Characterization and Comprehensive Evaluation of Phenotypic and Yield Traits in Salt-Stress-Tolerant Peanut Germplasm for Conservation and Breeding
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Experimental Design
2.2.1. Screening of Salt Tolerance of Peanuts during Germination
2.2.2. Screening of Salt Tolerance of Peanuts at the Seedling Stage
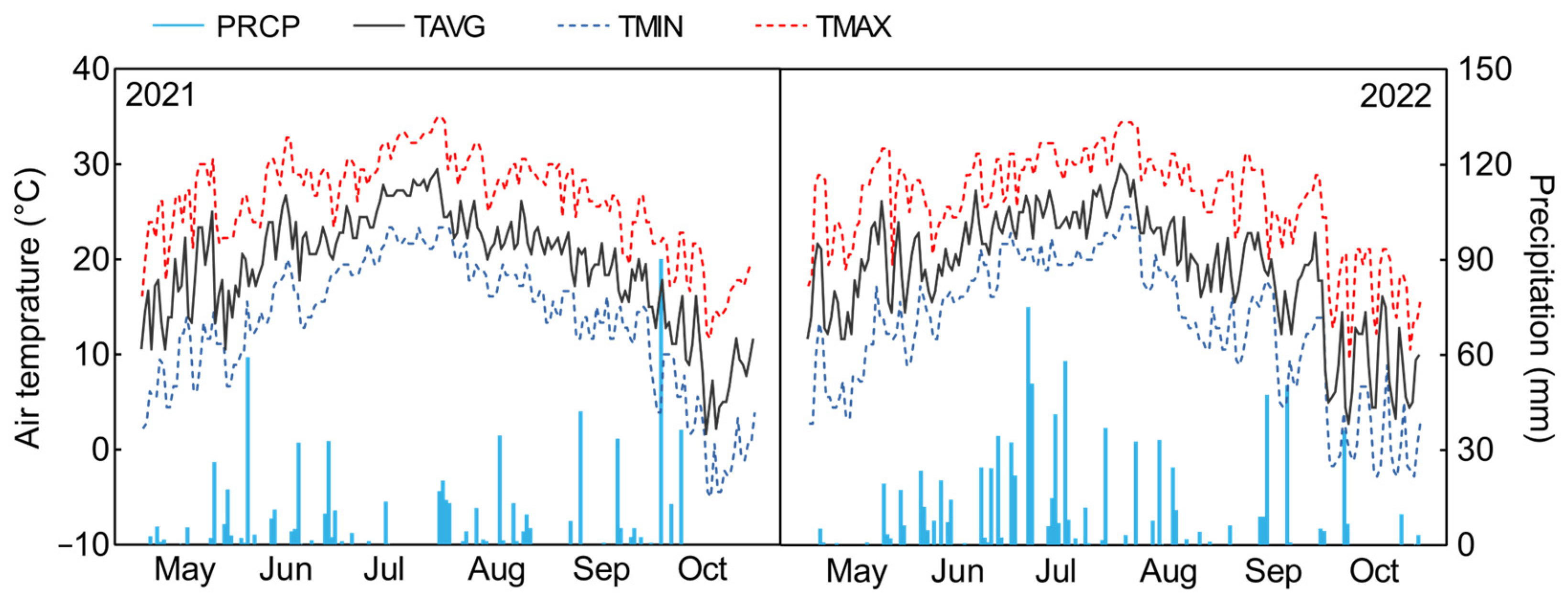
2.2.3. Screening of Salt Tolerance of Peanuts in the Field
2.3. Index and Method
2.3.1. Determination of Seed Vitality Index
2.3.2. Determination of Seed Morphological Index
2.3.3. Determination of Seedling Morphological Index
2.3.4. Investigation of Agronomic Characteristics in the Field
2.3.5. Determination of Yield Characteristics
2.4. Data Analysis and Statistics
3. Results
3.1. Evaluation of Stress Tolerance during Peanut Germination
3.1.1. Determination of NaCl Stress Concentration during Germination
3.1.2. STCs of Germination Characteristics under 75 mM NaCl
3.1.3. Analysis of Membership Function during Germination
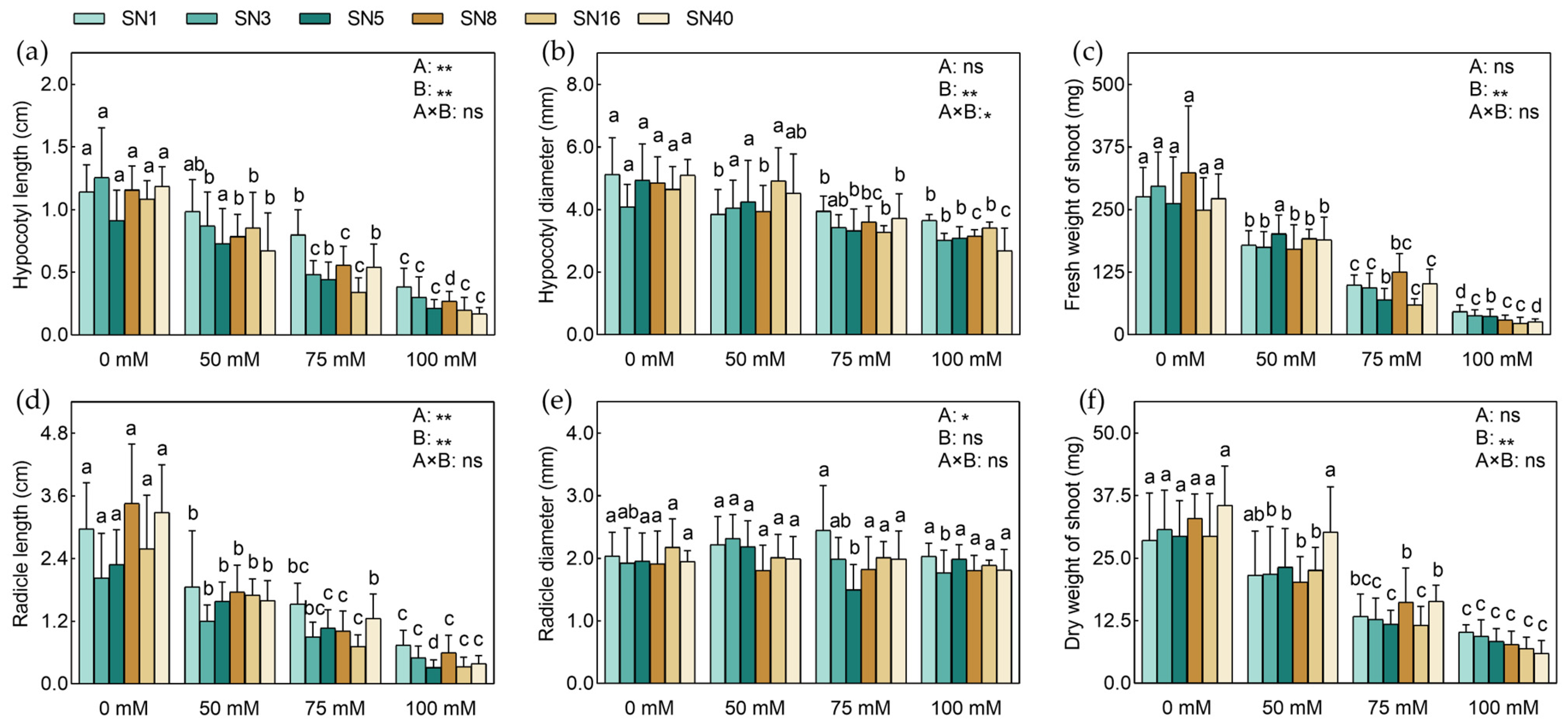
3.2. Evaluation of Stress Tolerance at the Seedling Stage
3.2.1. Grade Salt Damage to Peanut Leaves under NaCl Stress at the Seedling Stage
3.2.2. STCs of Peanuts under NaCl Stress at the Seedling Stage
3.3. Evaluation of Stress Tolerance of Peanuts in the Field
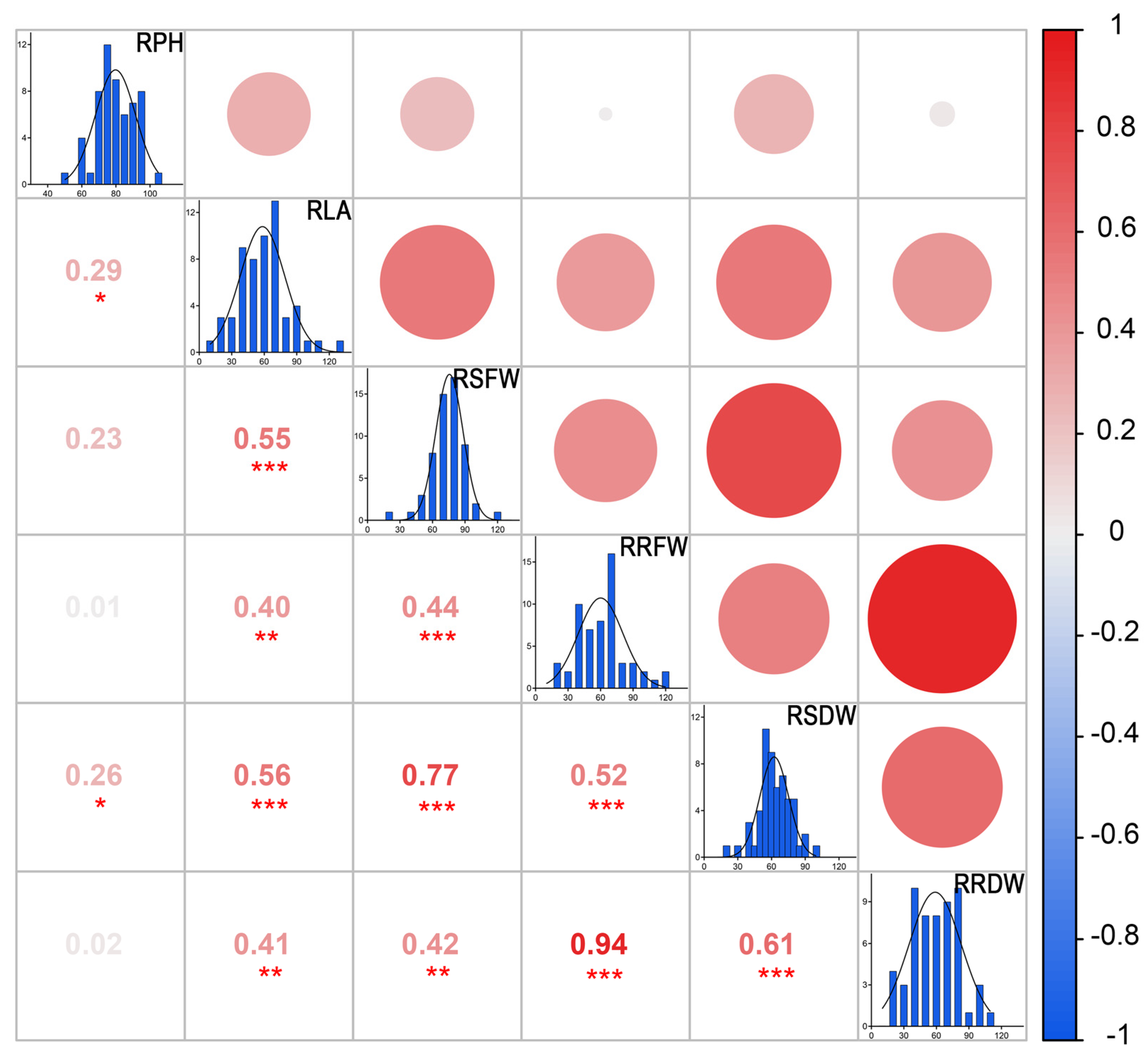
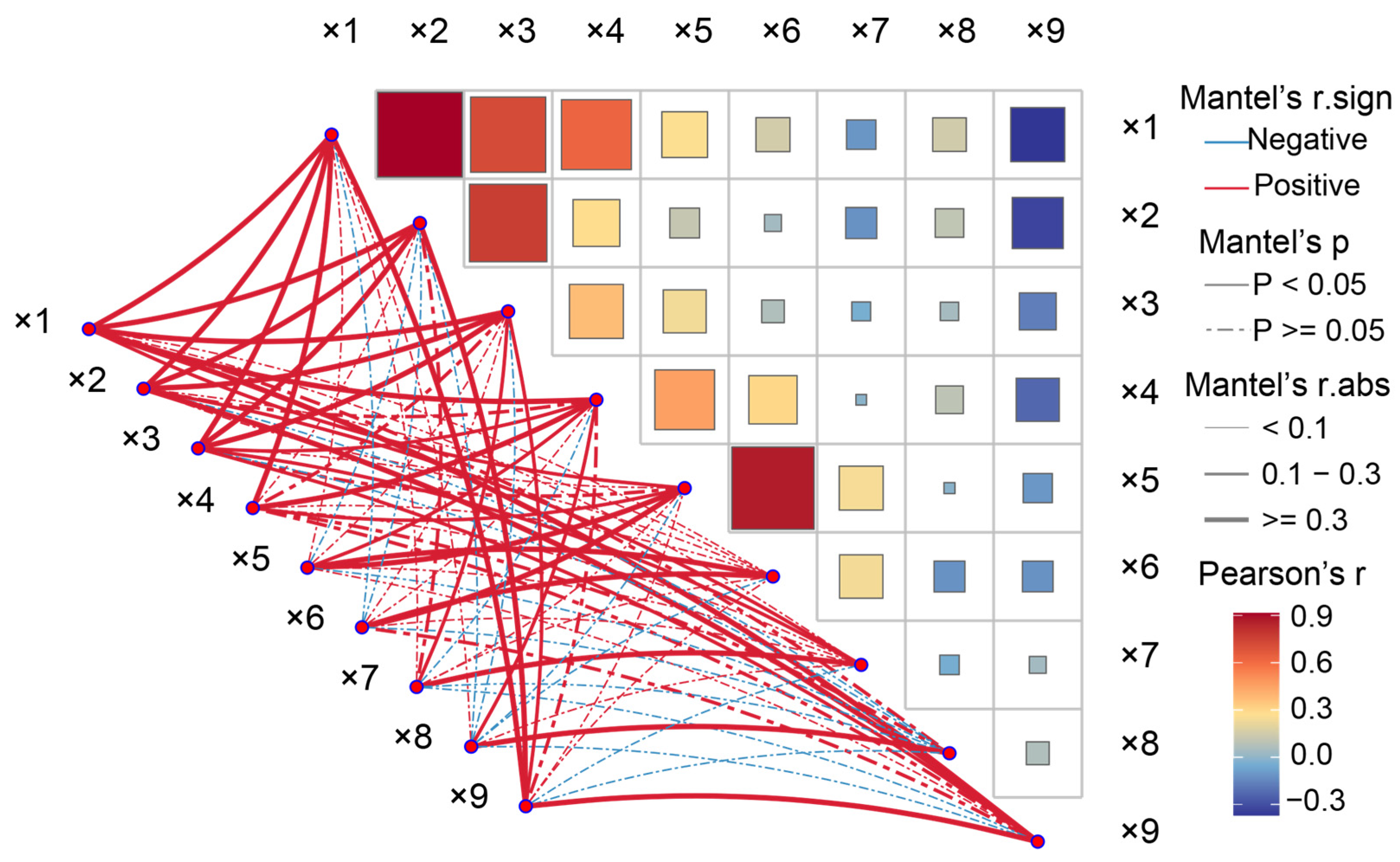
3.3.1. Correlation Analysis of Nine Indicators of STCs of Peanut Cultivars under NaCl Stress in the Field
3.3.2. Eigenvectors, Contribution Rate, and Weight of Principal Components of Peanut Cultivars under NaCl Stress in the Field
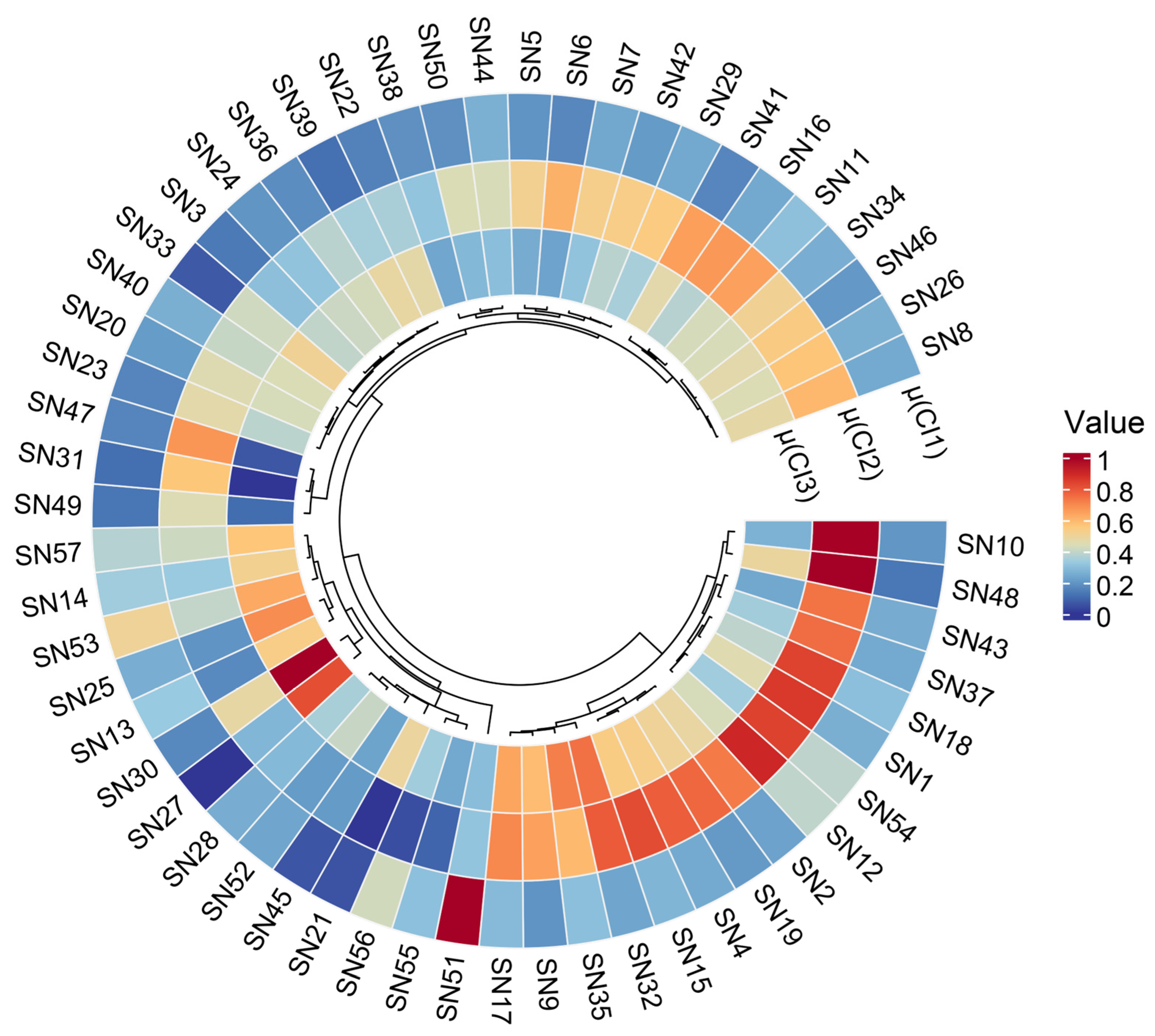
3.3.3. Comprehensive Evaluation of Peanut Cultivars under NaCl Stress in the Field
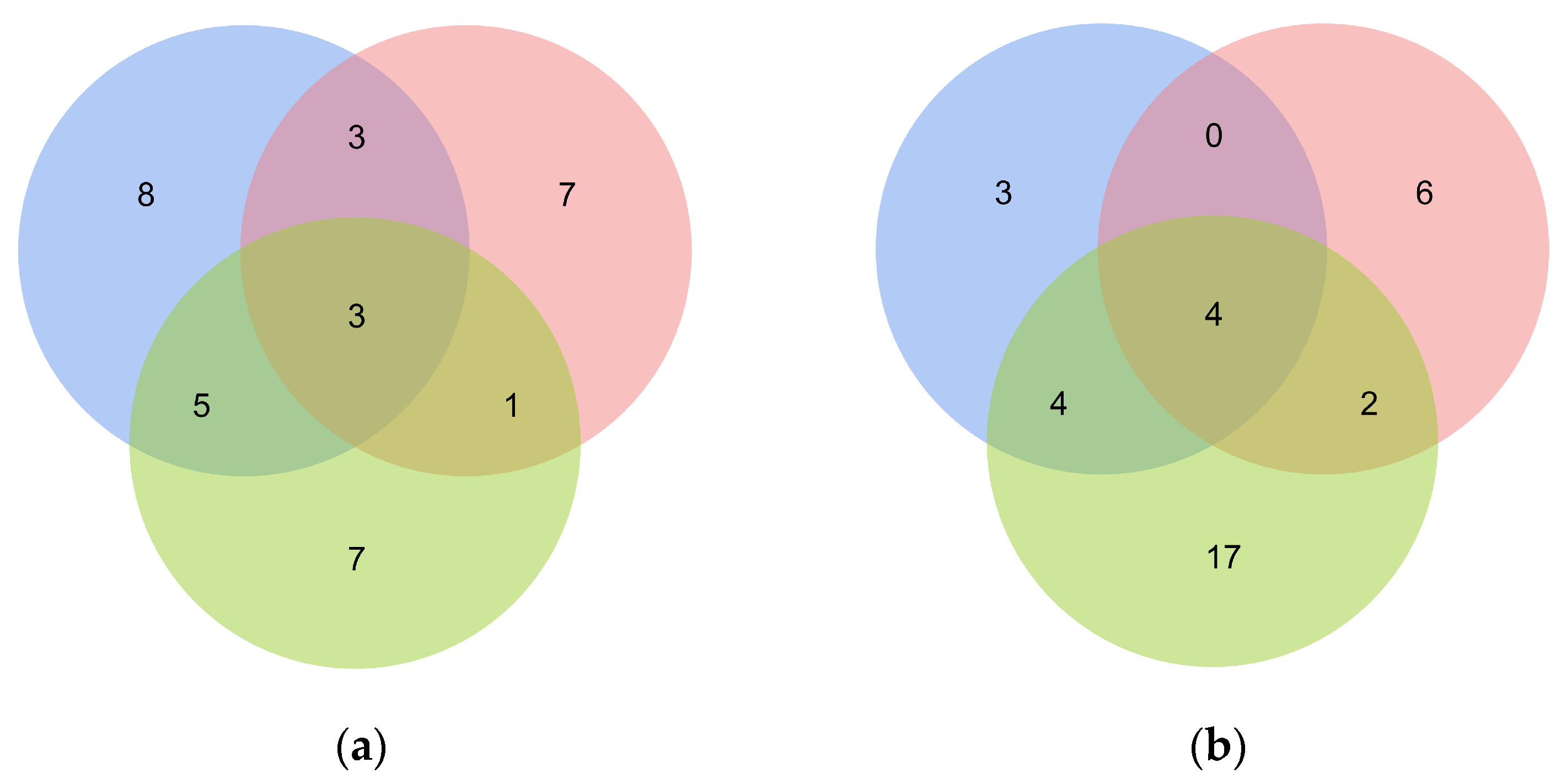
3.4. Evaluation and Comparison of Salt Tolerance of Different Peanut Cultivars at the Germination and Seedling Stages and in the Field
4. Discussion
4.1. Screening of Peanut Salt Tolerance
4.2. Evaluation Index of Peanut Salt Tolerance
4.3. Evaluation of Peanut Salt Tolerance and Grouping of Cultivars
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bonku, R.; Yu, J. Health aspects of peanuts as an outcome of its chemical composition. Food Sci. Hum. Wellness 2020, 9, 21–30. [Google Scholar] [CrossRef]
- Zhang, C.; Selvaraj, J.N.; Yang, Q.; Liu, Y. A Survey of Aflatoxin-Producing Aspergillus sp. from Peanut Field Soils in Four Agroecological Zones of China. Toxins 2017, 9, 40. [Google Scholar] [CrossRef]
