Abstract
The ERF subfamily, a significant part of the APETALA2/ethylene-responsive element-binding factor (AP2/ERF) transcription family, plays a crucial role in plant growth, development, and stress responses. Despite its importance, research on this gene family in sweet cherry (Prunus avium L.) is limited. This study identified and analyzed the sweet cherry ERF subfamily in terms of classification, physicochemical properties, structural characteristics, chromosome distribution, gene replication and collinearity, Cis-acting elements, and potential protein interactions. Preliminary investigations of transcription during fruit cracking and normal development were also conducted. Fifty ERFs (PatiERF1~50) were identified, distributed unevenly across eight chromosomes and classified into ten groups with nineteen conserved motifs. Collinearity analysis with other plant species revealed homology, with the highest number of ERF orthologous genes found in apple (Malus domestica L.). Cis-acting elements, particularly abscisic acid response factor, were abundant in PatiERF promoters. Weighted gene co-expression network analysis (WGCNA) and quantitative real-time PCR (RT-qPCR) analysis indicated the involvement of PatiERFs in sweet cherry fruit development and cracking, and nine and four significant candidates related to these processes were speculated, respectively. Furthermore, four other classes of transcription factors (TFs), namely MYB, GRAS, BHLH, and BZIP, as well as 23 structure genes, were predicted to have co-expression and interaction relationships with PatiERFs during fruit development. This suggests their potential synergistic regulation with ERFs in the cherry fruit development process. Our study represents the first comprehensive genome-wide analysis of the ERF subfamily in sweet cherry, laying a crucial foundation for a deeper understanding of the molecular mechanisms correlated with fruit growth, development, and cracking mediated by ERF genes.
1. Introduction
Sweet cherry (Prunus avium L.) stands out as an economically significant fruit species in the Prunus genus, known for its attractive appearance and taste. In recent years, the surge in cherry prices and significant consumer market potential have fueled an increasing interest in cherry cultivation, resulting in a rapid expansion of cultivation areas. However, this expansion has introduced a set of challenges, encompassing issues such as deformed fruit, diminished sweetness and flavor, fruit cracking, color discrepancies, vulnerability to cold and drought damage, and suboptimal post-harvest storage and preservation. These challenges have evolved into critical factors impeding the healthy progression of the cherry industry. Therefore, a thorough investigation into the regulatory network and mechanisms of key genes associated with fruit development becomes essential. Such a study holds significant implications for enhancing fruit quality and mitigating avoidable losses, ultimately contributing to the sustainable and prosperous development of the cherry industry.
Ethylene-related pathways are crucial for the growth, development, post-harvest, and stress responses in both climacteric and non-climacteric fruits [1]. The ethylene response factor (ERF), as the final response gene in the ethylene signaling pathway, is believed to regulate the expression of ethylene-related genes [2] and could feedback modulate phytohormone biosynthesis, including ethylene, cytokinin, gibberellin, and abscisic acid [3]. Thus, studying the ERFs in sweet cherry is essential for unraveling the mechanisms behind fruit development and stress responses associated with ethylene (ETH).
ERF is an important plant-specific transcription factor (TF). It comprises multiple groups and gene members [4]. It is also the largest subfamily belonging to the AP2/ERF gene family, characterized by a single AP2/ERF domain. Due to the vastness of this gene subfamily, scientists are filled with great curiosity about the diversity and uniqueness of the associated biological functions, leading to a series of research efforts. Studies have demonstrated that the ERF gene family plays a pivotal role in a wide array of regulatory functions throughout the plant life cycle, including seed germination, flowering, fruit ripening, organ senescence, programmed cell death, and responses to pathogen attacks and abiotic stresses [5,6,7]. Further classification within the ERF subfamily segregates it into the ERF and the C-repeat-binding factor/dehydration-responsive element-binding (CBF/DREB) groups based on discrepancies in the 14th and 19th amino acids within the AP2 domain [8]. In sweet cherry, several studies have examined the regulation of ERF genes in development. For instance, PaWINA, PaWINB, Pa_02691 (ERF023), Pa_00639 (ERF071), and Pa_00973 were found to be involved in exocarp development [9,10,11]. PavRAV2 was found to affect fruit size [12], and ERF-TFs were found to act during flowering transition [13]. Despite these findings, the understanding of the ERF gene’s role in developmental regulation in sweet cherry remains limited compared to other plant species.
Fruit cracking is a significant challenge during the development of sweet cherry fruit. While various chemical reagents have been employed in production to reduce cracking, their effectiveness is limited, and they often come with significant costs. The fundamental solution lies in breeding varieties with inherent resistance to cracking. Several ERFs, including Arabidopsis AtWIN1, AtSHN1, barley HvNUD [14,15,16], cucumber CsCER4 [17], watermelon ERF4 [18], and grape VviERF045 [19], have been linked to fruit cracking. Research indicates that these ERF genes primarily participate in the regulation of cell wall synthesis and metabolism [18]. However, knowledge concerning ERF genes with similar functions in sweet cherry is notably scarce. Moreover, the precise number of family members remains unknown. Thus, given that ERF genes constitute a large gene family, comprehensive studies to elucidate their members and characteristics are imperative for the exploration and exploitation of their functionality. These research efforts are of significant importance in formulating effective strategies to address issues such as fruit cracking and other developmental challenges in fruit development.
With the release of plant genome sequences, the ERF gene families have been extensively identified and characterized in numerous plants [20,21,22,23,24,25,26]. High-quality whole-genome sequences [27] facilitate this work in sweet cherry. Consequently, this study aimed to identify the ERF genes in sweet cherry at the whole-genome level, then explore their classification groups, establish collinearity relationships with other plant species, analyze the Cis-acting elements possessed in the promoter region, and investigate transcription and expression during normal fruit development at different developmental stages. Furthermore, we investigated their expression in cracked fruits from cultivars exhibiting varying degrees of resistance to fruit cracking. This comprehensive examination allowed us to gain a preliminary understanding of the characteristics of ERF genes, identifying several potential genes associated with fruit development and cultivar cracking. Our findings offer valuable insights into the mechanisms underlying cherry fruit development and stress-induced cracking mediated by ERFs, contributing to the development of cultivars with enhanced resistance to cracking.
2. Materials and Methods
2.1. Plant Material and Sample Collection
The experiment was conducted on two sweet cherry cultivars, Prunus avium L. ‘Tieton’ and ‘Mzhu’, planted in the garden of the sweet cherry germplasm at the Yantai Academy of Agricultural Sciences in Yantai, China (121°16′28″ E, 37°29′9″ N; altitude, 6 m above sea level) (Figure 1). Ten trees of each sweet cherry cultivar were used.

Figure 1.
Examined sweet cherry cultivars. (A) Tieton. (B) Mzhu.
The ‘Tieton’ is a cultivar with firm flesh that is susceptible to cracking; in contrast, the ‘Mzhu’ cultivar is less firm and more tolerant to cracking. The former is the most popular cultivar at present in China; the latter is a potentially good cultivar that is expected to be extended in the future. These two cultivars have basically the same ripening time. Both are grafted on ‘Daqingye’ rootstock.
Three sweet cherry fruit trees with favorable growth conditions and the same developmental stage were chosen. The fruits at four key stages of development, namely the initial fruit setting stage (the tenth day after full bloom; DAFB 10), stone hardening stage (DAFB 24), color transition stage (DAFB 38), and maturity stage (DAFB 52), were collected for the validation of the expression of nine crucial fruit development genes predicted based on transcriptome data. Fruits of ‘Tieto’ and ‘Mzhu’ harvested 52 days after flowering were subjected to water immersion-induced fruit-cracking experiments. Each sampling was performed between 9:00 AM and 10:00 AM. After harvesting, the fruits were promptly brought to the laboratory for photography. Subsequently, the flesh and skin were rapidly separated, frozen in liquid nitrogen, and stored at −80 °C for later use. Three biological replicates were set for each time point. A portion of the samples from the maturity stage was subjected to subsequent experiments involving warm-water cracking.
2.2. In Vitro Cracking Assay
A total of 140 ‘Tieton’ and ‘Mzhu’ fruits were randomly hand-picked from around 15-year-old trees at their commercial ripening stage. The initial fruit weight (g) was determined with an electronic balance (EW2200-2NM, Kern, Germany) before immersion; then, fruits with stems were completely immersed in distilled water at 22 °C. Fruit weight and the number of cracked fruits were recorded at 3 h and 6 h, respectively. At each time point, cracked cherries were removed and counted, and fruits without cracks were re-incubated. The following formula was used to calculate the fruit-cracking rate and water absorption of each variety at different treatment times.
Fruit cracking rate = Number of fruit cracking/total number of initially treated fruit × 100%
Water absorption rate = (Weight after water absorption-weight before water absorption)/weight before water absorption × 100%.
Before each weighing, the surface moisture was wiped with filter paper. Three duplicates of each treatment were utilized. At each treatment time, the skins of cracking and non-cracking cherry fruits were collected to analyze the expression of the ERF gene. After the samples were frozen in liquid nitrogen, they were immediately transferred to a −80 °C refrigerator until use.
