Spatial Variability of Grape Berry Maturation Program at the Molecular Level
Abstract
1. Introduction
2. Materials and Methods
2.1. Vineyard
2.2. Experimental Design
2.3. Vine Measurements
2.3.1. Vine Water Status
2.3.2. Canopy Size
2.3.3. Yield and Pruning Weight
2.4. Grape Composition Analysis
2.4.1. Basic Chemistry
2.4.2. Color and Phenolic Maturity
2.4.3. Free and Bound Aroma Compounds
2.5. Berry Sampling
2.6. Gene Expression Analysis
2.7. Remote Sensing
2.8. Statistical Analysis
2.9. Molecular Phenology Scale
3. Results and Discussion
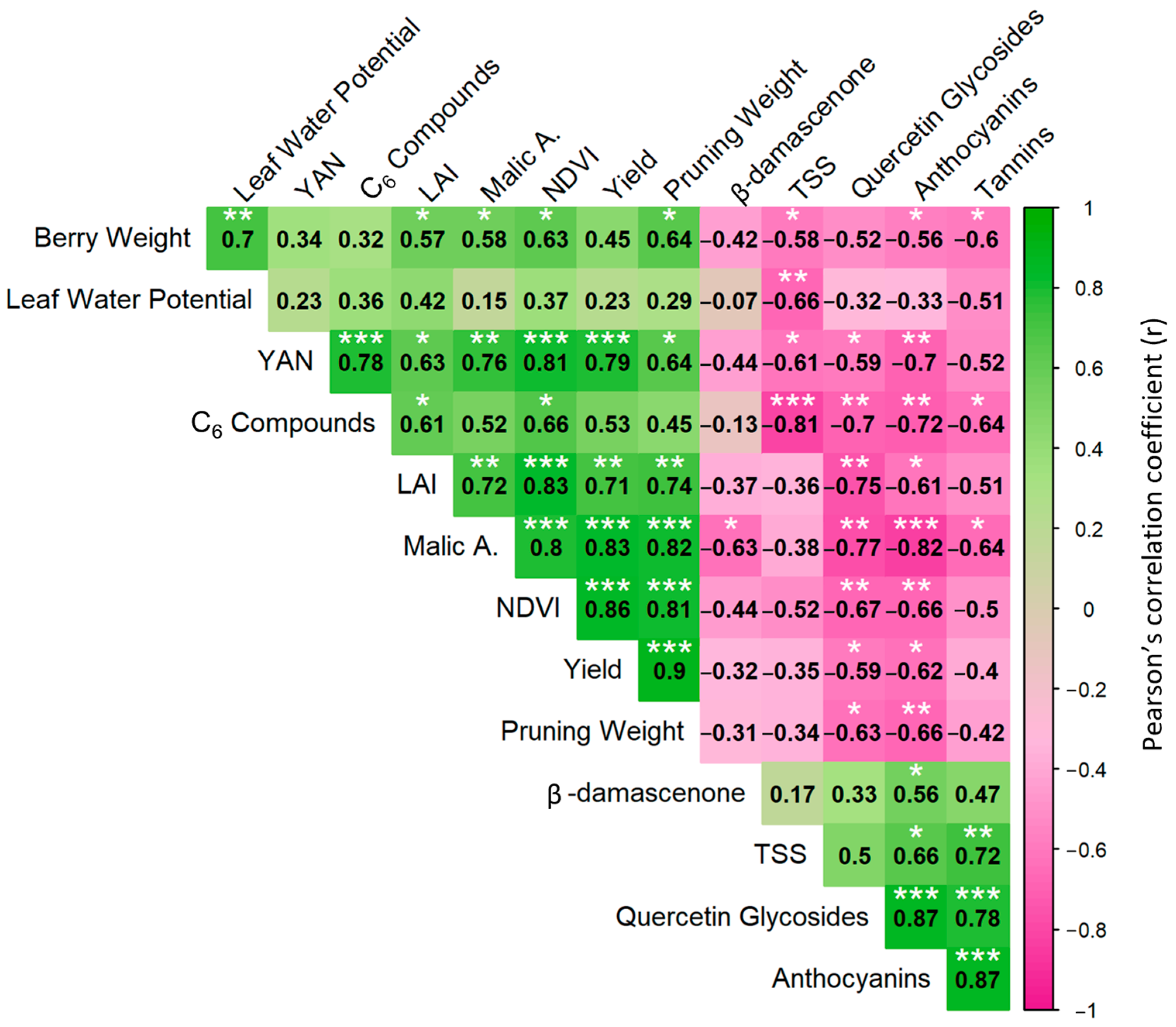
3.1. Vineyard Spatial Variability in Vigor and Quality Traits
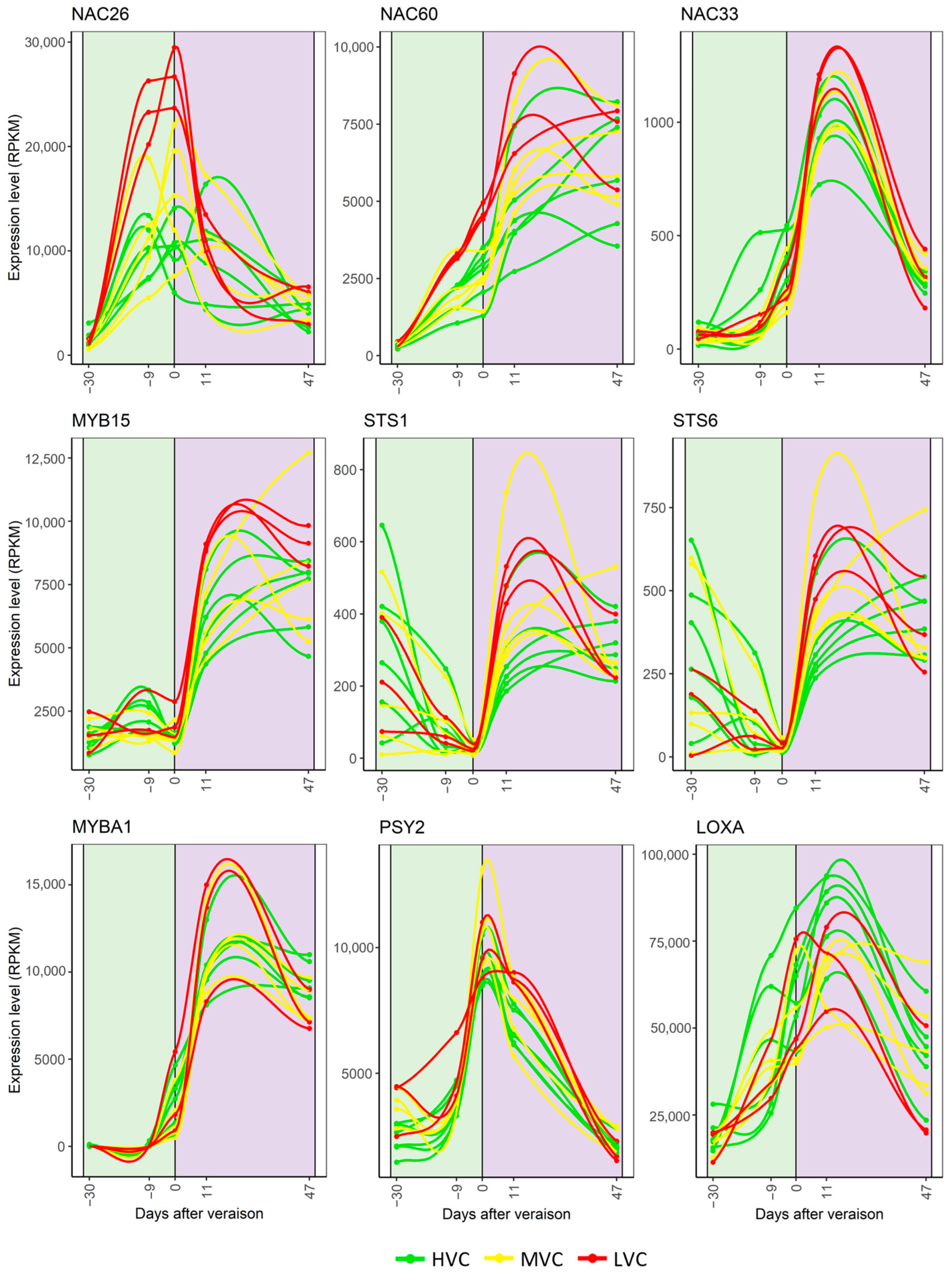
3.2. Spatial Variability in the Expression of Key Genes Involved in Berry Maturation
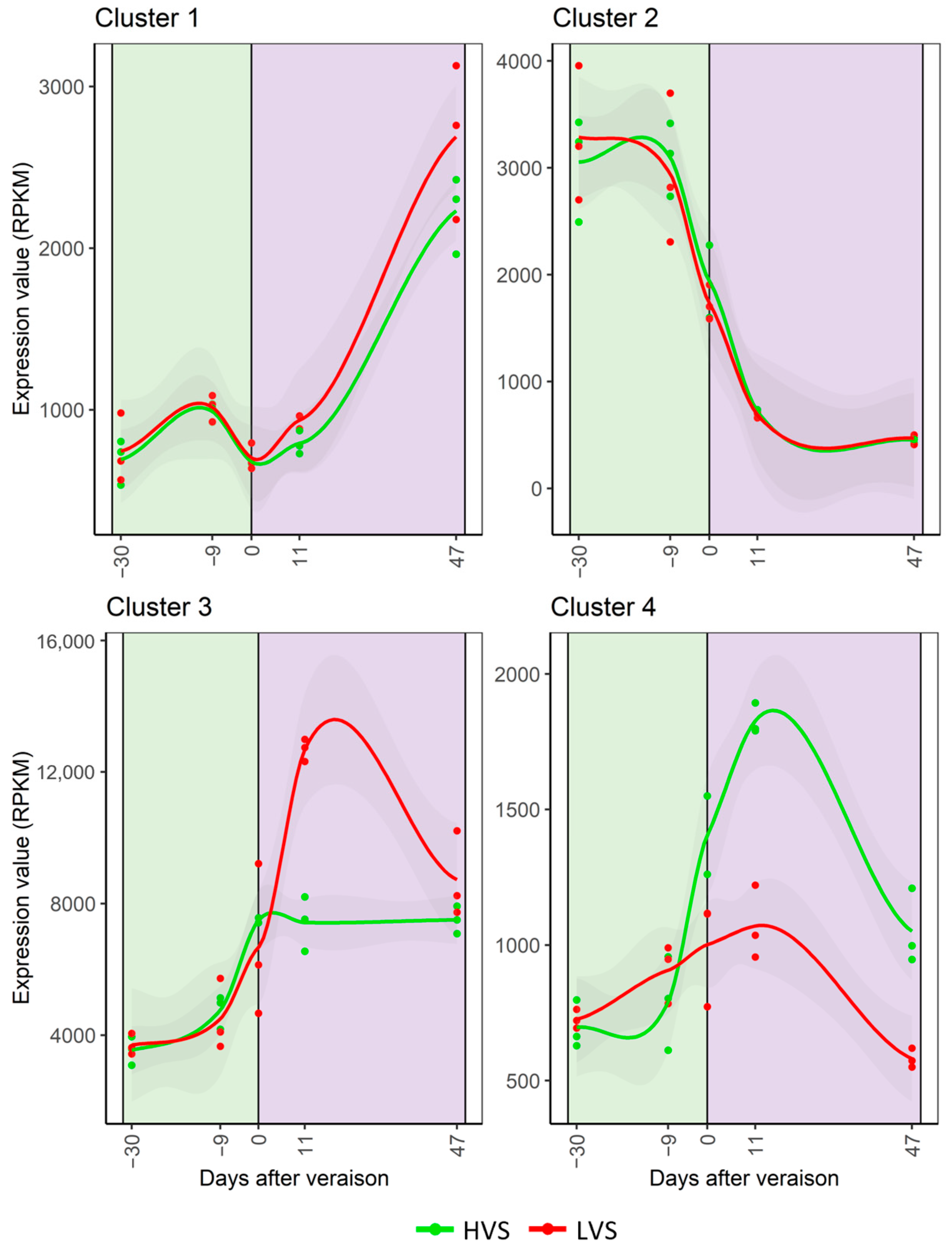
3.3. Spatial Variability in Berry Ripening Transcriptomic Program
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tardaguila, J.; Baluja, J.; Arpon, L.; Balda, P.; Oliveira, M. Variations of soil properties affect the vegetative growth and yield components of “Tempranillo” grapevines. Precis. Agric. 2011, 12, 762–773. [Google Scholar] [CrossRef]
- van Leeuwen, C.; Trégoat, O.; Choné, X.; Bois, B.; Pernet, D.; Gaudillère, J.-P. Vine water status is a key factor in grape ripening and vintage quality for red Bordeaux wine. How can it be assessed for vineyard management purposes? OENO One 2009, 43, 121–134. [Google Scholar] [CrossRef]
- Pallas, B.; Christophe, A.; Lecoeur, J. Are the common assimilate pool and trophic relationships appropriate for dealing with the observed plasticity of grapevine development? Ann. Bot. 2009, 105, 233–247. [Google Scholar] [CrossRef]
- Andrew, G.R.; Justine, E.V.H. Influence of Grapevine Training Systems on Vine Growth and Fruit Composition: A Review. Am. J. Enol. Vitic. 2009, 60, 251. [Google Scholar] [CrossRef]
- Bramley, R.G.V.; Ouzman, J.; Boss, P.K. Variation in vine vigour, grape yield and vineyard soils and topography as indicators of variation in the chemical composition of grapes, wine and wine sensory attributes. Aust. J. Grape Wine Res. 2011, 17, 217–229. [Google Scholar] [CrossRef]
- dos Santos Costa, D.; Oliveros Mesa, N.F.; Santos Freire, M.; Pereira Ramos, R.; Teruel Mederos, B.J. Development of predictive models for quality and maturation stage attributes of wine grapes using vis-nir reflectance spectroscopy. Postharvest Biol. Technol. 2019, 150, 166–178. [Google Scholar] [CrossRef]
