Genome-Wide Identification of the PEBP Family Gene in Three Cymbidium Species and Its Expression Patterns Analysis in C. ensifolium
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. Identification and Physicochemical Properties of PEBP Genes from Three Cymbidium Species
2.3. Phylogenetic Analysis of PEBP Genes
2.4. Chromosome Distribution and Collinear Correlation of PEBP Genes in Three Cymbidium Species
2.5. Gene Structure and Conserved Motif Analysis of PEBP Gene
2.6. Sequence Analysis of PEBP Gene Promoter
2.7. Expression Pattern and qRT-PCR Analysis
3. Results
3.1. Identification and Sequence Analysis of PEBP Genes in Three Cymbidium Species
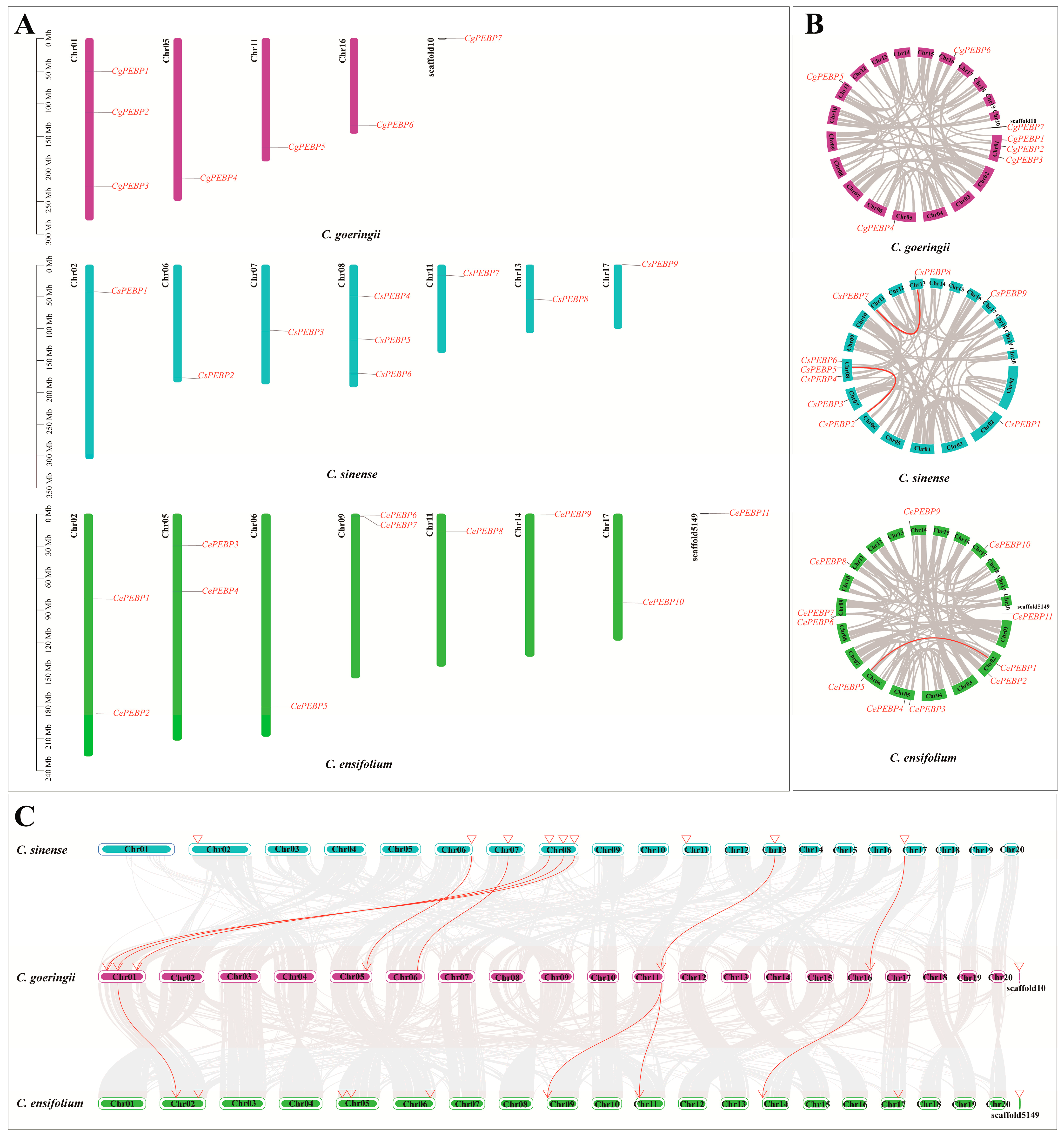
3.2. Chromosome Localization and Collinearity Analysis of PEBP Genes in Three Cymbidium Species
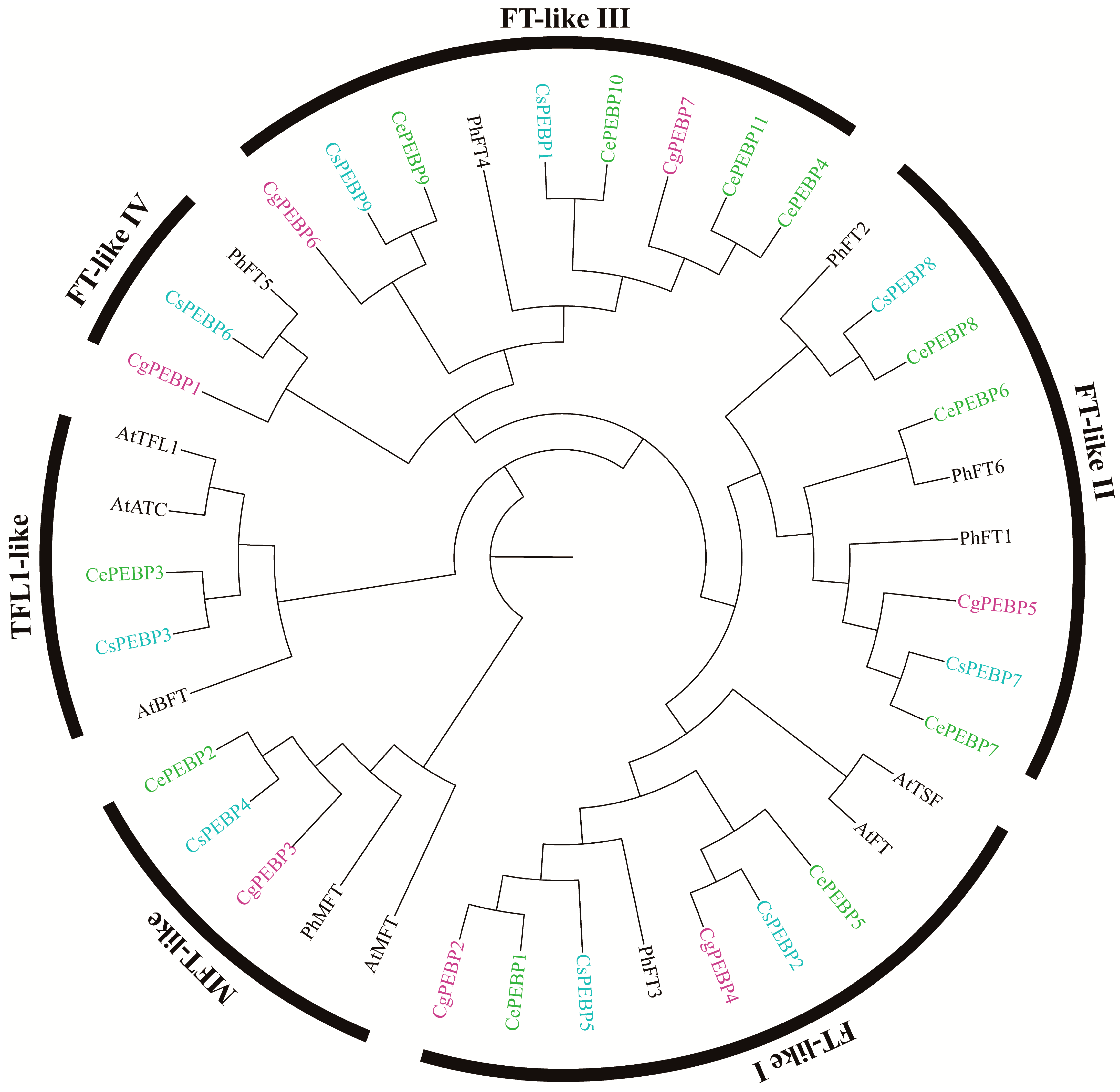
3.3. Phylogenetic Relationship Analysis of PEBP Genes
3.4. Conserved Motif and Gene Structure Analysis of PEBP Gene Family
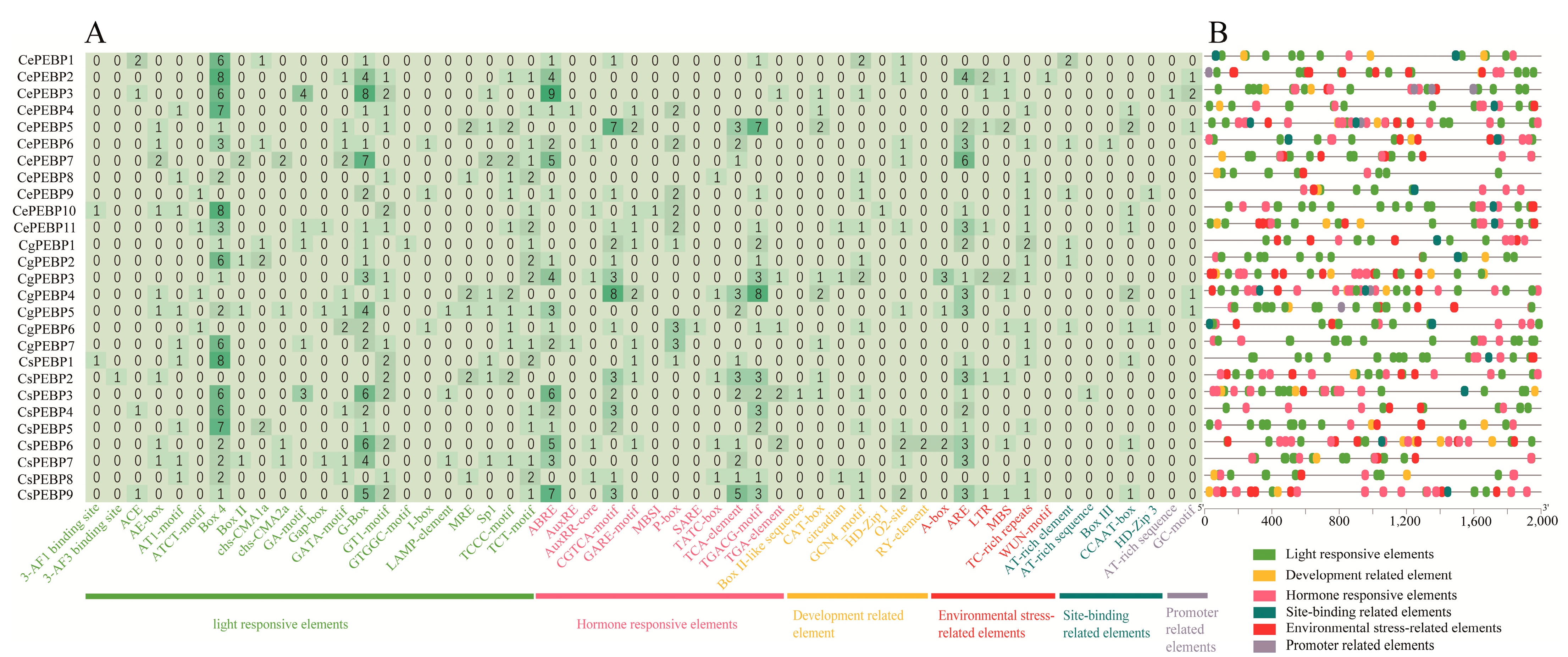
3.5. Cis-Element Analysis of Three Cymbidium Species
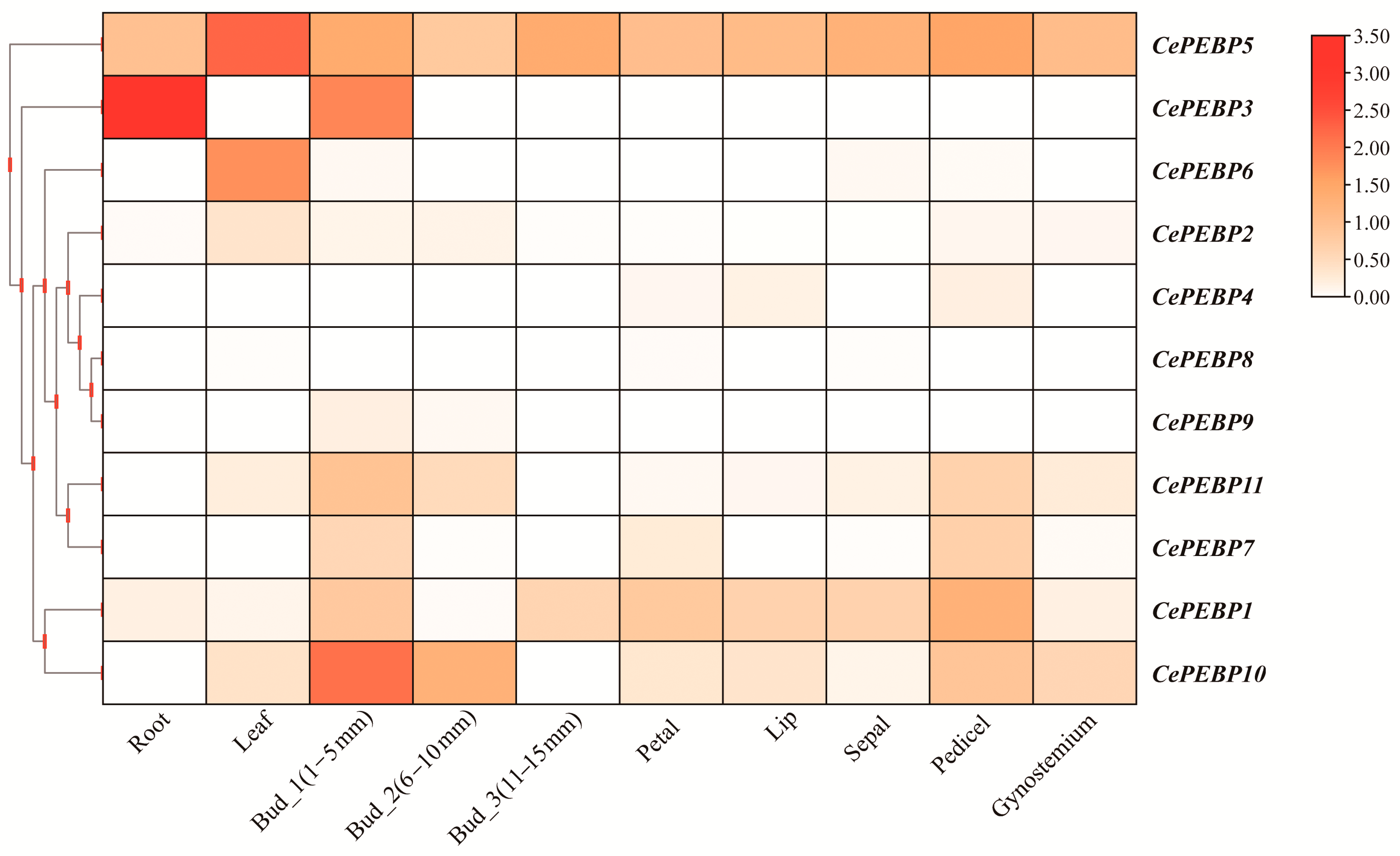
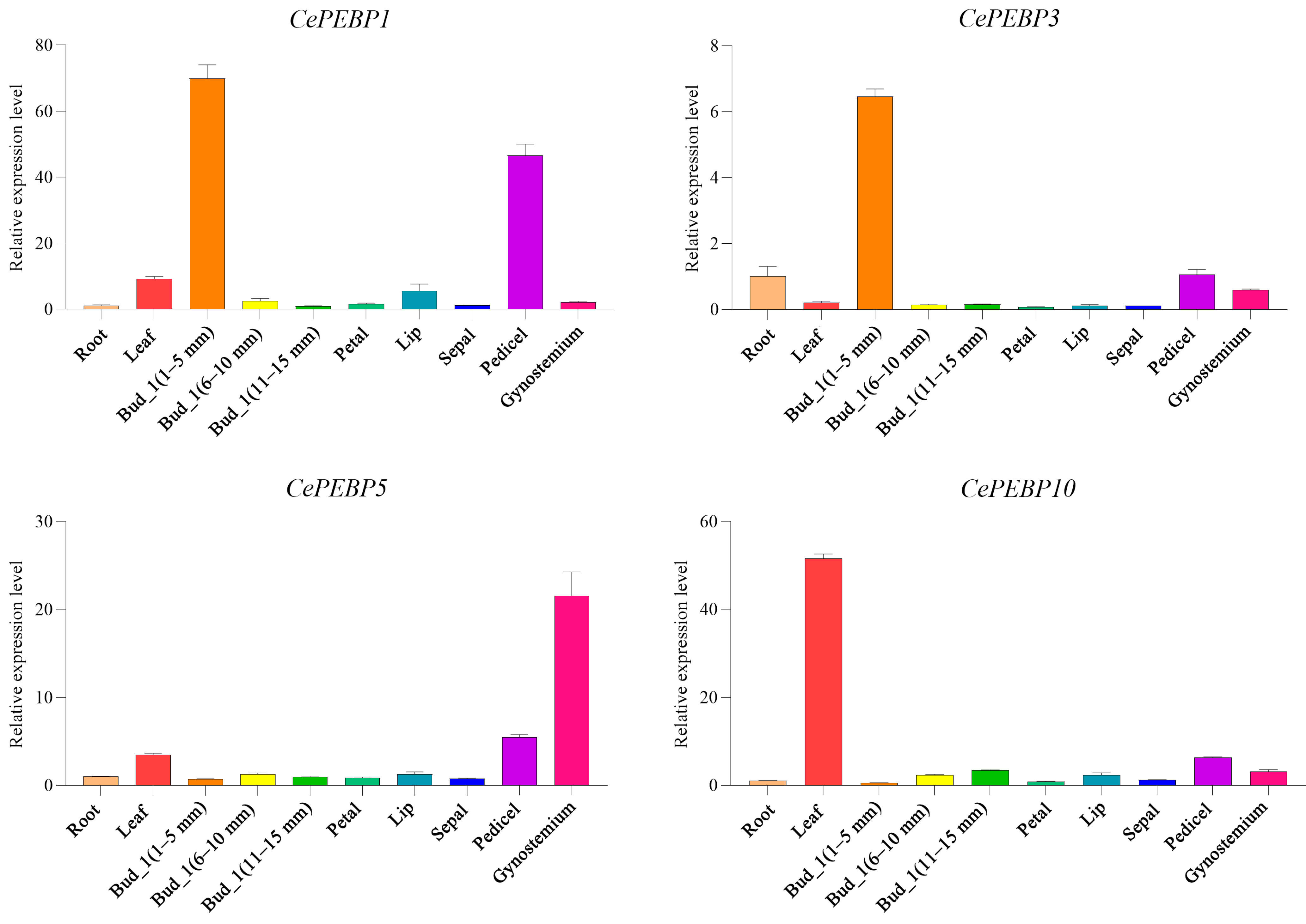
3.6. Expression Patterns of PEBP Genes in C. ensifolium
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, M.; Li, P.; Yan, X.; Wang, J.; Cheng, T.; Zhang, Q. Genome-wide characterization of PEBP family genes in nine Rosaceae tree species and their expression analysis in P. mume. BMC Ecol. Evol. 2021, 21, 32. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Liu, H.; Sang, N.; Li, Y.; Zhang, T.; Sun, J.; Huang, X. Identification of cotton MOTHER OF FT AND TFL1 homologs, GhMFT1 and GhMFT2, involved in seed germination. PLoS ONE 2019, 14, e0215771. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Li, G.; Krom, N.; Tang, Y.; Wen, J. Genetic regulation of flowering time and inflorescence architecture by MtFDa and MtFTa1 in Medicago truncatula. Plant Physiol. 2021, 185, 161–178. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Guo, X.; Hao, Y.; Lu, G.; Li, D.; Lu, J.; Zhang, T. Genome-wide characterization of PEBP gene family in Perilla frutescens and PfFT1 promotes flowering time in Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 1026696. [Google Scholar] [CrossRef]
- Zheng, X.-M.; Wu, F.-Q.; Zhang, X.; Lin, Q.-B.; Wang, J.; Guo, X.-P.; Lei, C.-L.; Cheng, Z.-J.; Zou, C.; Wan, J.-M. Evolution of the PEBP gene family and selective signature on FT-like clade. J. Syst. Evol. 2016, 54, 502–510. [Google Scholar] [CrossRef]
- Hedman, H.; Källman, T.; Lagercrantz, U. Early evolution of the MFT-like gene family in plants. Plant Mol. Biol. 2009, 70, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Xi, W.; Liu, C.; Hou, X.; Yu, H. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 2010, 22, 1733–1748. [Google Scholar] [CrossRef]
- Nakamura, S.; Abe, F.; Kawahigashi, H.; Nakazono, K.; Tagiri, A.; Matsumoto, T.; Utsugi, S.; Ogawa, T.; Handa, H.; Ishida, H.; et al. A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell 2011, 23, 3215–3229. [Google Scholar] [CrossRef]
- Li, Q.; Fan, C.; Zhang, X.; Wang, X.; Wu, F.; Hu, R.; Fu, Y. Identification of a soybean MOTHER OF FT AND TFL1 homolog involved in regulation of seed germination. PLoS ONE 2014, 9, e99642. [Google Scholar] [CrossRef]
- Eshed, Y.; Lippman, Z.B. Revolutions in agriculture chart a course for targeted breeding of old and new crops. Science 2019, 366, eaax0025. [Google Scholar] [CrossRef]