2.3. Genome-Wide Identification of ERFs in Sweet Cherry
The whole genome of ‘Tieton’ (Prunus avium Tieton v2.0 genome) was used as the sweet cherry genome data in this study [27]. Genome sequence and annotation files were downloaded from the GDR databases (https://www.rosaceae.org/ accessed on 26 February 2024). The amino acid sequences of Arabidopsis thaliana ERF genes (AthERFs) were obtained from the TAIR database (https://www.arabidopsis.org/index.jsp accessed on 16 January 2022). Using AthERFs as a reference [8], ERF genes in the sweet cherry genome were obtained by a local BLASTP algorithm program. The obtained protein sequences were further tested for conservative structure and compared by Batch SMART, and the sequences containing the AP2 conserved domain were retained. A total of 50 non-redundant ERFs were obtained, which were renamed PatiERF01~PatiERF50 according to their positions on chromosomes. Jalview 2 software [28] was used to display the result of multiple sequence alignment and the conserved structure of the AP2 motifs of 50 PatiERFs. The identified PatiERFs were mapped onto sweet cherry chromosomes using TBtools software [29]. The similarity of the amino acid sequences between the previously published ERF genes of sweet cherry and our 50 PatiERFs genes was also determined using TBtools I software [29].
2.4. Basic Physicochemical Properties, Conserved Domain, and Gene Structure Analysis
The physical and chemical properties of PatiERFs were predicted by ProtParam (https://web.expasy.org/protparam/, accessed on 8 February 2022). The secondary structure, structure of the transmembrane domain, and subcellular location were investigated using PRABI-Gerland (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html, accessed on 8 February 2022), TMHMM (http://www.cbs.dtu.dk/services/TMHMM/, accessed on 8 February 2022), and the Plant-mPloc website (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/, accessed on 8 February 2022), respectively. Conserved motifs of PatiERFs were identified using MEME [30] (http://meme-suite.org, accessed on 8 February 2022). The maximum number of motifs found was set as 20. Gene structure analysis was performed using Tbtools software [29].
2.5. Phylogenetic Analysis
Based on the amino acid sequences of ERFs, an ML phylogenetic tree was constructed using MEGA11 [31] software, with a p-distance model. The MUSCLE method was adopted for multiple sequence alignment. The amino acid replacement model was JTT. Gap/missing data were possessed by complete deletion. The bootstrap value was 1000. Meanwhile, to determine the relations between the number of orthologous gene pairs (OGPs) of ERFs and the relative phylogenetic distances among different species, another phylogenetic tree was built based on the amino acid sequences of genes from the whole genomes of other plant species. The details of the additional nine plant species, genome versions, and sources are shown in Table 1.

Table 1.
Genome versions of plant species used in this study and their sources.
2.6. PPI Analysis
The STRING database was used for protein–protein interaction prediction. Species parameter was selected for A. thaliana plant. Cytoscape 3.8.2 was utilized to visualize the results of PPI [32].
2.7. Prediction of Cis-Acting Elements
The upstream 2000 bp of PatiERFs was extracted as promoter region. PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ accessed on 26 February 2024) [33] was used for Cis-acting element prediction.
2.8. Collinearity Analysis
MCScan [34] was used for collinearity analysis of the ERF genes within P. avium, as well as across different species (Table 1). The E value was set to 1 × 10−10, and the first five results were extracted by blast. Ka/Ks were calculated by TBtools software [29].
2.9. WGCNA Analysis
The standardized RNA-seq data (Reads/kb/Million, RPKM) for sweet cherry fruits at different growth stages were presented by Alkio et al. [10] (the accession number is GAJZ00000000 in GenBank). Contigs with predicted full-length open reading frames were used in this analysis. The WGCNA v1.69 package [35] was used to construct the co-expression network.
2.10. RT-qPCR Analysis
Two experiments were conducted using RT-qPCR. One involved the validation of the gene expression of PatiERFs predicted to be significantly associated with the developmental process through WGCNA based on transcriptome data. The other comprised a preliminary analysis of whether PatiERFs were involved in the water-induced cracking process. For the first experiment, the materials examined included fruits from different developmental stages of cv. ‘Tieton’. The second experiment used the fruit skins of two sweet cherry cultivars, ‘Tieto’ and ‘Mzhu’, subjected to water-induced cracking treatment for 3 h and 6 h, with non-induced fruits as controls. In total, 16 PatiERF genes underwent RT-qPCR experiments.
RT-qPCR was performed using SYBR-Green dye in a LightCycler 480 Real-Time PCR System (Roche). The qPCR experimental conditions were: 20 μL reactions, including 50~300 ng template cDNA, 0.5 μM forward and reverse primers, and 10 μL 2 × SYBR-Green premix. Each sample was analyzed in triplicate. The PCR procedure was 95 °C for 15 s, and 40 cycles of two-step amplification were performed (60 °C for 15 s, 68 °C for 20 s, and fluorescence signals were collected). Three biological replicates were performed for each RT-qPCR analysis. Fluorescence signals were collected to draw dissolution curves, and the relative expression of target genes was quantified by 2−ΔΔCT [36]. For extraction of total RNA, an RNAprep Pure Plant Plus Kit (Polysaccharides & Polyphenolics-rich) was used. First-strand complementary DNA (cDNA) synthesis was performed using a FastKing gDNA Dispelling RT SuperMix Kit. qPCR was performed using a SuperReal PreMix Plus (SYBR Green) kit. All the above kits were purchased from TIANGEN BIOTECH (Beijing) Co., LTD., China, and were used according to the manufacturer’s instructions. The Actin gene from sweet cherry (CACT1, GenBank FJ560908) was taken for normalization, and the primer sequence was referred to Balbontín et al. [11]. Specific primer sequences for the genes were designed using Primer Premier software [37]. The primers and sequences are shown in Table S2.
2.11. Statistical Analysis
The datasets of water absorption rate, fruit-cracking rate, and expression levels of PatiERFs obtained by RT-qPCR were subjected to statistical analyses. Analysis of variance was conducted using GraphPad Prism 8.0 on the data from three replicates. p values less than 0.01 were considered statistically significant.
3. Results
3.1. Genome-Wide Identification of ERF Gene Family and Basic Physicochemical Properties
The conserved AP2 domains of 50 PatiERFs are illustrated in Figure 2A, displaying a highly conserved structure with YGR (N-terminal) and RAYD (C-terminal) motifs, as well as three conserved β-sheet and α-helix regions. These features align with the typical structure of AP2/ERF proteins [8]. Since all PatiERFs exhibited a single AP2 conserved domain, they were classified as members of the ERF subfamily. The identified PatiERFs represented 0.124% of the genes annotated in the sweet cherry genome.
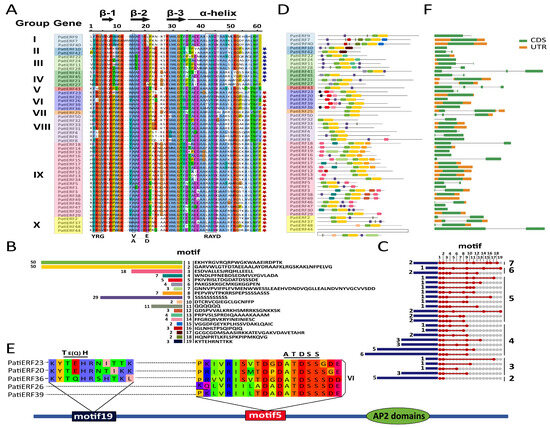
Figure 2.
Characteristics of PatiERFs. (A) The alignment of the AP2 domain, with diverse shading on gene names corresponding to different gene groups. Black bars and arrows indicate putative α-helix and β-sheet regions within the AP2 domain, respectively [38]. The blue and red stars represent the DREB subgroup and ERF subgroup, respectively. (B) 19 conserved motifs and their sequences, with varying color frames indicating distinct motifs. The frame length difference mirrors the prevalence of the motif in the PatiERF gene: longer columns indicate a higher number of genes sharing the motif. The numbers on the left denote the count of genes with the motif. (C) Patterns of different motif combinations concurrently present in PatiERFs and the number of corresponding PatiERFs for each pattern. Motifs within the same PatiERF are depicted by solid red dots connected by solid red lines. The numerical value on the leftmost side denotes the count of genes exhibiting a specific motif pattern, while the rightmost number signifies the number of motifs within the associated motif combination pattern. (D) MEME analysis on conserved domains within PatiERFs. (E) Characteristic structural sequences and comparison of five CRF-PatiERFs. (F) PatiERF structure. CDS: CODING Sequence; UTR: untranslated region.
Analysis of basic physicochemical properties and subcellular localization predictions (Table S1) indicated that PatiERF proteins had an average length of 287 amino acids, a molecular weight of 31,675 Dalton, and an average isoelectric point value of 6.75. Approximately 66% of PatiERFs were predicted to be localized in the nucleus. Significantly, the overall grand average of hydropathy for PatiERFs was predicted to be negative, indicative of their hydrophilic nature. The instability index ranged from 35.08 to 77.59, with an average of 54.36. No transmembrane region was anticipated.
