li, L.; Cui, S.; Hou, M.; Wang, L.; Mu, G.; Liu, L.; Yang, X. Identification and expression analysis of WRKY gene family under drought stress in peanut (Arachis hypogaea L.). PLoS ONE 2020, 15, e0231396. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Clevenger, J.; Chu, Y.; Scheffler, B.; Ozias-Akins, P. A developmental transcriptome map for allotetraploid Arachis hypogaea. Front. Plant Sci. 2016, 7, 1446. [Google Scholar] [CrossRef]
- Li, J.; Ma, Y.; Hu, M.; Zhao, Y.; Liu, B.; Wang, C.; Zhang, M.; Zhang, L.; Yang, X.; Mu, G. Multi-omics and miRNA interaction joint analysis highlight new insights into anthocyanin biosynthesis in peanuts (Arachis hypogaea L.). Front. Plant Sci. 2022, 13, 818345. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Moon, J.; Parry, G.; Estelle, M. The ubiquitin-proteasome pathway and plant development. Plant Cell. 2004, 16, 3181–3195. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, Y.; Luo, W.; Li, W.; Chen, N.; Zhang, D.; Chong, K. The F-box protein OsFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in rice. Plant Physiol. 2013, 163, 1673–1685. [Google Scholar] [CrossRef]
- Zhang, X.; Gou, M.; Liu, C.J. Arabidopsis Kelch repeat F-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell. 2013, 25, 4994–5010. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.P.; Lee, C.L.; Chen, P.H.; Wu, S.H.; Yang, C.C.; Shaw, J.F. Molecular analyses of the Arabidopsis TUBBY-like protein gene family. Plant Physiol. 2004, 134, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Swamy, M.J.; Mondal, S. Subunit association, and thermal and chemical unfolding of Cucurbitaceae phloem exudate lectins. A review. Int. J. Biol. Macromol. 2023, 233, 123434. [Google Scholar] [CrossRef]
- Dinant, S.; Clark, A.M.; Zhu, Y.; Vilaine, F.; Palauqui, J.C.; Kusiak, C.; Thompson, G.A. Diversity of the superfamily of phloem lectins (phloem protein 2) in angiosperms. Plant Physiol. 2003, 131, 114–128. [Google Scholar] [CrossRef] [PubMed]
- DeFalco, T.A.; Anne, P.; James, S.R.; Willoughby, A.C.; Schwanke, F.; Johanndrees, O.; Genolet, Y.; Derbyshire, P.; Wang, Q.; Rana, S.; et al. A conserved module regulates receptor kinase signalling in immunity and development. Nat. Plants 2022, 8, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Young, N.D.; Debellé, F.; Oldroyd, G.E.; Geurts, R.; Cannon, S.B.; Udvardi, M.K.; Benedito, V.A.; Mayer, K.F.; Gouzy, J.; Schoof, H.; et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 2011, 480, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Gilmartin, P.M.; Sarokin, L.; Memelink, J.; Chua, N.H. Molecular light switches for plant genes. Plant Cell 1990, 2, 369–378. [Google Scholar]
- Schindler, U.; Menkens, A.E.; Beckmann, H.; Ecker, J.R.; Cashmore, A.R. Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO J. 1992, 11, 1261–1273. [Google Scholar] [CrossRef]
- Liu, T.L.; Newton, L.; Liu, M.J.; Shiu, S.H.; Farré, E.M. A G-Box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol. 2016, 170, 528–539. [Google Scholar] [CrossRef]
- Ahmad, N.; Hou, L.; Ma, J.; Zhou, X.; Xia, H.; Wang, M.; Leal-Bertioli, S.; Zhao, S.; Tian, R.; Pan, J.; et al. Bulk RNA-Seq analysis reveals differentially expressed genes associated with lateral branch angle in peanut. Genes 2022, 13, 841. [Google Scholar] [CrossRef]
- Lama, S.; Broda, M.; Abbas, Z.; Vaneechoutte, D.; Belt, K.; Säll, T.; Vandepoele, K.; Van Aken, O. Neofunctionalization of mitochondrial proteins and incorporation into signaling networks in plants. Mol. Biol. Evol. 2019, 36, 974–989. [Google Scholar] [CrossRef]
- Schwager, K.M.; Calderon-Villalobos, L.I.; Dohmann, E.M.; Willige, B.C.; Knierer, S.; Nill, C.; Schwechheimer, C. Characterization of the VIER F-box PROTEINE genes from Arabidopsis reveals their importance for plant growth and development. Plant Cell 2007, 19, 1163–1178. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Abrahan, C.; Colquhoun, T.A.; Liu, C.J. A proteolytic regulator controlling chalcone synthase stability and flavonoid biosynthesis in Arabidopsis. Plant Cell 2017, 29, 1157–1174. [Google Scholar] [CrossRef]
- Li, Y.; Xue, S.; He, Q.; Wang, J.; Zhu, L.; Zou, J.; Zhang, J.; Zuo, C.; Fan, Z.; Yue, J.; et al. Arabidopsis F-BOX STRESS INDUCED 4 is required to repress excessive divisions in stomatal development. J. Integr. Plant Biol. 2022, 64, 56–72. [Google Scholar] [CrossRef]
- Li, L.; Yang, X.; Cui, S.; Meng, X.; Mu, G.; Hou, M.; He, M.; Zhang, H.; Liu, L.; Chen, C.Y. Construction of high-density genetic map and mapping quantitative trait loci for growth habit-related traits of peanut (Arachis hypogaea L.). Front. Plant Sci. 2019, 10, 745. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Guo, L.; Wang, W.; Miao, P.; Mu, G.; Chen, C.Y.; Meng, C.; Yang, X. Genome-Wide Identification, Characterization and Expression Profile of F-Box Protein Family Genes Shed Light on Lateral Branch Development in Cultivated Peanut (Arachis hypogaea L.). Horticulturae 2024, 10, 255. https://doi.org/10.3390/horticulturae10030255
Li C, Guo L, Wang W, Miao P, Mu G, Chen CY, Meng C, Yang X. Genome-Wide Identification, Characterization and Expression Profile of F-Box Protein Family Genes Shed Light on Lateral Branch Development in Cultivated Peanut (Arachis hypogaea L.). Horticulturae. 2024; 10(3):255. https://doi.org/10.3390/horticulturae10030255
Chicago/Turabian StyleLi, Chuheng, Liguo Guo, Weiqian Wang, Penghui Miao, Guojun Mu, Charles Y. Chen, Chengsheng Meng, and Xinlei Yang. 2024. "Genome-Wide Identification, Characterization and Expression Profile of F-Box Protein Family Genes Shed Light on Lateral Branch Development in Cultivated Peanut (Arachis hypogaea L.)" Horticulturae 10, no. 3: 255. https://doi.org/10.3390/horticulturae10030255
APA StyleLi, C., Guo, L., Wang, W., Miao, P., Mu, G., Chen, C. Y., Meng, C., & Yang, X. (2024). Genome-Wide Identification, Characterization and Expression Profile of F-Box Protein Family Genes Shed Light on Lateral Branch Development in Cultivated Peanut (Arachis hypogaea L.). Horticulturae, 10(3), 255. https://doi.org/10.3390/horticulturae10030255