- Karlgren, A.; Gyllenstrand, N.; Kallman, T.; Sundstrom, J.F.; Moore, D.; Lascoux, M.; Lagercrantz, U. Evolution of the PEBP gene family in plants: Functional diversification in seed plant evolution. Plant Physiol. 2011, 156, 1967–1977. [Google Scholar] [CrossRef] [PubMed]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- Bradley, D.; Carpenter, R.; Copsey, L.; Vincent, C.; Rothstein, S.; Coen, E. Control of inflorescence architecture in Antirrhinum. Nature 1996, 379, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Bradley, D.; Ratcliffe, O.; Vincent, C.; Carpenter, R.; Coen, E. Inflorescence commitment and architecture in Arabidopsis. Science 1997, 275, 80–83. [Google Scholar] [CrossRef]
- Kaneko-Suzuki, M.; Kurihara-Ishikawa, R.; Okushita-Terakawa, C.; Kojima, C.; Nagano-Fujiwara, M.; Ohki, I.; Tsuji, H.; Shimamoto, K.; Taoka, K.I. TFL1-Like Proteins in Rice Antagonize Rice FT-Like Protein in Inflorescence Development by Competition for Complex Formation with 14-3-3 and FD. Plant Cell Physiol. 2018, 59, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Freytes, S.N.; Canelo, M.; Cerdan, P.D. Regulation of Flowering Time: When and Where? Curr. Opin. Plant Biol. 2021, 63, 102049. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.; Xiang, W.; Zhang, L.; Gao, C.; An, N.; Xing, L.; Ma, J.; Zhao, C.; Zhang, D. Identification of apple TFL1-interacting proteins uncovers an expanded flowering network. Plant Cell Rep. 2021, 40, 2325–2340. [Google Scholar] [CrossRef] [PubMed]
- Hanzawa, Y.; Money, T.; Bradley, D. A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. USA 2005, 102, 7748–7753. [Google Scholar] [CrossRef]
- Ho, W.W.; Weigel, D. Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 2014, 26, 552–564. [Google Scholar] [CrossRef]