3.2. Multiple Alignment and Classification of PatiERFs
To elucidate the sequence consistency and conservation of feature regions among PatiERFs, a comprehensive alignment was conducted for a set of 50 PatiERFs, and subsequently, a phylogenetic tree was constructed in conjunction with Arabidopsis ERFs (AthERFs). The role of AthERFs primarily served as a benchmark for the classification and functional prediction of PatiERFs. Following the classification scheme of AthERFs [8], sweet cherry PatiERFs could be categorized into ten groups (groups I~X), as illustrated in Figure 3.

Figure 3.
Phylogenetic analysis of PatiERFs and AthERFs. Distinct groups/subgroups are marked with different colored arcs. PatiERFs in sweet cherry are highlighted with solid red dots in the inner circle.
The Gly-4 (G), Arg-6 (R), Glu-18 (E), Trp-30 (W), Leu-31 (L), Ala-40 (A), Arg-8 (R), Gly-11 (G), Ile-19 (I), Arg-28 (R), Leu-31 (L), Gly-32 (G), Asp-45 (D), Ala-48 (A), Gly-53 (G), and Asn-59 (N) residues were found to be completely conserved (100%) across all PatiERFs (Figure 2A). Among the 50 PatiERFs, nine sequences displayed residues V and E at positions 16 and 21, adhering to the classical classification criteria of DREB, warranting assignment to the DREB subgroup. Conversely, 36 sequences featured residues A and D at these positions, aligning with the ERF subgroup classification [8]. The remaining five sequences possessed (V, L), (V, A), or (V, D) residue combinations, positioning them outside the delineations of both the ERF and DREB subgroups.
3.3. Conserved Motif Analysis and Gene Structure Analysis
Conserved motifs in TFs, typically associated with DNA binding, protein interactions, and transcriptional activity, are often shared within subgroups of extensive TF families in plants [8]. To explore this, we conducted an analysis of conserved motifs, examining their distribution in individual genes, the conservation within gene groups, and the commonalities and specificities between different groups.
A total of 19 conserved motifs were predicted. Motif 1 and motif 2 emerged as the primary motifs, as depicted in Figure 2B. Motif9 and motif3, presented in 58% (29/50) and 36% (18/50) of PatiERFs, respectively, exhibited prominence. The few motifs that were the least present were: motifs 10, 15, 17, and 18. PatiERFs exhibited variability in motif composition and abundance across species. Sequences with seven different motifs were the least frequent (Figure 2C).
The specificity and positional variability of each motif were observed among different group/subgroup members, resulting in diverse distribution patterns (Figure 2D). Additionally, within the same group of PatiERFs, motifs displayed diversity in their locations, with some fixed on the left part of the gene sequence and others distributed across both ends of the gene sequence (Figure 2D; Table S3). Furthermore, motifs specific to individual gene sequences or families were identified (Table S4). For instance, motif 3 was exclusive to group IX. Among the 19 motifs, motif 9 stood out as the most extensive beyond the AP2 domain, with a higher likelihood of appearing multiple times within the same sequence.
In Arabidopsis, cytokinin response factors (CRFs) were situated within the group VI and group VI-L phylogenetic branches of AthERF proteins. The conserved CRF protein structure comprises an N-terminal CRF domain (ATDSS) crucial for protein interactions. Some CRFs featured [THE (Thr-Glu-His)] regions, which are located at the left end of the AP2 domain [39]. In our PatiERFs, a total of five putative CRFs were identified, three of which exhibited both ATDSS and THE conserved domains, all classified within group VI. (Figure 2E).
To understand structural diversity, the exon and intron organization of PatiERF genes were also analyzed. The open reading frames of 25 PatiERFs contained one or more introns (Table S1; Figure 2F), ranging from one to three, among which 42% of genes contained a single intron, while 50% were intronless. Genes with minimal or fewer introns tend to exhibit efficient expression in response to stress conditions [40]. As a result, intronless ERF genes may rapidly respond to changes in the external environment [41]. Consequently, the overall lower number of introns in sweet cherry implied that these PatiERFs might effectively respond to environmental changes. Furthermore, we observed that PatiERFs within the same group/subgroup shared similar intron–exon organizations, suggesting the significance of intron–exon organization in gene evolution.
3.4. Distribution, Collinearity Analysis, and the Calculation of Ka/Ks
To unveil the relationship between the genetic divergence within the ERF subfamily and gene duplication in sweet cherry, the chromosomal distributions of PatiERFs were determined. Fifty PatiERFs were detected across all eight chromosomes (Figure 4A). Chromosome 2 (Chr2) harbored the highest number of PatiERFs, whereas Chromosome 7 (Chr7) had the fewest. The distribution profiles of PatiERFs on different chromosomes exhibited variations. For example, PatiERFs on Ch2 and Chr8 were concentrated in the arms of chromosomes, while those on Chr1 were evenly distributed. Different chromosomes contained different numbers of gene groups (Figure 4B). Chr1 and Chr6 possessed the largest number of PatiERF gene groups (six groups), while Chr4 and Chr7 had the smallest numbers of gene families. Additionally, different gene groups, as well as subgroups within the same group, displayed distinct preferences on different chromosomes (Figure 4B). For instance, group IV and subgroup Xa contained the same number of genes, but group IV was localized on Chr2, 3, 6, and 7, while subgroup Xa was distributed on Chr1, 5, 7, and 8.
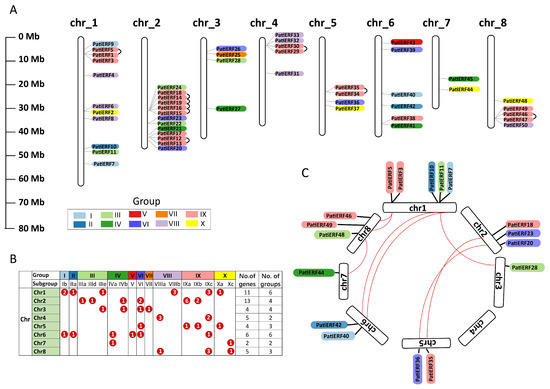
Figure 4.
Distribution and homology of PatiERFs on chromosomes (Chr). (A) Distribution of PatiERFs across the sweet cherry genome. The chromosome number is marked on the top of each chromosome. The black arc represents pairs of ERF genes with tandem duplication. (B) Groups/subgroups and the number of PatiERFs on each chromosome. (C) Collinearity of PatiERFs on different chromosomes. The red arc represents pairs of PatiERFs with tandem duplication.
Tandem duplication and segmental duplication are pivotal processes in the generation of new gene functions and the expansion of gene families. Nine pairs of tandem duplications were detected, forming six clusters distributed on five chromosomes, with the largest numbers (five pairs) distributed on Chr2, implying a hot area of PatiERF distribution (Figure 4A). No tandem duplicates were detected on Chr3, 6, or 7. Apart from tandem duplication, ERF genes in sweet cherry also underwent segmental duplication events. Eight segmental duplication events were detected on seven chromosomes of sweet cherry (Figure 4C).
Ka/Ks ratios are frequently employed to depict the selection pressure on genes and the pace of evolution. A Ka/Ks > 1 suggests positive selection and implies accelerated evolution. Conversely, a Ka/Ks < 1 suggests purifying selection, indicating a functional constraint. When Ka/Ks = 1, it signifies that genes are undergoing neutral migration [42]. Our further calculation of Ka/Ks values for homologous genes revealed values of less than 1 for all duplicated PatiERF gene pairs (Table S5), indicating that PatiERFs primarily evolved under the influence of purifying selection.
3.5. Collinearity Analysis of PatiERFs between Plants Species
To further explore the evolutionary relationships among ERFs in distinct plant species, collinearity analysis was conducted between sweet cherry and nine other plant species (Figure S1). The number of orthologous gene pairs (OGPs) across different species, to some extent, can reflect the variation and conservation of genes during evolution. In our study, PatiERFs OGPs were detected in all nine other plant species, revealing a variable distribution of OGPs, with the highest count observed in apples (114). PatiERFs distributed on Chr1, Chr2, and Chr6 in sweet cherry had the most OGPs with AthERFs (Figure 5B).

Figure 5.
Collinearity analysis between sweet cherry and other plant species. (A) Correlation (r) between the ratio of PatiERF orthologous gene pairs (OGPs) in nine other plant species and the corresponding genome size of each species. The left Y-axis represents the genome size, depicted in the chart using a bar graph; the right Y-axis represents the proportion of PatiERFs OGPs in each species to the corresponding genome size, presented in the chart using a line graph. (B) Distribution of PatiERFs with OGPs in other plant species on the sweet cherry chromosome. The numerical values in the cells, color-coded for depth, represent the percentage calculated from collinear ortholog pairs/the number of total ortholog pairs detected between sweet cherry and the respective plant species. (C) Word cloud generated from the total OGPs of PatiERFs compared with other plant species. Gene name size reflects the number of homologous pairs; larger names indicate more orthologous genes. PatiERF genes with orthologous genes in other plant species exceeding 20 are highlighted in red. (D) Evolutionary relationships of ten species (including sweet cherry) based on their whole genomes. (E) Number of homologous pairs for each PatiERF gene with other species. Sweet cherry-specific PatiERFs are highlighted with a pink shading bar, while light-blue shading indicates genes with homologous pairing relationships unique to individual species.