- Huang, R.; Xiao, D.; Wang, X.; Zhan, J.; Wang, A.; He, L. Genome-wide identification, evolutionary and expression analyses of LEA gene family in peanut (Arachis hypogaea L.). BMC Plant Biol. 2022, 22, 155. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.; Khandwal, D.; Choudhary, B.; Ardeshana, D.; Jha, R.K.; Tanna, B.; Yadav, S.; Mishra, A.; Varshney, R.K.; Siddique, K.H.M. Differential Physio-Biochemical and Metabolic Responses of Peanut (Arachis hypogaea L.) under Multiple Abiotic Stress Conditions. Int. J. Mol. Sci. 2022, 23, 660. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Wang, S.; Zhou, L.; Sun, S.; Zhang, J.; Zhuang, L. Combination of Biochar and Functional Bacteria Drives the Ecological Improvement of Saline-Alkali Soil. Plants 2023, 12, 284. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, F.; Ma, H.; Li, J. Evaluation of the Saline-Alkaline Tolerance of Rice (Oryza sativa L.) Mutants Induced by Heavy-Ion Beam Mutagenesis. Biology 2022, 11, 126. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Zhao, X.; Ren, J.; Dong, J.; Zhang, H.; Dong, Q.; Jiang, C.; Zhong, C.; Zhou, Y.; Yu, H. Influence of Peanut, Sorghum, and Soil Salinity on Microbial Community Composition in Interspecific Interaction Zone. Front. Microbiol. 2021, 12, 678250. [Google Scholar] [CrossRef] [PubMed]
- Zeeshan, M.; Lu, M.; Naz, S.; Sehar, S.; Cao, F.; Wu, F. Resemblance and Difference of Seedling Metabolic and Transporter Gene Expression in High Tolerance Wheat and Barley Cultivars in Response to Salinity Stress. Plants 2020, 9, 519. [Google Scholar] [CrossRef] [PubMed]
- Razzaq, A.; Saleem, F.; Wani, S.H.; Abdelmohsen, S.A.M.; Alyousef, H.A.; Abdelbacki, A.M.M.; Alkallas, F.H.; Tamam, N.; Elansary, H.O. De-novo Domestication for Improving Salt Tolerance in Crops. Front. Plant Sci. 2021, 12, 681367. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Li, J.; Yang, Y.; Jiang, C.; Guo, Y. Designing salt stress-resilient crops: Current progress and future challenges. J. Integr. Plant Biol. 2023. Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Chugh, V.; Kaur, D.; Purwar, S.; Kaushik, P.; Sharma, V.; Kumar, H.; Rai, A.; Singh, C.M.; Kamaluddin; Dubey, R.B. Applications of Molecular Markers for Developing Abiotic-Stress-Resilient Oilseed Crops. Life 2022, 13, 88. [Google Scholar] [CrossRef]
- Ertiro, B.T.; Olsen, M.; Das, B.; Gowda, M.; Labuschagne, M. Genetic Dissection of Grain Yield and Agronomic Traits in Maize under Optimum and Low-Nitrogen Stressed Environments. Int. J. Mol. Sci. 2020, 21, 543. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Yu, X.; Wang, H.; Yu, Z.; Zhang, X.; Zhao, Y. Quantitative trait locus mapping for important yield traits of a sorghum-sudangrass hybrid using a high-density single nucleotide polymorphism map. Front. Plant Sci. 2022, 13, 1098605. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Jagendorf, A.; Zhu, J. Understanding and Improving Salt Tolerance in Plants. Crop Sci. 2005, 45, 437–448. [Google Scholar] [CrossRef]