- Sassu, A.; Gambella, F.; Ghiani, L.; Mercenaro, L.; Caria, M.; Pazzona, A.L. Advances in Unmanned Aerial System Remote Sensing for Precision Viticulture. Sensors 2021, 21, 956. [Google Scholar] [CrossRef]
- Lyu, H.; Grafton, M.; Ramilan, T.; Irwin, M.; Wei, H.-E.; Sandoval, E. Using Remote and Proximal Sensing Data and Vine Vigor Parameters for Non-Destructive and Rapid Prediction of Grape Quality. Remote Sens. 2023, 15, 5412. [Google Scholar] [CrossRef]
- Moral, F.J.; Rebollo, F.J.; Serrano, J. Using a Non-Contact Sensor to Delineate Management Zones in Vineyards and Validation with the Rasch Model. Sensors 2023, 23, 9183. [Google Scholar] [CrossRef]
- Gatti, M.; Schippa, M.; Garavani, A.; Squeri, C.; Frioni, T.; Dosso, P.; Poni, S. High potential of variable rate fertilization combined with a controlled released nitrogen form at affecting cv. Barbera vines behavior. Eur. J. Agron. 2020, 112, 125949. [Google Scholar] [CrossRef]
- Pereyra, G.; Pellegrino, A.; Gaudin, R.; Ferrer, M. Evaluation of site-specific management to optimise Vitis vinifera L. (cv. Tannat) production in a vineyard with high heterogeneity. OENO One 2022, 56, 397–412. [Google Scholar] [CrossRef]
- Bramley, R.G.V.; Ouzman, J.; Thornton, C. Selective harvesting is a feasible and profitable strategy even when grape and wine production is geared towards large fermentation volumes. Aust. J. Grape Wine Res. 2011, 17, 298–305. [Google Scholar] [CrossRef]
- Baca-Bocanegra, B.; Hernández-Hierro, J.M.; Nogales-Bueno, J.; Heredia, F.J. Feasibility study on the use of a portable micro near infrared spectroscopy device for the “in vineyard” screening of extractable polyphenols in red grape skins. Talanta 2019, 192, 353–359. [Google Scholar] [CrossRef]
- Di Gennaro, S.F.; Matese, A. Evaluation of novel precision viticulture tool for canopy biomass estimation and missing plant detection based on 2.5D and 3D approaches using RGB images acquired by UAV platform. Plant Methods 2020, 16, 91. [Google Scholar] [CrossRef]
- Anastasiou, E.; Balafoutis, A.; Darra, N.; Psiroukis, V.; Biniari, A.; Xanthopoulos, G.; Fountas, S. Satellite and Proximal Sensing to Estimate the Yield and Quality of Table Grapes. Agriculture 2018, 8, 94. [Google Scholar] [CrossRef]
- Squeri, C.; Poni, S.; Di Gennaro, S.F.; Matese, A.; Gatti, M. Comparison and Ground Truthing of Different Remote and Proximal Sensing Platforms to Characterize Variability in a Hedgerow-Trained Vineyard. Remote Sens. 2021, 13, 2056. [Google Scholar] [CrossRef]