In assessing the broader context of PatiERF OGPs across sweet cherry and other plant species, we calculated the proportion relative to the genome size of each species. Remarkably, a statistically significant negative correlation (r = −0.810) was observed between the number of OGPs and the size of the respective genomes under comparison. Notably, species characterized by larger genomes displayed a comparatively smaller proportion of PatiERF OGPs, suggesting a potential lack of conspicuous differentiation of PatiERFs following species divergence (Figure 5A).
Our study found a connection between PatiERF OGPs and evolutionary distances among species (Figure 5D). Notably, Prunus armeniaca, Prunus persica (P. persica), Prunus avium (P. avium), and Malus domestica (M. domestica) belong to the Rosaceae family, with P. armeniaca, P. persica, and P. avium sharing the genus Prunus, having a close phylogenetic relationship. In comparison to more distantly related species like Citrullus lanatus, Solanum lycopersicum, and Actinidia chinensis, our results showed a larger proportion of OGPs to the total genome in closely related species, emphasizing the significance of these pairings in understanding evolutionary relationships. Additionally, M. domestica, which underwent genome-wide duplication around 5 million years ago [42], exhibited a genome size approximately twice that of P. armeniaca/P. persica. Despite this, the proportion of ERF OGPs in sweet cherry was significantly less in M. domestica (about half). This suggested that the ERF gene family did not undergo significant expansion or contraction after genome duplication, highlighting the conservative nature of their evolutionary process in Rosaceae. This observation aligned with our Ka/Ks analyses, indicating that the evolution of PatiERF genes primarily underwent purifying selection.
We also found varying numbers of OGPs (1 to 5) among different species (Figure 5C,E). Some genes, like PatiERF9, had a single ortholog across all species; others, such as PatiERF11, PatiERF23, PatiERF20, and PatiERF49, commonly had more orthologs across multiple species. Meanwhile, some genes, including PatiERF15 (0), PatiERF19 (0), PatiERF14 (1), and PatiERF16 (1), had minimal orthologs in other species. These findings highlighted the complex evolutionary processes of ERF genes, encompassing expansion and contraction. Genes appearing in multiple copies likely played crucial roles in plant adaptive evolution, while those appearing as single copies across species might be vital for shared plant functions or traits. Unique cherry genes might be linked to species-specific traits emerging after differentiation from other species.
3.6. Cis-Acting Element Analysis
TFs regulate gene expression by interacting with Cis-acting elements [43]. To understand the role of PatiERF genes in stress responses and hormone actions, we analyzed Cis elements in the promoters of 50 PatiERFs. These elements fell into four categories: light-responsive (647), phytohormones-responsive (410), stress-responsive, and growth- and development-responsive (90) (Table S6; Figure 6A). Abscisic acid- (ABA) (170) and methyl jasmonate (MJ) (112)-responsive elements were more abundant than salicylic acid (SA), gibberellin (GA), and auxin (IAA). Within stress-responsive elements, hypoxia induction-responsive elements were most prevalent (145).

Figure 6.
Cis-acting elements predicted in the promoters of PatiERFs. (A) The proportion of different Cis elements in the promoters of PatiERFs. (B) The proportion of genes with different Cis elements in the promoter region.
The proportion of different Cis-acting elements detected in the promoter region was varied (Figure 6B). The most widely distributed were light-responsive Cis-acting elements, followed by hypoxia induction- and abscisic acid (ABA)-responsive factors. The least presented Cis-acting elements were root-specific, organ meristem, dehydration, low-temperature, and salt stress-responsive elements. The types and number of Cis-acting elements in each PatiERF varied (from 3 to 12). Some were unique to a certain gene, such as PatiERF7, PatiERF36, and PatiERF46. The details of Cis-acting elements and quantities contained in each PatiERF promoter are shown in Table S6.
Each PatiERF displayed a different mix and quantity of Cis-acting elements (3 to 12). Some, like PatiERF7, PatiERF36, and PatiERF46, featured unique elements.
3.7. Protein–Protein Interaction (PPI) Analysis
To obtain preliminary information about the interaction relationship between PatiERFs and other proteins, we performed PPI analysis using orthology-based prediction in Arabidopsis. Out of the 50 PatiERFs, 33 were mapped to the A. thaliana genome. Functional predictions revealed four associated with root development, three with cell growth, and the rest primarily involved in stress induction and signaling. In 27 out of these 33 PatiERFs, protein interactions were observed, including interactions between PatiERFs and other non-PatiERF proteins (Figure 7). The involved proteins primarily function in crucial hormone signaling pathways such as jasmonic acid, cytokinin (CK), and ethylene. Additionally, they participate in stress-induced responses and transcriptional regulation. Most of the transcriptional regulation involves activators, with three genes (PatiERF31, PatiERF33, and PatiERF50) predicted to have repressor functions, interacting with HD1 and SNL3 as predicted functional partners. Only one KEGG pathway was predicted: ath04016, representing the MAPK plant signaling pathway.

Figure 7.
Protein–protein interaction (PPI) analysis of PatiERFs. 27 PatiERFs mapped to the Arabidopsis genome and their putative interacting proteins. The dots with magenta shadow denote PatiERFs; the dots with turquoise shadow represent their putatively interactive proteins. The dot size correlates with the number of interacting proteins; larger dots indicate a higher count of interacting proteins.
In the molecular function annotations of Gene Ontology (GO), fatty acyl-CoA binding (GO:0000062) was predicted. In terms of biological processes, the false discovery rate (FDR) of intercellular signal processing ranked third (1.26 × 10−97), following ethylene-activated signal transduction and ethylene response. Subcellular localization predictions identified three compartments associated with the cell membrane: the GOCC:0043231 intracellular membrane-bounded organelle, GOCC:0043227 membrane-bounded organelle, and GOCC:0043229 intracellular organelle. Additionally, the Myb complex (GOCC:0031523) was discovered.
3.8. PatiERF Expression and Weighted Gene Co-Expression Network Analysis (WGCNA) in Normally Developing Fruits
To investigate the transcription and expression patterns of PatiERFs during sweet cherry fruit development, we utilized published RNA-seq datasets [10]. Through WGCNA analysis, we identified modules correlated with distinct developmental stages, then analyzed PatiERF transcription within key modules and obtained co-expression gene sets associated with PatiERFs at various stages. By annotating homologous proteins and predicting interactions, we deduced structural genes, transcription factors, and potential transcriptional regulatory networks interacting with PatiERFs at each developmental stage. Our results showed that 15 PatiERFs were significantly differentially expressed during normal fruit development. They belonged to eight groups, with group III and group IX accounting for the largest proportion (Figure 8A). Group III ERFs are widely believed to regulate cell wall synthesis genes [18,44,45]. WXP1 is involved in activating wax production in the acyl reduction pathway [46]. Among our developmentally related PatiERF genes, PatiERF11, PatiERF22, and PatiERF28 belonged to group III. PatiERF7 and PatiERF40 showed high homology to the WXP1 protein in Medicago sativa. Therefore, considering that genes in the same group or with close homologs often share similar functions [8], we speculated that PatiERF7, 11, 22, 28, and 40 might play important roles during sweet cherry development by influencing cell wall synthesis or metabolism.

Figure 8.
The expression of PatiERF genes during the normal development of sweet cherry fruits. (A) Significantly expressed PatiERFs and their group affiliations. (B) Heat map of the correlations between modules and traits. The corresponding correlation and p value (in brackets) displayed in each cell. Each row corresponds to a module, and each column represents a different characteristic. A pentagram denotes a significant correlation, with yellow highlighting the module containing PatiERFs. (C) PatiERFs in key modules, the number of proteins interacting in each module, the group/subgroup affiliations, and associated developmental stage. (D) The putative TFs interacting with PatiERFs in the key modules. (E) PPI network between PatiERFs and the proteins corresponding to their putative co-expression genes in five key modules. Genes annotated with ‘Pa_’ are detailed in Table S7. (E.1–E.5) Heat maps representing the expression levels of PatiERFs and their co-expression genes at different developmental stages within specific modules. Darker red hues indicate higher gene expression levels, while lighter blue hues signify lower expression.
We further utilized the Good Samples Genes MS function in the WGCNA package to detect and filter genes with excessive missing values. A total of 6500 transcripts with full-length ORF were obtained for subsequent analysis. A weighted gene network with a soft threshold power of 7 and a cutoff of 0.85 was constructed, defining modules with a minimum of 30 genes. Merging parameters mergeCutHeight = 0.25 and maxBlockSize = 5000 produced 18 modules. Based on the premise of dividing the sequencing samples into three developmental stages (Stage I~Stage III) according to the cherry S-shaped growth curve [10], seven modules were detected to have a high correlation with the developmental stages. Among the 15 PavERF genes, four were filtered out, while nine were found to be distributed among five key modules (blue, brown, red, turquoise, and yellow). They were involved in all three developmental stages of the fruit (Figure 8B). Both positive and negative correlations exist in the relationship between modules and traits.