- Quan, R.; Wang, J.; Hui, J.; Bai, H.; Lyu, X.; Zhu, Y.; Zhang, H.; Zhang, Z.; Li, S.; Huang, R. Improvement of Salt Tolerance Using Wild Rice Genes. Front. Plant Sci. 2018, 8, 2269. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, Z.; Cheng, J.; Lai, Y.; Wang, J.; Bao, Y.; Huang, J.; Zhang, H. QTL analysis of Na+ and K+ concentrations in roots and shoots under different levels of NaCl stress in rice (Oryza sativa L.). PLoS ONE 2012, 7, e51202. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.; Zhu, Z.; Wang, B.; Chen, M. Recent Progress on the Salt Tolerance Mechanisms and Application of Tamarisk. Int. J. Mol. Sci. 2022, 23, 3325. [Google Scholar] [CrossRef]
- Lu, P.; Magwanga, R.O.; Kirungu, J.N.; Hu, Y.; Dong, Q.; Cai, X.; Zhou, Z.; Wang, X.; Zhang, Z.; Hou, Y.; et al. Overexpression of Cotton a DTX/MATE Gene Enhances Drought, Salt, and Cold Stress Tolerance in Transgenic Arabidopsis. Front. Plant Sci. 2019, 10, 299. [Google Scholar] [CrossRef]
- Zhang, X.; Cheng, Z.; Yao, W.; Gao, Y.; Fan, G.; Guo, Q.; Zhou, B.; Jiang, T. Overexpression of PagERF072 from Poplar Improves Salt Tolerance. Int. J. Mol. Sci. 2022, 23, 10707. [Google Scholar] [CrossRef]
- Hamwieh, A.; Xu, D. Conserved salt tolerance quantitative trait locus (QTL) in wild and cultivated soybeans. Breed. Sci. 2008, 58, 355–359. [Google Scholar] [CrossRef]
- Li, D.; Dossa, K.; Zhang, Y.; Wei, X.; Wang, L.; Zhang, Y.; Liu, A.; Zhou, R.; Zhang, X. GWAS Uncovers Differential Genetic Bases for Drought and Salt Tolerances in Sesame at the Germination Stage. Genes 2018, 9, 87. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Q.; Wang, H.; Tao, S.; Cao, H.; Shi, Y.; Bakirov, A.; Xu, A.; Huang, Z. Discovery of common loci and candidate genes for controlling salt-alkali tolerance and yield-related traits in Brassica napus L. Plant Cell Rep. 2023, 42, 1039–1057. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Zhu, Y.; Liu, H.; Liang, Z.; Zhang, M.; Zou, C.; Yuan, G.; Gao, S.; Pan, G.; Shen, Y.; et al. A Combination of a Genome-Wide Association Study and a Transcriptome Analysis Reveals circRNAs as New Regulators Involved in the Response to Salt Stress in Maize. Int. J. Mol. Sci. 2022, 23, 9755. [Google Scholar] [CrossRef] [PubMed]