- Bramley, R.G.V.; Hamilton, R.P. Understanding variability in winegrape production systems. Aust. J. Grape Wine Res. 2004, 10, 32–45. [Google Scholar] [CrossRef]
- Fiorillo, E.; Crisci, A.; De Filippis, T.; Di Gennaro, S.F.; Di Blasi, S.; Matese, A.; Primicerio, J.; Vaccari, F.P.; Genesio, L. Airborne high-resolution images for grape classification: Changes in correlation between technological and late maturity in a Sangiovese vineyard in Central Italy. Aust. J. Grape Wine Res. 2012, 18, 80–90. [Google Scholar] [CrossRef]
- Kasimati, A.; Espejo-Garcia, B.; Vali, E.; Malounas, I.; Fountas, S. Investigating a selection of methods for the prediction of total soluble solids among wine grape quality characteristics using normalized difference vegetation index data from proximal and remote sensing. Front. Plant Sci. 2021, 12, 683078. [Google Scholar] [CrossRef]
- Lamb, D.W.; Weedon, M.M.; Bramley, R.G.V. Using remote sensing to predict grape phenolics and colour at harvest in a Cabernet Sauvignon vineyard: Timing observations against vine phenology and optimising image resolution. Aust. J. Grape Wine Res. 2004, 10, 46–54. [Google Scholar] [CrossRef]
- García-Estévez, I.; Quijada-Morín, N.; Rivas-Gonzalo, J.C.; Martínez-Fernández, J.; Sánchez, N.; Herrero-Jiménez, C.M.; Escribano-Bailón, M.T. Relationship between hyperspectral indices, agronomic parameters and phenolic composition of Vitis vinifera cv Tempranillo grapes. J. Sci. Food Agric. 2017, 97, 4066–4074. [Google Scholar] [CrossRef]
- Bonilla, I.; Martinez de Toda, F.; Martínez-Casasnovas, J.A. Vine vigor, yield and grape quality assessment by airborne remote sensing over three years: Analysis of unexpected relationships in cv. Tempranillo. Span. J. Agric. Res. 2015, 13, e0903. [Google Scholar] [CrossRef]
- Savoi, S.; Santiago, A.; Orduña, L.; Matus, J.T. Transcriptomic and metabolomic integration as a resource in grapevine to study fruit metabolite quality traits. Front. Plant Sci. 2022, 13, 937927. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.L.; Price, S.F.; McCord, J.D. Reversed-phase high-performance liquid chromatography methods for analysis of wine polyphenols. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1999; Volume 299, pp. 113–121. [Google Scholar]