Within the five key modules significantly related to fruit development, we identified 9 transcripts of PatiERFs and 31 putative proteins interacting with them (Figure 8C; Table S7). Among the predicted interacting proteins, eight TFs were identified, categorized into four kinds of TF family, namely zinc finger, MYB, GRAS, and BHLH (Figure 8D). We observed that TFs interacting with PatiERFs were most diverse at Stage I. MYB was detected at both Stage I and Stage III. Additional proteins, such as ATL2, RING/U-box, ATPDI12, ETO1-like 1, ADH, ATBCAT-2, SUB1 calcium ion-binding, and ACBP2 acyl-CoA-binding protein 2, were predicted to interact with PatiERFs. Further analyzing the expression heat map (Figure 8E) revealed that PatiERFs exhibited high expression mainly in the middle and late stages of fruit development, indicating their role in these phases. PatiERF28 stood out with high expression at the earliest stage, suggesting involvement in fruit initiation. Both positive synergistic and negative inhibitory expressions were observed.
To validate RNA-seq accuracy, we used RT-qPCR to confirm the expression of nine PatiERFs correlated with cherry fruit development, as identified via WGCNA. Sample collection spanned four key development stages, focusing on fruit peel tissue. RT-qPCR results closely matched transcriptional predictions, as shown in Figure S2, affirming RNA-seq reliability.
3.9. Analysis of PatiERFs Expression in Simulated Cracked Fruit Treated with Water
To investigate PatiERF transcription during fruit cracking, we conducted an in vitro cracking assay on varieties with differing susceptibility. We measured water absorption and fruit cracking, crucial indicators of cracking characteristics, and obtained PatiERF expression profiles. Rapid water absorption, a key factor in fruit cracking, was observed within 0 to 3 h (h), peaking at 3 h and slowing thereafter (Figure 9A). Correspondingly, fruit-cracking rates increased with water absorption (Figure 9B). In general, both examined varieties followed a similar trend, but ‘Tieton’ exhibited higher values than ‘MZhu’.

Figure 9.
Water absorption rate (A), fruit-cracking rate (B), and expression levels of seven PatiERFs in ‘Tieton’ and ‘Mzhu’ fruits under immersion treatments. (C) Statistical analysis of the relative expression levels of PatiERFs between different cultivars and under various treatment times. Bars represent the mean of replicates ± standard error. Asterisks (*, **, ***, and ****) denote statistically significant differences at p < 0.05, 0.01, 0.001, and 0.0001, respectively.
In RT-qPCR analysis of seven PatiERFs, diverse expression profiles were observed across different cultivars and water treatment durations (Figure 9C). Specifically, PatiERF20 showed increasing expression in ‘Tieton’ over immersion time, contrasting with a decreasing trend in ‘Mzhu’. Significant expression differences between the two cultivars were noted at 3 h and 6 h (p < 0.01). PatiERF42 exhibited significant down-regulation in both cultivars (p < 0.01), with a more pronounced decrease in ‘Tieton’. PatiERF39 displayed a decrease–increase pattern in both cultivars, reaching significance in ‘Tieton’ after 3 h (p < 0.05) and becoming highly significant after 6 h. In ‘Mzhu’, significance was reached within 3 h, with a slight increase approaching control levels at 6 h. PatiERF13 expression increased in both cultivars, peaking in ‘Tieton’ at 3 h and remaining significantly different from the control in ‘Mzhu’ at 6 h. PatiERF25 in ‘Tieton’ showed significant differences at 3 h, decreasing to control levels afterward, while no significant difference was exhibited in ‘Mzhu’. Initially, PatiERF36 showed no differentially induced expression in either cultivar within the first 3 h of immersion treatment. However, significant differential expression emerged between the treatment and the control in both cultivars after 6 h. Notably, ‘Tieton’ exhibited up-regulated expression of PatiERF36, whereas ‘Mzhu’ showed down-regulation.
Except for PatiERF10 (in both ‘Tieton’ and ‘Mzhu’) and PatiERF25 (in ‘Mzhu’), all other PatiERFs exhibited significant differential expression in the two cultivars following water-induced cracking treatment. In ‘Tieton’, five were up-regulated, and one showed significant down-regulation (Figure S3A). Meanwhile, in ‘Mzhu’, one PatiERF was up-regulated, and three were significantly down-regulated (Figure S3B).
4. Discussion
The ERF genes play crucial and versatile roles in various plant species, but research on these genes in sweet cherry has been limited. Consequently, the functions of many ERF gene members in this plant species remain unclear. Recognizing the need for a comprehensive understanding, we conducted a detailed and systematic identification and characterization analysis of the ERF gene family in sweet cherry at the whole-genome level. Our investigation also included the preliminary examination of their transcripts and expression patterns during normal fruit development and in response to fruit-cracking conditions. This study represents the first comprehensive genome-wide report on the ERF gene family in sweet cherry, providing essential foundational data for future research on the functions of ERF genes in this context.
4.1. Sequence and Structural Characteristics of ERF Genes in Sweet Cherry
The amino acid sequences of the PatiERF gene family obtained in this study commonly feature the AP2 characteristic domain, crucial components or regions such as α-helix and β-folds, and conserved bases at specific positions. These results align well with the criteria for defining typical characteristics of ERF subfamily members as outlined by previous studies [47,48,49], indicating our successful identification of members within the cherry ERF gene subfamily. In order to investigate the relationships between the ERFs previously studied and mentioned in the Introduction and our PatiERF genes, a comparison of gene similarities based on their amino acid sequences was also conducted. The detected amino acid similarity between genes ranged from 25.45% to 97.94% (Table S9). No genes that were completely identical to PatiERFs were found. PavRV2 showed the lowest similarity to all tested gene sequences, as it belongs to another subfamily, namely RV2 within the AP2/ERF gene family.
Our study revealed that 11 PatiERFs might have transcriptional inhibitory effects. Transcriptionally repressive ERFs typically exhibit two structural features [50]. Feature 1 includes the EAR conserved motif [D(L/M)NXXP or DLNXXP], and feature 2 comprises the B3 repression domain (BRD, R/KLVGV). In our results, PatiERFs satisfying feature 1 were found in subgroup IIa (PatiERF42 and PatiERF10) and group VIII (PatiERF32, PatiERF50, PatiERF31, and PatiERF33). Those satisfying feature 2 criteria included PatiERF9 (Ib), PatiERF20 (VI), and PatiERF6 (VIIIb). Additionally, the conserved B3 motif DML mentioned in A. thaliana [8] was also present in PatiERF15 (IXa) and PatiERF44 (Xc), designating them as EAR-ERF genes. Studies have reported that ERF genes containing the EAR motif act as negative regulators of ETH, JA, and ABA hormone-responsive genes [51,52,53]. For instance, the EAR motif in wheat Q protein and rice OsERF3 protein represses transcriptional activity [54]. Therefore, we speculated that the identified 11 EAR-PatiERFs might function in the transcriptional inhibitory activity of their target proteins. Another study reported that the inhibitory function of EAR-ERF genes could be overridden in ERF genes with the EDLL motif [50]. The eligible genes in our study were PatiERF47 and PatiERF1 in subgroup IXc.
4.2. PatiERFs Potentially Involved in Sweet Cherry Fruit Development
In this study, we identified 15 differentially transcribed PatiERFs from sweet cherry fruits based on transcriptome datasets [10]. Through WGCA, we found that nine of them were significantly correlated with development. Notably, PatiERFs associated with development were detected at each developmental stage, indicating their involvement in sweet cherry fruit development. Unlike the majority of previous studies focusing on ERF roles during fruit ripening [55,56,57], our findings revealed that ERFs function in early developmental stages, aligning with observations in plum, tomato, and kiwi [58,59,60]. This suggested the importance of extending ERF gene research beyond fruit maturation stages to include early development.
In our study, 23 structural genes and four kinds of TFs, including MYB, BZIP, BHLH, and GRAS, were obtained. These TFs have been widely validated, playing important regulatory roles in many aspects of plant growth and development, signal transduction, and secondary metabolism [61]. Thus, our result shows that the development of sweet cherry fruit is a very complex process involving the regulation of abundant genes. The predicted interaction between ERF and these TFs could serve as a starting point for further investigation of the regulating mechanism of fruit development at various stages.
TFs affect plants in several ways. In terms of ERFs, they exert their influence by directly acting on structural genes [62,63,64,65], as well as forming diverse TF complexes [56,66,67,68]. In this study, 23 structural genes, such as E3 ubiquitin ligase (RNF217), alcohol dehydrogenase1(AtADH1), aldo-keto reductase (AT2G37770.2), ABC protein (AtRABC1), membrane lipoprotein (ATTIL), and lipid transporter (ACBP), were identified for co-expression and potential cooperation with PatiERFs. These genes have been demonstrated to be involved in a variety of life processes, like cell wall synthesis and metabolism, energy metabolism, secondary metabolite metabolism, cell composition, flavor formation (alcohol dehydrogenase and aldo-keto reductase), and fruit color transformation (E3 ubiquitin ligase). Therefore, based on previous literature highlighting the regulatory effects of ERFs on fruit growth and ripening, we inferred a close association between our nine PatiERFs and sweet cherry fruit development. These PatiERFs, along with 4 major TF families and 23 structural genes identified in this study, serve as crucial candidates for the exploration of the molecular mechanisms underlying ERF-mediated sweet cherry fruit development.