- Ahn, J.H.; Miller, D.; Winter, V.J.; Banfield, M.J.; Lee, J.H.; Yoo, S.Y.; Henz, S.R.; Brady, R.L.; Weigel, D. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 2006, 25, 605–614. [Google Scholar] [CrossRef]
- Ramya, M.; Park, P.H.; Chuang, Y.C.; Kwon, O.K.; An, H.R.; Park, P.M.; Baek, Y.S.; Kang, B.C.; Tsai, W.C.; Chen, H.H. RNA sequencing analysis of Cymbidium goeringii identifies floral scent biosynthesis related genes. BMC Plant Biol. 2019, 19, 337. [Google Scholar] [CrossRef] [PubMed]
- Ai, Y.; Li, Z.; Sun, W.H.; Chen, J.; Zhang, D.; Ma, L.; Zhang, Q.H.; Chen, M.K.; Zheng, Q.D.; Liu, J.F.; et al. Correction: The Cymbidium genome reveals the evolution of unique morphological traits. Hortic. Res. 2021, 8, 264. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.X.; Gao, J.; Wei, Y.L.; Ren, R.; Zhang, G.Q.; Lu, C.Q.; Jin, J.P.; Ai, Y.; Wang, Y.Q.; Chen, L.J.; et al. The genome of Cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 2021, 19, 2501–2516. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Chen, G.-Z.; Huang, J.; Liu, D.-K.; Xue, F.; Chen, X.-L.; Chen, S.-Q.; Liu, C.-G.; Liu, H.; Ma, H.; et al. The Cymbidium goeringii genome provides insight into organ development and adaptive evolution in orchids. Ornam. Plant Res. 2021, 1, 1–13. [Google Scholar] [CrossRef]
- Jiang, L.; Jiang, X.; Li, Y.; Gao, Y.; Wang, S.; Ma, Y.; Wang, G. FT-like paralogs are repressed by an SVP protein during the floral transition in Phalaenopsis orchid. Plant Cell Rep. 2022, 41, 233–248. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Peng, Y.; Chen, J.; Zhu, X.; Xie, K.; Ahmad, S.; Zhao, K.; Peng, D.; Liu, Z.J.; Zhou, Y. The Genome-Level Survey of the WOX Gene Family in Melastoma dodecandrum Lour. Int. J. Mol. Sci. 2023, 24, 17349. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Zhao, X.; Huang, Y.; Zhang, M.M.; He, X.; Ke, S.; Li, Y.; Zhang, C.; Ahmad, S.; Lan, S.; et al. Genome-Wide Identification of the YABBY Gene Family in Dendrobium Orchids and Its Expression Patterns in Dendrobium chrysotoxum. Int. J. Mol. Sci. 2023, 24, 10165. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Rombauts, S.; Déhais, P.; Montagu, M.V.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef]
- Chen, G.Z.; Huang, J.; Zhou, X.Q.; Hao, Y.; Chen, J.L.; Zhou, Y.Z.; Ahmad, S.; Lan, S.; Liu, Z.J.; Peng, D.H. Comprehensive Analysis for GRF Transcription Factors in Sacred Lotus (Nelumbo nucifera). Int. J. Mol. Sci. 2022, 23, 6673. [Google Scholar] [CrossRef]
- Huang, W.; Fang, Z.; Zeng, S.; Zhang, J.; Wu, K.; Chen, Z.; Teixeira da Silva, J.A.; Duan, J. Molecular cloning and functional analysis of Three FLOWERING LOCUS T (FT) homologous genes from Chinese Cymbidium. Int. J. Mol. Sci. 2012, 13, 11385–11398. [Google Scholar] [CrossRef]
- Ospina-Zapata, D.A.; Madrigal, Y.; Alzate, J.F.; Pabon-Mora, N. Evolution and Expression of Reproductive Transition Regulatory Genes FT/TFL1 With Emphasis in Selected Neotropical Orchids. Front. Plant Sci. 2020, 11, 469. [Google Scholar] [CrossRef]
- Gao, J.; Huang, B.H.; Wan, Y.T.; Chang, J.; Li, J.Q.; Liao, P.C. Functional divergence and intron variability during evolution of angiosperm TERMINAL FLOWER1 (TFL1) genes. Sci. Rep. 2017, 7, 14830. [Google Scholar] [CrossRef]