- Previtali, P.; Dokoozlian, N.K.; Pan, B.S.; Wilkinson, K.L.; Ford, C.M. Crop Load and Plant Water Status Influence the Ripening Rate and Aroma Development in Berries of Grapevine (Vitis vinifera L.) cv. Cabernet Sauvignon. J. Agric. Food Chem. 2021, 69, 7709–7724. [Google Scholar] [CrossRef] [PubMed]
- Kotseridis, Y.; Baumes, R.L.; Skouroumounis, G.K. Quantitative determination of free and hydrolytically liberated beta-damascenone in red grapes and wines using a stable isotope dilution assay. J. Chromatogr. A 1999, 849, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Fasoli, M.; Richter, C.L.; Zenoni, S.; Bertini, E.; Vitulo, N.; Dal Santo, S.; Dokoozlian, N.; Pezzotti, M.; Tornielli, G.B. Timing and Order of the Molecular Events Marking the Onset of Berry Ripening in Grapevine. Plant Physiol. 2018, 178, 1187–1206. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Payá, D.; Santiago, A.; Orduña, L.; Zhang, C.; Amato, A.; D’Inca, E.; Fattorini, C.; Pezzotti, M.; Tornielli, G.B.; Zenoni, S.; et al. The Grape Gene Reference Catalogue as a Standard Resource for Gene Selection and Genetic Improvement. Front. Plant Sci. 2022, 12, 803977. [Google Scholar] [CrossRef] [PubMed]
- Scrucca, L.; Fraley, C.; Murphy, T.; Raftery, A.E. Model-Based Clustering, Classification, and Density Estimation Using Mclust in R, 1st ed.; Chapman and Hall/CRC: New York, NY, USA, 2023. [Google Scholar]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2019, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, L.; Rousseeuw, P. Finding Groups in Data: An Introduction to Cluster Analysis; John Wiley & Sons: Hoboken, NJ, USA, 1990. [Google Scholar]
- Tornielli, G.B.; Sandri, M.; Fasoli, M.; Amato, A.; Pezzotti, M.; Zuccolotto, P.; Zenoni, S. A molecular phenology scale of grape berry development. Hortic. Res. 2023, 10, uhad048. [Google Scholar] [CrossRef]
- Smart, R.E.; Dick, J.K.; Gravett, I.M.; Fisher, B.M. Canopy Management to Improve Grape Yield and Wine Quality—Principles and Practices. S. Afr. J. Enol. Vitic. 2017, 11, 3–17. [Google Scholar] [CrossRef]
- Dobrowski, S.Z.; Ustin, S.L.; Wolpert, J.A. Grapevine dormant pruning weight prediction using remotely sensed data. Aust. J. Grape Wine Res. 2003, 9, 177–182. [Google Scholar] [CrossRef]
- Conde, C.; Silva, P.; Fontes, N.; Dias, A.C.P.; Tavares, R.M.; Sousa, M.J.; Agasse, A.; Delrot, S.; Gerós, H. Biochemical Changes throughout Grape Berry Development and Fruit and Wine Quality. Food 2007, 1, 1–22. [Google Scholar]