4.3. Potential Involvement of PatiERFs in Sweet Cherry Fruit Cracking
In this study, 3 h and 6 h after immersing cherry fruits in water were chosen as two crucial experimental time points. The selection was based on the time adopted in previous literature reports [69,70]. We also considered crucial time points observed during our actual experimental process when characteristics exhibited particularly noticeable changes.
ERFs have been implicated in fruit cracking. For instance, ClaERF4 directly contributes to crack resistance in various watermelon varieties [18], and in apples, MdSHN3 regulates cuticle formation, a factor linked to fruit cracking [71]. Nonetheless, in general, there is a limited number of reports on ERF genes functionally connected to fruit cracking. In sweet cherry, only a few cases have been reported [11,72]. qCrack-LG1.1m was concluded to be more reliable for elucidating the phenotype variation of fruit-cracking traits [72]. Therefore, intrigued by this, we conducted an analysis to predict the genes harbored by it. Fortunately, three ERF genes were discovered (Table S8). Furthermore, through BlastP and RT-qPCR analysis, we found that four (PatiERF13, 20, 36, 39) out of our five PatiERF genes (PatiERF13, 20, 25, 36, and 39) with high homology to ERFs in this interval exhibited significant gene expression differences in varieties with varying susceptibility to cracking. Therefore, in general, this discovery provided support for the speculated functional role of the qCrack-LG1.1m QTL, and the PatiERFs, including PatiERF13, PatiERF20, PatiERF36, and PatiERF39, identified within its interval can offer valuable insights in terms of isolating candidate ERF genes. Further research on the function of these genes could contribute to understanding the mechanisms of fruit cracking in different varieties mediated by ERFs.
It is noteworthy that among the aforementioned five PatiERFs genes, PatiERF20, PatiERF36, and PatiERF39 were also ERF genes containing specific CRF domains (ERF-PatiERF). In addition, two other PatiERF genes, PatiERF10 and PatiERF42, selected for RT-qPCR analysis of fruit cracking also included unique domains known as EAR (EAR-PatiERF). Our results showed that PatiERF10 and PatiERF42 had no significantly differentl expressions in different varieties, suggesting a weaker association with fruit-cracking variations. In contrast, PatiERF20, PatiERF36, and PatiERF39 (CRF-ERFs) exhibited significant differences between varieties, suggesting a potential role in varying susceptibility to fruit cracking. Therefore, the preliminary result of this study suggested that CRF-ERFs might contribute more significantly to fruit cracking traits in different cherry varieties. Interestingly, the expression trend of these three CRF-ERFs differed notably between the two varieties, implying their distinct involvement in fruit-cracking disparities. This raises questions about whether these differences could be attributed to varying modes of expression regulation and how they are influenced. Additionally, it remains unclear whether these genes will exhibit consistent expression patterns when more varieties with fruit-cracking differences are included. The extent of the correlation between these genes and variability in cherry varieties regarding fruit-cracking differences requires further investigation.
Cytokinin response factors (CRFs) belong to the AP2/ERF gene family and are named for the conserved CRF domain that they contain [39]. Among the ERF genes, CRF genes constitute a relatively small proportion, with approximately 10 identified in Arabidopsis [73], 11 in S. lycopersicum [74], and 21 in Brassica napus [75]. In our study, a total of five CRFs were identified in sweet cherry. As crucial responders to CK [39], CRFs can undergo CK-induced treatments and play a role in regulating the CK signaling pathway, influencing various life processes [76,77,78]. Building on this, our study also preliminarily identified three CRFs that may respond to CK and play a role in fruit cracking.
In agricultural practices, CK has been employed as a management measure to reduce the rain-induced fruit-cracking incidence [79]. Therefore, we hypothesize that CK treatments might reduce fruit-cracking events by affecting the expression of CRF-ERF genes or by influencing their related metabolic pathways. Although CK has been historically used for fruit-cracking prevention, the molecular mechanisms associated with fruit cracking are not well understood. Consequently, our study results inspire future investigations. CRF genes could serve as entry points to explore the mechanism by which CK reduces cherry fruit cracking, shedding light on the involved signal transduction pathways. This, in turn, could provide a basis for understanding the occurrence of fruit cracking, isolating genes related to crack resistance, and breeding crack-resistant varieties. However, it is crucial to note that our gene expression investigation was preliminary, based on a limited sample size. Further detailed work is needed to validate these findings across a wider range of varieties.
5. Conclusions
This study presented the first systematic identification of the ERF gene family in the entire genome of sweet cherry, followed by a detailed investigation of its characteristics. The expression patterns of the ERF genes were also explored across all developmental stages of fruits, both under normal conditions and when exposed to water-induced cracking. We obtained comprehensive information on the grouping, structural characteristics, evolutionary relationships with other ERFs, Cis-acting elements, and gene expression patterns. Additionally, we found nine ERF genes to be significantly associated with different developmental stages of normal cherry fruits. Moreover, we speculated on 4 major classes of TFs and 23 potentially crucial structural genes that might interact with these ERF genes during normal fruit development. Furthermore, we detected four ERF candidate genes potentially correlated with distinct cracking traits in different sweet cherry varieties. This study provides an important foundation for further understanding of the molecular mechanisms of fruit growth, development, and cracking mediated by ERFs.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae10030247/s1. Figure S1: The syntenic relationships of ERF genes in P. avium with P. armeniaca, P. persica, M. domestica, Z. jujuba, C. lanatus, A. thaliana, V. vinifera, A. chinensis, and S. lycopersicum. Syntenic gene pairs are connected by red-colored lines. Figure S2: Expression profiles of PatiERF genes in normally developing fruits. The bar charts represent the results of qRT-PCR for nine presumed crucial genes in fruit development. The expression patterns of each PatiERF gene from RNA-seq data are depicted by groups of four connected colored circles. The x-axis shows different sampling dates, and S1, S2, S3, and S4 represent DAFB10, DAFB24, DAFB38, and DAFB52, respectively. The y-axis indicates the relative expression levels of genes. The data are the average of three biological replicates, with error bars representing the standard deviation from three independent techniques. Figure S3: Relative expression levels of the seven PatiERFs at 3 h and 6 h after treatments in ‘Tieton’ (A) and ‘Mzhu’ (B). Table S1: Primers and sequences used in this study. Table S2: List of all PatiERFs and their basic information. Table S3: Primer distribution characteristics of motifs in PatiERFs. Table S4: Motifs unique to specific PatiERFs/groups/subgroups. Table S5: Estimated Ka/Ks ratios of duplicated PatiERFs in sweet cherry. Table S6: Cis-acting elements predicted in PatiERFs. The cell-fill color transitions from green through yellow to red, indicating a gradual increase in the number of Cis-acting elements present. Table S7: Annotations of proteins with interactive relationships with PatiERFs. Table S8: The ERF genes in the qCrack-LG.1m interval and their highest homologous PatiERFs obtained using BLASTP 2.12.0+. Table S9: The similarity of the amino acid sequences between the ERF genes of sweet cherry already published and mentioned in the Introduction of this study and our 50 PatiERFs.
Author Contributions
Conceptualization, X.D. and X.Z.; methodology, Y.L. (Yanju Li) and Y.W. (Yilin Wang); software, P.L., J.Z. and Z.B.; validation, X.D. and X.Z.; formal analysis, Y.W. (Yanbo Wang) and J.L.; investigation, X.D. and Y.W. (Yanbo Wang); resources, X.D. and Y.L. (Yunfen Liu); data curation, X.Y. and Y.W. (Yanbo Wang); writing—original draft preparation, Y.W. (Yanbo Wang) and X.D.; writing—review and editing, X.Y., L.Z. and S.Z.; visualization, X.D. and Y.W. (Yilin Wang); supervision, M.L. and Z.S.; project administration, M.L. and X.L.; funding acquisition, X.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Key R&D Projects in Tibet Autonomous Region (XZ202001ZY0022N) and the earmarked fund for CARS (CARS -30).
Data Availability Statement
All relevant data are presented within the manuscript and its Supporting Information files.