- Hou, C.J.; Yang, C.H. Functional analysis of FT and TFL1 orthologs from orchid (Oncidium Gower Ramsey) that regulate the vegetative to reproductive transition. Plant Cell Physiol. 2009, 50, 1544–1557. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Liu, X.; Vanneste, K.; Proost, S.; Tsai, W.C.; Liu, K.W.; Chen, L.J.; He, Y.; Xu, Q.; Bian, C.; et al. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 2015, 47, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Banfield, M.J.; Brady, R.L. The structure of Antirrhinum centroradialis protein (CEN) suggests a role as a kinase regulator. J. Mol. Biol. 2000, 297, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Si, Z.; Liu, H.; Zhu, J.; Chen, J.; Wang, Q.; Fang, L.; Gao, F.; Tian, Y.; Chen, Y.; Chang, L.; et al. Mutation of SELF-PRUNING homologs in cotton promotes short-branching plant architecture. J. Exp. Bot. 2018, 69, 2543–2553. [Google Scholar] [CrossRef]
- Zhou, S.; Jiang, L.; Guan, S.; Gao, Y.; Gao, Q.; Wang, G.; Duan, K. Expression profiles of five FT-like genes and functional analysis of PhFT-1 in a Phalaenopsis hybrid. Electron. J. Biotechnol. 2018, 31, 75–83. [Google Scholar] [CrossRef]
- Amasino, R. Seasonal and developmental timing of flowering. Plant J. 2010, 61, 1001–1013. [Google Scholar] [CrossRef]
- Teotia, S.; Tang, G. To bloom or not to bloom: Role of microRNAs in plant flowering. Mol. Plant 2015, 8, 359–377. [Google Scholar] [CrossRef]
- Song, Y.H.; Shim, J.S.; Kinmonth-Schultz, H.A.; Imaizumi, T. Photoperiodic flowering: Time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 2015, 66, 441–464. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, L.; Song, S.; Li, Y.; Shen, L.; Yu, H. DOFT and DOFTIP1 affect reproductive development in the orchid Dendrobium Chao Praya Smile. J. Exp. Bot. 2017, 68, 5759–5772. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Gene ID | Protein Length (AA) | Isoelectric Point (pI) | Molecular Weight (Mw) | Grand Average of Hydropathicity (GRAVY) | Aliphatic Index (AI) | Instability Index (II) |