- Kalua, C.M.; Boss, P.K. Evolution of Volatile Compounds during the Development of Cabernet Sauvignon Grapes (Vitis vinifera L.). J. Agric. Food Chem. 2009, 57, 3818–3830. [Google Scholar] [CrossRef] [PubMed]
- Castellarin, S.D.; Matthews, M.A.; Di Gaspero, G.; Gambetta, G.A. Water deficits accelerate ripening and induce changes in gene expression regulating flavonoid biosynthesis in grape berries. Planta 2007, 227, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Rienth, M.; Vigneron, N.; Darriet, P.; Sweetman, C.; Burbidge, C.; Bonghi, C.; Walker, R.P.; Famiani, F.; Castellarin, S.D. Grape Berry Secondary Metabolites and Their Modulation by Abiotic Factors in a Climate Change Scenario—A Review. Front. Plant Sci. 2021, 12, 643258. [Google Scholar] [CrossRef] [PubMed]
- Palai, G.; Caruso, G.; Gucci, R.; D’Onofrio, C. Berry flavonoids are differently modulated by timing and intensities of water deficit in Vitis vinifera L. cv. Sangiovese. Front. Plant Sci. 2022, 13, 1040899. [Google Scholar] [CrossRef]
- Cortell, J.M.; Halbleib, M.; Gallagher, A.V.; Righetti, T.L.; Kennedy, J.A. Influence of Vine Vigor on Grape (Vitis vinifera L. Cv. Pinot Noir) Anthocyanins. 1. Anthocyanin Concentration and Composition in Fruit. J. Agric. Food Chem. 2007, 55, 6575–6584. [Google Scholar] [CrossRef]
- Cortell, J.M.; Halbleib, M.; Gallagher, A.V.; Righetti, T.L.; Kennedy, J.A. Influence of Vine Vigor on Grape (Vitis vinifera L. Cv. Pinot Noir) and Wine Proanthocyanidins. J. Agric. Food Chem. 2005, 53, 5798–5808. [Google Scholar] [CrossRef]
- Yu, R.; Brillante, L.; Martínez-Lüscher, J.; Kurtural, S.K. Spatial Variability of Soil and Plant Water Status and Their Cascading Effects on Grapevine Physiology Are Linked to Berry and Wine Chemistry. Front. Plant Sci. 2020, 11, 790. [Google Scholar] [CrossRef]
- Martínez-Lüscher, J.; Brillante, L.; Kurtural, S.K. Flavonol Profile Is a Reliable Indicator to Assess Canopy Architecture and the Exposure of Red Wine Grapes to Solar Radiation. Front. Plant Sci. 2019, 10, 10. [Google Scholar] [CrossRef]
- Verdenal, T.; Dienes-Nagy, Á.; Spangenberg, J.E.; Zufferey, V.; Spring, J.L.; Viret, O.; Marin-Carbonne, J.; van Leeuwen, C. Understanding and managing nitrogen nutrition in grapevine: A review. OENO One 2021, 55, 1–43. [Google Scholar] [CrossRef]
- Sams, B.; Bramley, R.G.V.; Sanchez, L.; Dokoozlian, N.; Ford, C.; Pagay, V. Remote Sensing, Yield, Physical Characteristics, and Fruit Composition Variability in Cabernet Sauvignon Vineyards. Am. J. Enol. Vitic. 2022, 73, 93–105. [Google Scholar] [CrossRef]