- Tsiupka, V.; Tsiupka, S.; Plugatar, Y.; Bulavin, I.; Komar-Tyomnaya, L. Assessment of the Drought-Tolerance Criteria for Screening Peach Cultivars. Horticulturae 2023, 9, 1045. [Google Scholar] [CrossRef]
- Hussain, M.I.; Muscolo, A.; Ahmed, M.; Asghar, M.A.; Al-Dakheel8, A.J. Agro-Morphological, Yield and Quality Traits and Interrelationship with Yield Stability in Quinoa (Chenopodium quinoa Willd.) Genotypes under Saline Marginal Environment. Plants 2020, 9, 1763. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.S.; Tester, M.; Fiene, G.; Mousa, M.A.A. Early Growth Stage Characterization and the Biochemical Responses for Salinity Stress in Tomato. Plants 2021, 10, 712. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Wang, G.; Yu, X.; Li, L.; Li, C.; Song, Y.; Xu, Z.; Zhang, J.; Guan, C. Assessing alfalfa (Medicago sativa L.) tolerance to salinity at seedling stage and screening of the salinity tolerance traits. Plant Biol. Stuttg. 2021, 23, 664–674. [Google Scholar] [CrossRef]
- Fu, B.; Liang, J.; Zhang, M.; Zhao, M.; Ji, X.; Wang, J. Influence of Genotypic and Environmental Factors on Tobacco Leaves Based on Metabolomics. Life 2022, 12, 590. [Google Scholar] [CrossRef]
- Xu, T.; Meng, S.; Zhu, X.; Di, J.; Zhu, Y.; Yang, X.; Yan, W. Integrated GWAS and transcriptomic analysis reveal the candidate salt-responding genes regulating Na+/K+ balance in barley (Hordeum vulgare L.). Front. Plant Sci. 2022, 13, 1004477. [Google Scholar] [CrossRef]
- Gustafsson, C.; Willforss, J.; Lopes-Pinto, F.; Ortiz, R.; Geleta, M. Identification of genes regulating traits targeted for domestication of field cress (Lepidium campestre) as a biennial and perennial oilseed crop. BMC Genom. 2018, 19, 36. [Google Scholar] [CrossRef] [PubMed]
- Laksana, C.; Sophiphun, O.; Chanprame, S. In vitro and in vivo screening for the identification of salt-tolerant sugarcane (Saccharum officinarum L.) clones: Molecular, biochemical, and physiological responses to salt stress. Saudi J. Biol. Sci. 2023, 30, 103655. [Google Scholar] [CrossRef] [PubMed]
- Hussain, N.; Ghaffar, A.; Zafar, Z.U.; Javed, M.; Shah, K.H.; Noreen, S.; Manzoor, H.; Iqbal, M.; Hassan, I.F.Z.; Bano, H.; et al. Identification of novel source of salt tolerance in local bread wheat germplasm using morpho-physiological and biochemical attributes. Sci. Rep. 2021, 11, 10854. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Oh, S.; Kang, Y.J.; Kim, K.; Moon, S.-K.; Moon, B.; Myung, S.; Kim, M.-S.; Lee, Y.K.; Ko, K. Effect of Oak Tree Sawdust Fermentation Period on Peanut Seed Germination, Seedling Biomass, and Morphology. Horticulturae 2021, 7, 182. [Google Scholar] [CrossRef]