|---|---|---|---|---|---|---|---|
| CePEBP1 | JL006795 | 176 | 6.42 | 19,848.39 | −0.311 | 80.74 | 43.37 |
| CePEBP2 | JL020923 | 173 | 7.74 | 19,256.21 | −0.118 | 83.29 | 58.26 |
| CePEBP3 | JL010014 | 173 | 9.06 | 19,607.53 | −0.202 | 82.14 | 45.42 |
| CePEBP4 | JL026838 | 189 | 6.73 | 21,260.35 | −0.151 | 89.15 | 46.08 |
| CePEBP5 | JL020421 | 174 | 6.42 | 19,926.58 | −0.355 | 75.52 | 35.95 |
| CePEBP6 | JL027939 | 101 | 5.62 | 10,995.31 | −0.265 | 73.27 | 41.88 |
| CePEBP7 | JL002228 | 174 | 9.03 | 19,523.09 | −0.375 | 77.82 | 48.29 |
| CePEBP8 | JL001165 | 177 | 9.18 | 20,180.03 | −0.227 | 83.11 | 45.55 |
| CePEBP9 | JL013430 | 178 | 8.48 | 20,082.78 | −0.39 | 74.44 | 42.64 |
| CePEBP10 | JL027407 | 112 | 5.27 | 12,308.11 | −0.14 | 86.88 | 52.12 |
| CePEBP11 | JL028740 | 183 | 6.12 | 20,580.41 | −0.248 | 81.97 | 48.35 |
| CgPEBP1 | GL13937 | 236 | 5.13 | 26,017.24 | −0.323 | 80.04 | 48.92 |
| CgPEBP2 | GL01335 | 176 | 6.42 | 19,848.39 | −0.311 | 80.74 | 43.37 |
| CgPEBP3 | GL28974 | 66 | 10.75 | 7806.99 | −0.577 | 66.52 | 54.87 |
| CgPEBP4 | GL14129 | 125 | 6.83 | 14,228.19 | −0.331 | 82.56 | 30.19 |
| CgPEBP5 | GL07645 | 174 | 9.03 | 19,522.14 | −0.333 | 80.06 | 48.67 |
| CgPEBP6 | GL00658 | 201 | 7.8 | 22,704.79 | −0.341 | 77.06 | 38.75 |
| CgPEBP7 | GL09595 | 181 | 6.73 | 20,394.24 | −0.229 | 82.87 | 49.19 |
| CsPEBP1 | cymsin_Mol026710 | 118 | 5.34 | 13,110.05 | −0.251 | 85.85 | 51.29 |
| CsPEBP2 | cymsin_Mol020839 | 189 | 6.42 | 21,659.53 | −0.355 | 79.31 | 37.43 |
| CsPEBP3 | cymsin_Mol012759 | 173 | 9.06 | 19,598.52 | −0.203 | 82.14 | 45.59 |
| CsPEBP4 | cymsin_Mol020552 | 187 | 7 | 20,394.34 | −0.184 | 73.48 | 57.35 |