- Coombe, B.G.; McCarthy, M.G. Dynamics of grape berry growth and physiology of ripening. Aust. J. Grape Wine Res. 2000, 6, 131–135. [Google Scholar] [CrossRef]
- Wang, N.; Zheng, Y.; Xin, H.; Fang, L.; Li, S. Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Rep. 2013, 32, 61–75. [Google Scholar] [CrossRef]
- Palumbo, M.C.; Zenoni, S.; Fasoli, M.; Massonnet, M.; Farina, L.; Castiglione, F.; Pezzotti, M.; Paci, P. Integrated network analysis identifies fight-club nodes as a class of hubs encompassing key putative switch genes that induce major transcriptome reprogramming during grapevine development. Plant Cell 2014, 26, 4617–4635. [Google Scholar] [CrossRef] [PubMed]
- Tello, J.; Torres-Pérez, R.; Grimplet, J.; Carbonell-Bejerano, P.; Martínez-Zapater, J.M.; Ibáñez, J. Polymorphisms and minihaplotypes in the VvNAC26 gene associate with berry size variation in grapevine. BMC Plant Biol. 2015, 15, 253. [Google Scholar] [CrossRef]
- D’Incà, E.; Cazzaniga, S.; Foresti, C.; Vitulo, N.; Bertini, E.; Galli, M.; Gallavotti, A.; Pezzotti, M.; Battista Tornielli, G.; Zenoni, S. VviNAC33 promotes organ de-greening and represses vegetative growth during the vegetative-to-mature phase transition in grapevine. New Phytol. 2021, 231, 726–746. [Google Scholar] [CrossRef] [PubMed]
- D’Incà, E.; Foresti, C.; Orduña, L.; Amato, A.; Vandelle, E.; Santiago, A.; Botton, A.; Cazzaniga, S.; Bertini, E.; Pezzotti, M.; et al. The transcription factor VviNAC60 regulates senescence- and ripening-related processes in grapevine. Plant Physiol. 2023, 192, 1928–1946. [Google Scholar] [CrossRef]
- Höll, J.; Vannozzi, A.; Czemmel, S.; D’Onofrio, C.; Walker, A.R.; Rausch, T.; Lucchin, M.; Boss, P.K.; Dry, I.B.; Bogs, J. The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell 2013, 25, 4135–4149. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.R.; Lee, E.; Bogs, J.; McDavid, D.A.J.; Thomas, M.R.; Robinson, S.P. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 2007, 49, 772–785. [Google Scholar] [CrossRef] [PubMed]
- Young, P.R.; Lashbrooke, J.G.; Alexandersson, E.; Jacobson, D.; Moser, C.; Velasco, R.; Vivier, M.A. The genes and enzymes of the carotenoid metabolic pathway in Vitis vinifera L. BMC Genom. 2012, 13, 243. [Google Scholar] [CrossRef] [PubMed]