- Wang, H.; Li, Z.; Ren, H.; Zhang, C.; Xiao, D.; Li, Y.; Hou, X.; Liu, T. Regulatory interaction of BcWRKY33A and BcHSFA4A promotes salt tolerance in non-heading Chinese cabbage [Brassica campestris (syn. Brassica rapa) ssp. chinensis]. Hort. Res. 2022, 9, uhac113. [Google Scholar] [CrossRef] [PubMed]
- Trejo-Téllez, L.I. Salinity Stress Tolerance in Plants. Plants 2023, 12, 3520. [Google Scholar] [CrossRef] [PubMed]
- Grozeva, S.; Kalapchieva, S.; Tringovska, I. In Vitro Screening for Salinity Tolerance in Garden Pea (Pisum sativum L.). Horticulturae 2023, 9, 338. [Google Scholar] [CrossRef]
- Khosravi, Z.; Pourmohammad, A.; Aliloo, A.-A.; Shahabivand, S.; Hassanpouraghdam, M.B.; Topcu, H. In vitro salinity stress mediates grass pea genotypes’ (Lathyrus sativus L.) responses. Turk. J. Agric. For. 2022, 46, 340–351. [Google Scholar] [CrossRef]
- Tokarz, B.; Wójtowicz, T.; Makowski, W.; Jędrzejczyk, R.J.; Tokarz, K.M. What is the Difference between the Response of Grass Pea (Lathyrus sativus L.) to Salinity and Drought Stress?—A Physiological Study. Agronomy 2020, 10, 833. [Google Scholar] [CrossRef]
- Liu, Y.; Shen, Y.; Chen, Z.; Wang, Z.; Yan, W. Identification of salt tolerance in peanut varieties/lines at the germination stage. Chin. J. Oil Crop Sci. 2012, 34, 168–173. [Google Scholar]
- Hoai, P.T.T.; Qiu, J.; Groszmann, M.; Rosa, A.D.; Tyerman, S.D.; Byrt, C.S. Arabidopsis plasma membrane intrinsic protein (AtPIP2;1) is implicated in a salinity conditional influence on seed germination. Funct. Plant Biol. 2023, 50, 633–648. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yang, B.; He, Y.; Zhan, C.; Cheng, Y.; Zhang, J.; Zhang, H.; Cheng, J.; Wang, Z. A quantitative trait locus, qSE3, promotes seed germination and seedling establishment under salinity stress in rice. Plant J. 2019, 97, 1089–1104. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Yang, Z.; Wei, X.; Yuan, F.; Yin, S.; Wang, B. Evaluation of salt-tolerant germplasm and screening of the salt-tolerance traits of sweet sorghum in the germination stage. Funct. Plant Biol. 2018, 45, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, A.; Kaur, N.; Sharma, A.; Kumar, A. Evaluation and screening of elite wheat germplasm for salinity stress at the seedling phase. Physiol. Plant 2021, 173, 2207–2215. [Google Scholar] [CrossRef] [PubMed]
- Puxuan, D.; Jun, L.; Ronghua, C.; Rouxian, W.; Chenggen, F.; Dandan, G.; Qing, L. Systematic Collection, Identification and Evaluation of Peanut Germplasm Resources in Guangdong Province. Plant Genet. Resour. 2023, 24, 671–679. [Google Scholar] [CrossRef]