Acknowledgments
The authors are grateful to S.S. Sun for help with the experiment.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gao, J.; Zhang, Y.; Li, Z.; Liu, M. Role of ethylene response factors (ERFs) in fruit ripening. Food Qual. Saf. 2020, 4, 15–20. [Google Scholar] [CrossRef]
- Liu, M.; Gomes, B.L.; Mila, I.; Purgatto, E.; Peres, L.E.; Frasse, P.; Maza, E.; Zouine, M.; Roustan, J.P.; Bouzayen, M. Comprehensive profiling of ethylene response factor expression identifies ripening-associated ERF genes and their link to key regulators of fruit ripening in tomato. Plant Physiol. 2016, 170, 1732–1744. [Google Scholar] [CrossRef]
- Gu, C.; Guo, Z.H.; Hao, P.P.; Wang, G.M.; Jin, Z.M.; Zhang, S.L. Multiple regulatory roles of AP2/ERF transcription factor in angiosperm. Bot. Stud. 2017, 58, 6. [Google Scholar] [CrossRef]
- Wessler, S.R. Homing into the origin of the AP2 DNA binding domain. Trends Plant Sci. 2005, 10, 54–56. [Google Scholar] [CrossRef]
- Jofuku, K.D.; Den Boer, B.; Van Montagu, M.; Okamuro, J.K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [PubMed]
- Hirota, A.; Kato, T.; Fukaki, H.; Aida, M.; Tasaka, M. The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of Arabidopsis. Plant Cell 2007, 19, 2156–2168. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Alkio, M.; Jonas, U.; Sprink, T.; van Nocker, S.; Knoche, M. Identification of putative candidate genes involved in cuticle formation in Prunus avium (sweet cherry) fruit. Ann. Bot. 2012, 110, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Alkio, M.; Jonas, U.; Declercq, M.; Van Nocker, S.; Knoche, M. Transcriptional dynamics of the developing sweet cherry (Prunus avium L.) fruit: Sequencing, annotation and expression profiling of exocarp-associated genes. Hortic. Res. 2014, 1, 11. [Google Scholar] [CrossRef]
- Balbontín, C.; Ayala, H.; Rubilar, J.; Cote, J.; Figueroa, C.R. Transcriptional analysis of cell wall and cuticle related genes during fruit development of two sweet cherry cultivars with contrasting levels of cracking tolerance. Chil. J. Agr. Res. 2014, 74, 162–169. [Google Scholar] [CrossRef]
- Qi, X.; Liu, L.; Liu, C.; Song, L.; Dong, Y.; Chen, L.; Li, M. Sweet cherry AP2/ERF transcription factor, PavRAV2, negatively modulates fruit size by directly repressing PavKLUH expression. Physiol. Plant. 2023, 175, e14065. [Google Scholar] [CrossRef]
- Villar, L.; Lienqueo, I.; Llanes, A.; Rojas, P.; Perez, J.; Correa, F.; Sagredo, B.; Masciarelli, O.; Luna, V.; Almada, R. Comparative transcriptomic analysis reveals novel roles of transcription factors and hormones during the flowering induction and floral bud differentiation in sweet cherry trees (Prunus avium L. cv. Bing). PLoS ONE 2020, 15, e0230110. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Dixit, S.; Jetter, R.; Thoenes, E.; Van Arkel, G.; Pereira, A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 2004, 16, 2463–2480. [Google Scholar] [CrossRef] [PubMed]
- Broun, P.; Poindexter, P.; Osborne, E.; Jiang, C.Z.; Riechmann, J.L. WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 4706–4711. [Google Scholar] [CrossRef]
- Taketa, S.; Amano, S.; Tsujino, Y.; Sato, T.; Saisho, D.; Kakeda, K.; Nomura, M.; Suzuki, T.; Matsumoto, T.; Sato, K. Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proc. Natl. Acad. Sci. USA 2008, 105, 4062–4067. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, S.; Li, M.; Hou, L. Cloning and expression analysis of Cucumis sativus L. CER4 involved in cuticular wax biosynthesis in cucumber. Biotechnol. Biotec. Eq. 2018, 32, 1113–1118. [Google Scholar] [CrossRef]
- Liao, N.; Hu, Z.; Li, Y.; Hao, J.; Chen, S.; Xue, Q.; Ma, Y.; Zhang, K.; Mahmoud, A.; Ali, A. Ethylene-responsive factor 4 is associated with the desirable rind hardness trait conferring cracking resistance in fresh fruits of watermelon. Plant Biotechnol. J. 2020, 18, 1066–1077. [Google Scholar] [CrossRef]
- Leida, C.; Dal Rì, A.; Dalla Costa, L.; Gómez, M.D.; Pompili, V.; Sonego, P.; Engelen, K.; Masuero, D.; Ríos, G.; Moser, C. Insights into the role of the berry-specific ethylene responsive factor VviERF045. Front. Plant Sci. 2016, 7, 1793. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, X. Genome-wide identification of AP2/ERF superfamily genes and their expression during fruit ripening of Chinese jujube. Sci. Rep. 2018, 8, 15612. [Google Scholar] [CrossRef]
- Girardi, C.L.; Rombaldi, C.V.; Dal Cero, J.; Nobile, P.M.; Laurens, F.; Bouzayen, M.; Quecini, V. Genome-wide analysis of the AP2/ERF superfamily in apple and transcriptional evidence of ERF involvement in scab pathogenesis. Sci. Hortic. 2013, 151, 112–121. [Google Scholar] [CrossRef]
- Zhang, C.; Shangguan, L.; Ma, R.; Sun, X.; Tao, R.; Guo, L.; Korir, N.; Yu, M. Genome-wide analysis of the AP2/ERF superfamily in peach (Prunus persica). Genet. Mol. Res. 2012, 11, 4789–4809. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, D.; Hu, S.; Liu, X.; Zeng, B.; Gao, W.; He, Y.; Qin, H.; Ma, X. Genome-wide analysis of the almond AP2/ERF superfamily and its functional prediction during dormancy in response to freezing stress. Biology 2022, 11, 1520. [Google Scholar] [CrossRef]
- Zhuang, J.; Peng, R.H.; Cheng, Z.M.M.; Zhang, J.; Cai, B.; Zhang, Z.; Gao, F.; Zhu, B.; Fu, X.Y.; Jin, X.F. Genome-wide analysis of the putative AP2/ERF family genes in Vitis vinifera. Sci. Hortic. 2009, 123, 73–81. [Google Scholar] [CrossRef]
- Xie, X.L.; Shen, S.L.; Yin, X.R.; Xu, Q.; Sun, C.D.; Grierson, D.; Ferguson, I.; Chen, K.S. Isolation, classification and transcription profiles of the AP2/ERF transcription factor superfamily in citrus. Mol. Biol. Rep. 2014, 41, 4261–4271. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Du, X.; Liu, M.; Liu, X.; Zhao, L.; Cao, L.; Zhang, S.; Song, L.; Sun, Y.; Liu, D. Genome-wide analysis of the AP2/ERF family in Oily Persimmon (Diospyros oleifera) and their preliminary roles exploration in response to polyamines for adventitious root formation in cultivated persimmon (D. kaki). Horticulturae 2023, 9, 191. [Google Scholar] [CrossRef]
- Wang, J.; Liu, W.; Zhu, D.; Hong, P.; Zhang, S.; Xiao, S.; Tan, Y.; Chen, X.; Xu, L.; Zong, X. Chromosome-scale genome assembly of sweet cherry (Prunus avium L.) cv. Tieton obtained using long-read and Hi-C sequencing. Hortic. Res. 2020, 7, 122. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.; Frank, M.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in bipolymers. In Proceedings of the International Conference on Intelligent Systems for Molecular Biology, Stanford, CA, USA, 14–17 August 1994. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models. Genome Res. 1971, 13, 426. [Google Scholar]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Singh, V.K.; Mangalam, A.; Dwivedi, S.; Naik, S. Primer premier: Program for design of degenerate primers from a protein sequence. Biotechniques 1998, 24, 318–319. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.D.; Yamasaki, K.; Ohme Takagi, M.; Tateno, M.; Suzuki, M. A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 1998, 17, 5484–5496. [Google Scholar] [CrossRef] [PubMed]
- Rashotte, A.; Goertzen, L. The CRF domain defines cytokinin response factor proteins in plants. BMC Plant Biol. 2010, 10, 74. [Google Scholar] [CrossRef]
- Heyn, P.; Kalinka, A.T.; Tomancak, P.; Neugebauer, K.M. Introns and gene expression: Cellular constraints, transcriptional regulation, and evolutionary consequences. Bioessays 2015, 37, 148–154. [Google Scholar] [CrossRef]
- Zafar, M.M.; Rehman, A.; Razzaq, A.; Parvaiz, A.; Mustafa, G.; Sharif, F.; Mo, H.; Youlu, Y.; Shakeel, A.; Ren, M. Genome-wide characterization and expression analysis of ERF gene family in cotton. BMC Plant Biol. 2022, 22, 134. [Google Scholar] [CrossRef]
- Fawcett, J.A.; Maere, S.; Van De Peer, Y. Plants with double genomes might have had a better chance to survive the Cretaceous–Tertiary extinction event. Proc. Natl. Acad. Sci. USA 2009, 106, 5737–5742. [Google Scholar] [CrossRef]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef]
- Saelim, L.; Akiyoshi, N.; Tan, T.T.; Ihara, A.; Yamaguchi, M.; Hirano, K.; Matsuoka, M.; Demura, T.; Ohtani, M. Arabidopsis Group IIId ERF proteins positively regulate primary cell wall-type CESA genes. J. Plant Res. 2019, 132, 117–129. [Google Scholar] [CrossRef]