- Lashbrooke, J.G.; Young, P.R.; Dockrall, S.J.; Vasanth, K.; Vivier, M.A. Functional characterisation of three members of the Vitis vinifera L. carotenoid cleavage dioxygenase gene family. BMC Plant Biol. 2013, 13, 156. [Google Scholar] [CrossRef] [PubMed]
- Matsui, K. Green leaf volatiles: Hydroperoxide lyase pathway of oxylipin metabolism. Curr. Opin. Plant Biol. 2006, 9, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Podolyan, A.; White, J.; Jordan, B.; Winefield, C. Identification of the lipoxygenase gene family from Vitis vinifera and biochemical characterisation of two 13-lipoxygenases expressed in grape berries of Sauvignon Blanc. Funct. Plant Biol. 2010, 37, 767–784. [Google Scholar] [CrossRef]
- Foresti, C.; Orduña, L.; Matus, J.T.; Vandelle, E.; Danzi, D.; Bellon, O.; Tornielli, G.B.; Amato, A.; Zenoni, S. NAC61 regulates late- and post-ripening osmotic, oxidative, and biotic stress responses in grapevine. J. Exp. Bot. 2023, erad507. [Google Scholar] [CrossRef]
- Pilati, S.; Malacarne, G.; Navarro-Payá, D.; Tomè, G.; Riscica, L.; Cavecchia, V.; Matus, J.T.; Moser, C.; Blanzieri, E. Vitis OneGenE: A Causality-Based Approach to Generate Gene Networks in Vitis vinifera Sheds Light on the Laccase and Dirigent Gene Families. Biomolecules 2021, 11, 1744. [Google Scholar] [CrossRef]
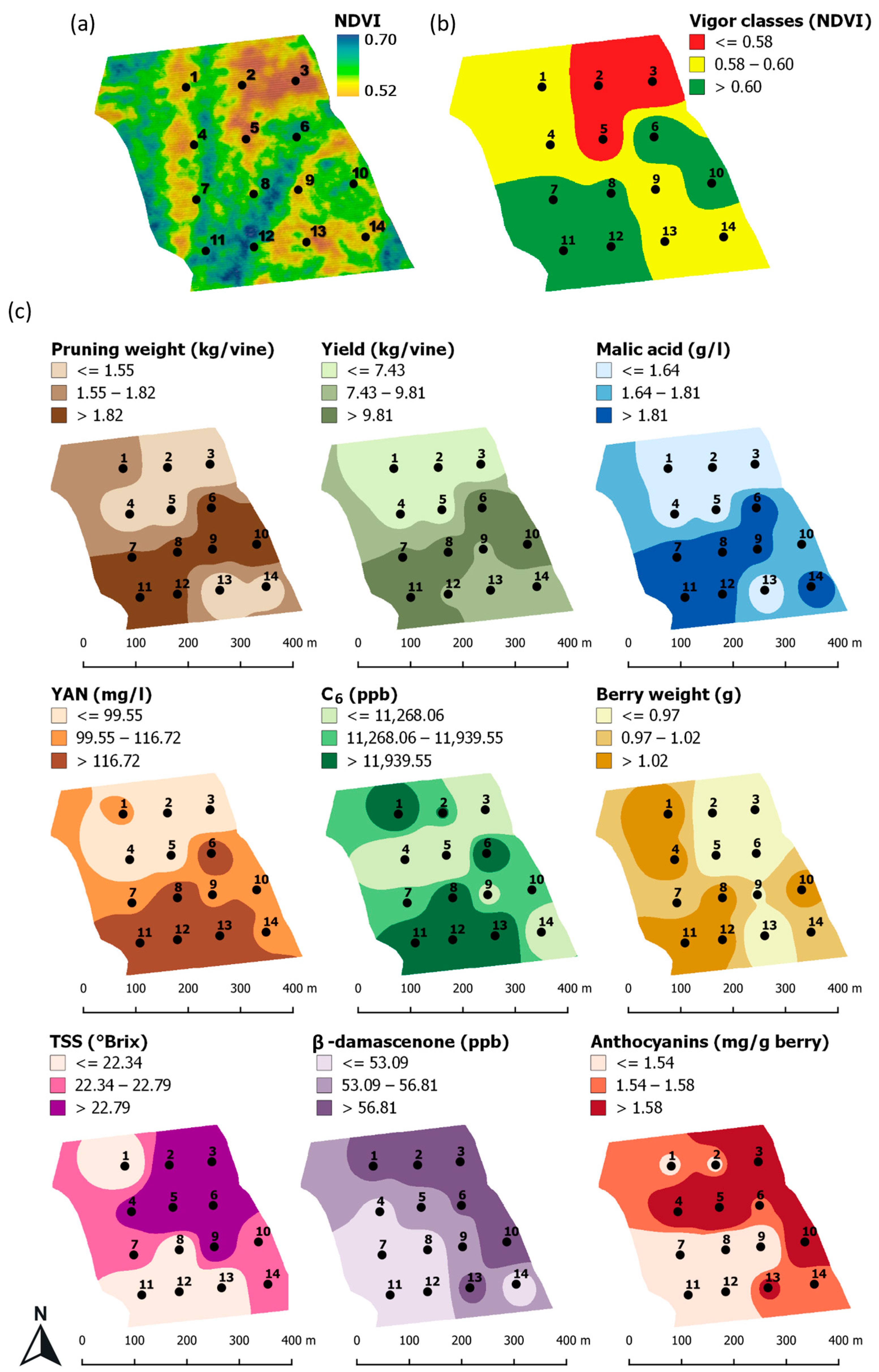


| Vigor | Block | NDVI | LAI | Leaf Water Potential (mPa) | Pruning Weight (kg/Vine) | Berry Weight (g) | Yield (kg/Vine) | TSS (°Brix) | Malic Acid (g/L) | Anthocyanins (mg/g Berry) | Tannins (ppm) | YAN (mg/L) | C6 Compounds (ppb) | Quercetin Glycosides (ppm) | β-Damascenone (ppb) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LVC | 3 | 0.540 | 3.2 | −0.78 | 0.83 | 0.91 | 3.88 | 23.8 | 1.00 | 2.02 | 3883 | 58 | 8315 | 159 | 62.49 |
| LVC | 5 | 0.554 | 3.0 | −1.22 | 1.22 | 0.85 | 5.36 | 25.5 | 1.50 | 2.00 | 4356 | 80 | 7045 | 163 | 54.55 |
| LVC | 2 | 0.567 | 3.8 | −0.85 | 1.44 | 0.90 | 5.74 | 23.1 | 1.50 | 1.51 | 3479 | 83 | 12,090 | 101 | 60.95 |
| Average | 0.554 a | 3.3 a | −0.95 | 1.16 a | 0.88 a | 4.99 a | 24.13 a | 1.33 a | 1.84 a | 3906 | 74 a | 9150 a | 141 a | 59.33 | |
| MVC | 9 | 0.581 | 3.7 | −0.94 | 2.03 | 0.97 | 9.66 | 23.5 | 1.88 | 1.38 | 3457 | 112 | 10,876 | 85 | 54.50 |
| MVC | 13 | 0.582 | 3.1 | −0.87 | 1.03 | 0.85 | 7.89 | 21.2 | 1.41 | 1.61 | 3623 | 139 | 13,897 | 130 | 59.36 |
| MVC | 14 | 0.582 | 3.8 | −0.73 | 1.42 | 1.00 | 8.46 | 22.8 | 1.88 | 1.54 | 3099 | 114 | 10,349 | 100 | 50.55 |
| MVC | 1 | 0.587 | 3.8 | −0.79 | 1.63 | 1.09 | 5.32 | 21.1 | 1.62 | 1.52 | 3203 | 102 | 13,238 | 101 | 58.48 |