- He, H.; Chen, B.; Yang, R.; Li, Y.; Ma, A.; Cao, L.; Wu, X.; Tian, H.; Gao, Y. Genotypic Variation in Nitrogen Utilization Efficiency of Oilseed Rape (Brassica napus) Under Contrasting N Supply in Pot and Field Experiments. Front. Plant Sci. 2017, 8, 1825. [Google Scholar] [CrossRef] [PubMed]
- Kunene, S.; Odindo, A.O.; Gerrano, A.S.; Mandizvo, T. Screening Bambara Groundnut (Vigna subterranea L. Verdc) Genotypes for Drought Tolerance at the Germination Stage under Simulated Drought Conditions. Plants 2022, 11, 3562. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Han, G.; Qiao, Z.; Li, Y.; Yang, Z.; Wang, B. Root Na+ Content Negatively Correlated to Salt Tolerance Determines the Salt Tolerance of Brassica napus L. Inbred Seedlings. Plants 2022, 11, 906. [Google Scholar] [CrossRef]
- Chakraborty, K.; Bhaduri, D.; Meena, H.N.; Kalariya, K. External potassium K+ application improves salinity tolerance by promoting Na+-exclusion, K+-accumulation and osmotic adjustment in contrasting peanut cultivars. Plant Physiol. Biochem. 2016, 103, 143–153. [Google Scholar] [CrossRef]
- Kumar, A.; Sheoran, P.; Mann, A.; Yadav, D.; Kumar, A.; Devi, S.; Kumar, N.; Dhansu, P.; Sharma, D.K. Deciphering trait associated morpho-physiological responses in pearlmillet hybrids and inbred lines under salt stress. Front. Plant Sci. 2023, 14, 1121805. [Google Scholar] [CrossRef]
- Kumar, A.; Sharma, C.L.; Kumar, P.; Devi, R.; Singh, K.; SL, K.; Kulshreshtha, N.; Yadav, R.K.; Sharma, S.K. Salinity and drought induced changes in gas exchange attributes and chlorophyll fluorescence characteristics of rice (Oryza sativa) varieties. Indian J. Agric. Sci. 2016, 86, 718–726. [Google Scholar] [CrossRef]
- Gu, H.; Huan, C.; Yang, F. Spatiotemporal Dynamics of Ecological Vulnerability and Its Influencing Factors in Shenyang City of China: Based on SRP Model. Int. J. Environ. Res. Public Health 2023, 20, 1525. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, J.; Prashanth, J.E.P.; Rajesh, N.; Reddy, S.M.; Pinjari, O.B. Principal component analysis approach for comprehensive screening of salt stress-tolerant tomato germplasm at the seedling stage. J. Biosci. 2020, 45, 141. [Google Scholar] [CrossRef] [PubMed]
- Chunthaburee, S.; Dongsansuk, A.; Sanitchon, J.; Pattanagul, W.; Theerakulpisut, P. Physiological and biochemical parameters for evaluation and clustering of rice cultivars differing in salt tolerance at seedling stage. Saudi J. Biol. Sci. 2016, 23, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, H.; Zeng, Y.; Xiang, L.; Lei, Z.; Huang, Q.; Li, T.; Shen, F.; Cheng, Q. A Salt Tolerance Evaluation Method for Sunflower (Helianthus annuus L.) at the Seed Germination Stage. Sci. Rep. 2020, 10, 10626. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Ma, X.; Cui, D.; Wang, Y.; Geng, L.; Cao, G.; Zhang, H.; Han, L. Comprehensive Evaluation and Analysis of the Mechanism of Cold Tolerance Based on the Transcriptome of Weedy Rice Seedlings. Rice 2020, 13, 12. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhao, Y.; Wang, W.; Xu, K.; Niu, L.; Yu, L.; Zou, J.W.; Wang, Z.; Zhang, Y.J.; Wang, F.; et al. Evaluation of Salt Tolerance Mechanism and Study on Salt Tolerance Relationship of Different Salt-Tolerant Wheat Varieties. Comput. Intell. Neurosci. 2022, 2022, 2395568. [Google Scholar] [CrossRef]


| Level | Plant Morphological Description |
|---|---|
| 1 | Seedlings grew normally and had no symptoms of salt injury |
| 2 | Seedling growth was normal; only one leaf showed slight chlorosis |
| 3 | Seedling growth was slightly inhibited, with 10% chlorosis of seedling leaves |
| 4 | 30% chlorosis of seedling leaves |
| 5 | 60% chlorosis of seedling leaves |
| 6 | The leaf tips of seedlings were withered and yellow |
| 7 | Seedlings leaves were completely withered or curled |
| 8 | Whole plant was dead or near death |
| Indicator | Relative Germination Rate | Relative Germination Potential | Relative Germination Index | Relative Vigor Index |
|---|---|---|---|---|
| Max | 1.000 | 1.000 | 0.883 | 0.876 |
| Min | 0.710 | 0.449 | 0.445 | 0.122 |
| Mean | 0.942 | 0.838 | 0.746 | 0.360 |
| CV (%) | 6.701 | 15.661 | 11.826 | 37.797 |
| Cultivar | GCD | Order | Cultivar | GCD | Order | Cultivar | GCD | Order |
|---|---|---|---|---|---|---|---|---|
| SN1 | 0.5077 | 4 | SN20 | 0.4076 | 25 | SN39 | 0.3850 | 53 |
| SN2 | 0.4011 | 34 | SN21 | 0.4888 | 6 | SN40 | 0.4098 | 24 |
| SN3 | 0.4363 | 13 | SN22 | 0.5075 | 5 | SN41 | 0.4123 | 21 |
| SN4 | 0.4695 | 8 | SN23 | 0.5361 | 3 | SN42 | 0.3854 | 51 |
| SN5 | 0.4333 | 14 | SN24 | 0.4029 | 30 | SN43 | 0.4301 | 16 |
| SN6 | 0.4102 | 23 | SN25 | 0.4067 | 26 | SN44 | 0.3923 | 42 |
| SN7 | 0.4420 | 12 | SN26 | 0.4496 | 9 | SN45 | 0.3954 | 41 |
| SN8 | 0.4813 | 7 | SN27 | 0.3858 | 50 | SN46 | 0.4126 | 20 |
| SN9 | 0.4322 | 15 | SN28 | 0.4200 | 17 | SN47 | 0.3830 | 55 |
| SN10 | 0.4047 | 29 | SN29 | 0.6471 | 1 | SN48 | 0.4025 | 32 |
| SN11 | 0.4478 | 10 | SN30 | 0.3850 | 52 | SN49 | 0.3819 | 57 |
| SN12 | 0.3901 | 44 | SN31 | 0.3967 | 39 | SN50 | 0.3824 | 56 |
| SN13 | 0.3887 | 47 | SN32 | 0.3955 | 40 | SN51 | 0.3893 | 45 |
| SN14 | 0.4116 | 22 | SN33 | 0.3891 | 46 | SN52 | 0.3984 | 37 |
| SN15 | 0.3832 | 54 | SN34 | 0.4472 | 11 | SN53 | 0.3862 | 48 |