| CsPEBP5 | cymsin_Mol006878 | 190 | 6.08 | 21,404.16 | −0.297 | 80.95 | 41.78 |
| CsPEBP6 | cymsin_Mol006013 | 243 | 6.08 | 27,504.27 | −0.363 | 80.16 | 42.56 |
| CsPEBP7 | cymsin_Mol018868 | 174 | 9.03 | 19,523.09 | −0.375 | 77.82 | 48.29 |
| CsPEBP8 | cymsin_Mol003216 | 178 | 9.18 | 20,292.16 | −0.23 | 83.2 | 42.1 |
| CsPEBP9 | cymsin_Mol017371 | 379 | 9.57 | 42,330.61 | −0.181 | 84.62 | 52.69 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Wang, F.; Zhang, Y.; Zheng, R.; Wu, X.; Ai, Y.; Ahmad, S.; Liu, Z.; Peng, D. Genome-Wide Identification of the PEBP Family Gene in Three Cymbidium Species and Its Expression Patterns Analysis in C. ensifolium. Horticulturae 2024, 10, 252. https://doi.org/10.3390/horticulturae10030252
Chen J, Wang F, Zhang Y, Zheng R, Wu X, Ai Y, Ahmad S, Liu Z, Peng D. Genome-Wide Identification of the PEBP Family Gene in Three Cymbidium Species and Its Expression Patterns Analysis in C. ensifolium. Horticulturae. 2024; 10(3):252. https://doi.org/10.3390/horticulturae10030252
Chicago/Turabian StyleChen, Jinliao, Fei Wang, Yangting Zhang, Ruiyue Zheng, Xiaopei Wu, Ye Ai, Sagheer Ahmad, Zhongjian Liu, and Donghui Peng. 2024. "Genome-Wide Identification of the PEBP Family Gene in Three Cymbidium Species and Its Expression Patterns Analysis in C. ensifolium" Horticulturae 10, no. 3: 252. https://doi.org/10.3390/horticulturae10030252
APA StyleChen, J., Wang, F., Zhang, Y., Zheng, R., Wu, X., Ai, Y., Ahmad, S., Liu, Z., & Peng, D. (2024). Genome-Wide Identification of the PEBP Family Gene in Three Cymbidium Species and Its Expression Patterns Analysis in C. ensifolium. Horticulturae, 10(3), 252. https://doi.org/10.3390/horticulturae10030252