| MVC | 4 | 0.589 | 3.4 | −0.76 | 1.27 | 1.07 | 6.03 | 22.9 | 1.54 | 1.66 | 3636 | 72 | 9385 | 118 | 51.52 |
| Average | 0.584 b | 3.6 ab | −0.82 | 1.48 a | 1.00 a | 7.47 a | 22.30 ab | 1.67 ab | 1.54 ab | 3404 | 108 ab | 11,549 ab | 107 ab | 54.88 | |
| HVC | 8 | 0.603 | 3.9 | −0.76 | 1.95 | 1.08 | 11.34 | 21.2 | 2.18 | 1.30 | 3073 | 134 | 13,439 | 96 | 49.73 |
| HVC | 7 | 0.607 | 3.6 | −0.96 | 1.83 | 0.99 | 10.12 | 22.5 | 1.92 | 1.37 | 3024 | 114 | 11,747 | 97 | 51.58 |
| HVC | 10 | 0.615 | 3.9 | −0.68 | 2.39 | 1.07 | 12.42 | 22.4 | 1.69 | 1.73 | 3763 | 115 | 11,334 | 114 | 67.48 |
| HVC | 6 | 0.628 | 4.5 | −0.97 | 2.12 | 0.93 | 14.41 | 23.3 | 2.02 | 1.56 | 3595 | 133 | 13,586 | 86 | 57.19 |
| HVC | 12 | 0.629 | 4.5 | −0.73 | 1.85 | 1.06 | 9.45 | 22.3 | 1.86 | 1.49 | 3414 | 143 | 12,897 | 103 | 44.66 |
| HVC | 11 | 0.635 | 4.3 | −0.70 | 3.08 | 1.13 | 16.58 | 21.5 | 2.27 | 1.20 | 3143 | 147 | 12,679 | 98 | 45.27 |
| Average | 0.620 c | 4.1 b | −0.80 | 2.20 b | 1.04 b | 12.38 b | 22.20 b | 1.99 b | 1.44 b | 3335 | 131 b | 12,614 b | 99 b | 52.65 | |
| CV % | 4.80 | 12.63 | 17.45 | 34.36 | 9.52 | 41.10 | 5.34 | 19.41 | 14.97 | 10.63 | 25.49 | 18.20 | 21.97 | 11.92 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shmuleviz, R.; Amato, A.; Previtali, P.; Green, E.; Sanchez, L.; Alsina, M.M.; Dokoozlian, N.; Tornielli, G.B.; Fasoli, M. Spatial Variability of Grape Berry Maturation Program at the Molecular Level. Horticulturae 2024, 10, 238. https://doi.org/10.3390/horticulturae10030238
Shmuleviz R, Amato A, Previtali P, Green E, Sanchez L, Alsina MM, Dokoozlian N, Tornielli GB, Fasoli M. Spatial Variability of Grape Berry Maturation Program at the Molecular Level. Horticulturae. 2024; 10(3):238. https://doi.org/10.3390/horticulturae10030238
Chicago/Turabian StyleShmuleviz, Ron, Alessandra Amato, Pietro Previtali, Elizabeth Green, Luis Sanchez, Maria Mar Alsina, Nick Dokoozlian, Giovanni Battista Tornielli, and Marianna Fasoli. 2024. "Spatial Variability of Grape Berry Maturation Program at the Molecular Level" Horticulturae 10, no. 3: 238. https://doi.org/10.3390/horticulturae10030238
APA StyleShmuleviz, R., Amato, A., Previtali, P., Green, E., Sanchez, L., Alsina, M. M., Dokoozlian, N., Tornielli, G. B., & Fasoli, M. (2024). Spatial Variability of Grape Berry Maturation Program at the Molecular Level. Horticulturae, 10(3), 238. https://doi.org/10.3390/horticulturae10030238