| SN16 | 0.3994 | 36 | SN35 | 0.3976 | 38 | SN54 | 0.4007 | 35 |
| SN17 | 0.6083 | 2 | SN36 | 0.4017 | 33 | SN55 | 0.3860 | 49 |
| SN18 | 0.4059 | 27 | SN37 | 0.4056 | 28 | SN56 | 0.4170 | 19 |
| SN19 | 0.4027 | 31 | SN38 | 0.3909 | 43 | SN57 | 0.4182 | 18 |
| Indicator | ×1 | ×2 | ×3 | ×4 | ×5 | ×6 | ×7 | ×8 | ×9 |
|---|---|---|---|---|---|---|---|---|---|
| Max | 0.952 | 0.988 | 0.970 | 0.972 | 1.078 | 1.176 | 1.034 | 1.231 | 1.060 |
| Min | 0.111 | 0.398 | 0.413 | 0.239 | 0.476 | 0.462 | 0.300 | 0.321 | 0.757 |
| Mean | 0.480 | 0.652 | 0.652 | 0.713 | 0.765 | 0.775 | 0.748 | 0.776 | 0.924 |
| CV (%) | 36.605 | 18.123 | 17.674 | 26.444 | 19.446 | 20.003 | 23.763 | 24.732 | 6.042 |
| Principle Factor | CI1 | CI2 | CI3 | |
|---|---|---|---|---|
| Eigenvalue | 3.295 | 1.892 | 1.052 | |
| Contribution ratio (%) | 36.612 | 21.020 | 11.690 | |
| Cumulative contribution ratio (%) | 36.612 | 57.632 | 69.323 | |
| Eigenvector | ×1 | 0.958 | 0.132 | 0.056 |
| ×2 | 0.931 | −0.079 | −0.015 | |
| ×3 | 0.833 | 0.049 | 0.023 | |
| ×4 | 0.539 | 0.489 | 0.142 | |
| ×5 | 0.181 | 0.932 | 0.033 | |
| ×6 | 0.040 | 0.920 | −0.105 | |
| ×7 | −0.196 | 0.327 | −0.226 | |
| ×8 | 0.127 | −0.041 | 0.861 | |
| ×9 | −0.460 | −0.060 | 0.498 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, N.; Zhang, H.; Ren, J.; Bai, B.; Guo, P.; Lv, Z.; Kang, S.; Zhao, X.; Yu, H.; Zhao, T. Characterization and Comprehensive Evaluation of Phenotypic and Yield Traits in Salt-Stress-Tolerant Peanut Germplasm for Conservation and Breeding. Horticulturae 2024, 10, 147. https://doi.org/10.3390/horticulturae10020147
Zhang N, Zhang H, Ren J, Bai B, Guo P, Lv Z, Kang S, Zhao X, Yu H, Zhao T. Characterization and Comprehensive Evaluation of Phenotypic and Yield Traits in Salt-Stress-Tolerant Peanut Germplasm for Conservation and Breeding. Horticulturae. 2024; 10(2):147. https://doi.org/10.3390/horticulturae10020147
Chicago/Turabian StyleZhang, Nan, He Zhang, Jingyao Ren, Baiyi Bai, Pei Guo, Zhenghao Lv, Shuli Kang, Xinhua Zhao, Haiqiu Yu, and Tianhong Zhao. 2024. "Characterization and Comprehensive Evaluation of Phenotypic and Yield Traits in Salt-Stress-Tolerant Peanut Germplasm for Conservation and Breeding" Horticulturae 10, no. 2: 147. https://doi.org/10.3390/horticulturae10020147
APA StyleZhang, N., Zhang, H., Ren, J., Bai, B., Guo, P., Lv, Z., Kang, S., Zhao, X., Yu, H., & Zhao, T. (2024). Characterization and Comprehensive Evaluation of Phenotypic and Yield Traits in Salt-Stress-Tolerant Peanut Germplasm for Conservation and Breeding. Horticulturae, 10(2), 147. https://doi.org/10.3390/horticulturae10020147