- Cao, S.; Wang, Y.; Li, X.; Gao, F.; Feng, J.; Zhou, Y. Characterization of the AP2/ERF transcription factor family and expression profiling of DREB subfamily under cold and osmotic stresses in Ammopiptanthus nanus. Plants 2020, 9, 455. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Broeckling, C.D.; Blancaflor, E.B.; Sledge, M.K.; Sumner, L.W.; Wang, Z.Y. Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J. 2005, 42, 689–707. [Google Scholar] [CrossRef]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Okamuro, J.K.; Caster, B.; Villarroel, R.; Van Montagu, M.; Jofuku, K.D. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 1997, 94, 7076–7081. [Google Scholar] [CrossRef] [PubMed]
- Ohme-Takagi, M.; Shinshi, H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 1995, 7, 173–182. [Google Scholar]
- He, S.; Hao, X.; He, S.; Hao, X.; Chen, X. Genome-wide identification, phylogeny and expression analysis of AP2/ERF transcription factors family in sweet potato. BMC Genom. 2021, 22, 748. [Google Scholar] [CrossRef] [PubMed]
- McGrath, K.C.; Dombrecht, B.; Manners, J.M.; Schenk, P.M.; Edgar, C.I.; Maclean, D.J.; Scheible, W.R.; Udvardi, M.K.; Kazan, K. Repressor-and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 2005, 139, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Tian, L.; Latoszek-Green, M.; Brown, D.; Wu, K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 2005, 58, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Song, C.P.; Agarwal, M.; Ohta, M.; Guo, Y.; Halfter, U.; Wang, P.; Zhu, J.K. Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 2005, 17, 2384–2396. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Liu, J.; Dong, H.; Sun, J. Functional regulation of Q by microRNA172 and transcriptional co-repressor TOPLESS in controlling bread wheat spikelet density. Plant Biotechnol. J. 2018, 16, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Giorgi, F.M.; Zenoni, S.; Osti, F.; Pezzotti, M.; Perata, P. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genom. 2010, 11, 719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yin, X.; Xiao, Y.; Zhang, Z.; Li, S.; Liu, X.; Zhang, B.; Yang, X.; Grierson, D.; Jiang, G. An ETHYLENE RESPONSE FACTOR-MYB transcription complex regulates furaneol biosynthesis by activating QUINONE OXIDOREDUCTASE expression in strawberry. Plant Physiol. 2018, 178, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guo, C.; Deng, M.; Li, S.; Chen, Y.; Gu, X.; Tang, G.; Lin, Y.; Wang, Y.; He, W. Genome-wide analysis of the ERF family and identification of potential genes involved in fruit ripening in octoploid strawberry. Int. J. Mol. Sci. 2022, 23, 10550. [Google Scholar] [CrossRef] [PubMed]
- El-Sharkawy, I.; Sherif, S.; Mila, I.; Bouzayen, M.; Jayasankar, S. Molecular characterization of seven genes encoding ethylene-responsive transcriptional factors during plum fruit development and ripening. J. Exp. Bot. 2009, 60, 907–922. [Google Scholar] [CrossRef]
- Chen, G.; Hu, Z.; Grierson, D. Differential regulation of tomato ethylene responsive factor LeERF3b, a putative repressor, and the activator Pti4 in ripening mutants and in response to environmental stresses. J. Plant. Physiol. 2008, 165, 662–670. [Google Scholar] [CrossRef]
- Yin, X.R.; Chen, K.S.; Allan, A.C.; Wu, R.M.; Zhang, B.; Lallu, N.; Ferguson, I.B. Ethylene-induced modulation of genes associated with the ethylene signalling pathway in ripening kiwifruit. J. Exp. Bot. 2008, 59, 2097–2108. [Google Scholar] [CrossRef]
- Chowdhary, A.A.; Mishra, S.; Mehrotra, S.; Upadhyay, S.K.; Bagal, D.; Srivastava, V. Plant transcription factors: An overview of their role in plant life. In Plant Transcription Factors; Academic Press: Cambridge, MA, USA, 2023; pp. 3–20. [Google Scholar]
- Wang, X.; Pan, L.; Wang, Y.; Meng, J.; Deng, L.; Niu, L.; Liu, H.; Ding, Y.; Yao, J.-L.; Nieuwenhuizen, N.J. PpIAA1 and PpERF4 form a positive feedback loop to regulate peach fruit ripening by integrating auxin and ethylene signals. Plant Sci. 2021, 313, 111084. [Google Scholar] [CrossRef]
- Wang, M.; Zhu, Q.; Deng, C.; Luo, Z.; Sun, N.; Grierson, D.; Yin, X.; Chen, K. Hypoxia-responsive ERF s involved in postdeastringency softening of persimmon fruit. Plant Biotechnol. J. 2017, 15, 1409–1419. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Min, D.; Ren, C.; Dong, L.; Shu, P.; Cui, X.; Zhang, X. Ethylene altered fruit cuticular wax, the expression of cuticular wax synthesis-related genes and fruit quality during cold storage of apple (Malus domestica Borkh. cv Starkrimson) fruit. Postharvest Biol. Technol. 2019, 149, 58–65. [Google Scholar] [CrossRef]
- Zhu, K.; Sun, Q.; Chen, H.; Mei, X.; Lu, S.; Ye, J.; Chai, L.; Xu, Q.; Deng, X. Ethylene activation of carotenoid biosynthesis by a novel transcription factor CsERF061. J. Exp. Bot. 2021, 72, 3137–3154. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Zhang, X.W.; Bi, S.Q.; You, C.X.; Wang, X.F.; Hao, Y.J. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 2020, 101, 573–589. [Google Scholar] [CrossRef]
- Ni, J.; Bai, S.; Zhao, Y.; Qian, M.; Tao, R.; Yin, L.; Gao, L.; Teng, Y. Ethylene response factors Pp4ERF24 and Pp12ERF96 regulate blue light-induced anthocyanin biosynthesis in ‘Red Zaosu’pear fruits by interacting with MYB114. Plant Mol. Biol. 2019, 99, 67–78. [Google Scholar] [CrossRef]
- Ning, Z.; Hu, K.; Zhou, Z.; Zhao, D.; Tang, J.; Wang, H.; Li, L.; Ding, C.; Chen, X.; Yao, G. IbERF71, with IbMYB340 and IbbHLH2, coregulates anthocyanin accumulation by binding to the IbANS1 promoter in purple-fleshed sweet potato (Ipomoea batatas L.). Plant Cell Rep. 2021, 40, 157–169. [Google Scholar] [CrossRef]
- Correia, S.; Schouten, R.; Silva, A.P.; Gonçalves, B. Sweet cherry fruit cracking mechanisms and prevention strategies: A review. Sci. Hortic. 2018, 240, 369–377. [Google Scholar] [CrossRef]
- Pereira, S.; Silva, V.; Bacelar, E.; Guedes, F.; Gonalves, B. Cracking in Sweet Cherry Cultivars Early Bigi and Lapins: Correlation with Quality Attributes. Plants 2020, 9, 1557. [Google Scholar] [CrossRef]
- Lashbrooke, J.; Aharoni, A.; Costa, F. Genome investigation suggests MdSHN3, an APETALA2-domain transcription factor gene, to be a positive regulator of apple fruit cuticle formation and an inhibitor of russet development. J. Exp. Bot. 2015, 66, 6579–6589. [Google Scholar] [CrossRef] [PubMed]
- Crump, W.W.; Peace, C.; Zhang, Z.; McCord, P. Detection of breeding-relevant fruit cracking and fruit firmness Quantitative Trait Loci in sweet cherry via pedigree-based and genome-wide association approaches. Front. Plant Sci. 2022, 13, 823250. [Google Scholar] [CrossRef] [PubMed]
- Zwack, P.; Shi, X.; Robinson, B.; Gupta, S.; Compton, M.; Gerken, D.; Goertzen, L.; Rashotte, A. Vascular expression and C-terminal sequence divergence of cytokinin response factors in flowering plants. Plant Cell Physiol. 2012, 53, 1683–1695. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Gupta, S.; Rashotte, A.M. Solanum lycopersicum cytokinin response factor (SlCRF) genes: Characterization of CRF domain-containing ERF genes in tomato. J. Exp. Bot. 2012, 63, 973–982. [Google Scholar] [CrossRef]
- Liu, Z.; Kong, L.; Zhang, M.; Lv, Y.; Liu, Y.; Zou, M.; Lu, G.; Cao, J.; Yu, X. Genome-wide identification, phylogeny, evolution and expression patterns of AP2/ERF genes and cytokinin response factors in Brassica rapa ssp. pekinensis. PLoS ONE 2013, 8, e83444. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Wang, L.; Guo, Y.; Li, Y.; Ümüt, H.; Wang, Y. An ERF transcription factor from Tamarix hispida, ThCRF1, can adjust osmotic potential and reactive oxygen species scavenging capability to improve salt tolerance. Plant Sci. 2017, 265, 154–166. [Google Scholar] [CrossRef] [PubMed]
- Keshishian, E. CRF2 and its Role in Cytokinin Response and Abiotic Stress. Ph.D. Thesis, Auburn University, Auburn, AL, USA, 2018. [Google Scholar]
- Brooks, M.D.; Cirrone, J.; Pasquino, A.V.; Alvarez, J.M.; Swift, J.; Mittal, S.; Juang, C.-L.; Varala, K.; Gutiérrez, R.A.; Krouk, G. Network Walking charts transcriptional dynamics of nitrogen signaling by integrating validated and predicted genome-wide interactions. Nat. Commun. 2019, 10, 1569. [Google Scholar] [CrossRef]
- Ginzberg, I.; Stern, R.A. Control of fruit cracking by shaping skin traits–apple as a model. CRC. Crit. Rev. Plant Sci. 2019, 38, 401–410. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).