Abstract
Background: Citrus yellow vein-clearing virus (CYVCV) is a Mandarivirus that causes great economic losses in lemon production. CYVCV infection is associated with obvious yellow vein-clearing disease symptoms and is directly regulated by plant hormone responses. Methods: To understand how lemon plants respond to CYVCV infection, we performed transcriptomic and phytohormone metabolomics. Results: A total of 936 differentially expressed genes were identified, and 773 were downregulated. Salicylic acid and auxin levels increased after CYVCV infection, and phytohormone regulatory systems were also explored. Jasmonic acid and auxin levels decreased after the CYVCV challenge, and jasmonic acid and auxin signaling pathway components were mostly downregulated. The differentially expressed genes (DEGs) involved in the immune response to viral infection, including those related to cell wall integrity, lectin, microtubules, and mildew resistance locus O (MLO), may also provide new candidate targets for CYVCV control. Conclusions: Our findings provide new insights into the molecular changes underlying the pathogenesis of CYVCV in lemon plants.
1. Introduction
Citrus yellow vein clearing disease (CYVCD), caused by Citrus yellow vein clearing virus (CYVCV), is a devastating viral disease in Citrus limon (lemon) production [1,2,3,4]. CYVCD was first discovered in lemons and limes in Pakistan in 1988 and subsequently in limes, citrons, and lemons in India; moreover, it has spread rapidly in the citrus-producing regions of Pakistan and Turkey [5,6,7,8,9]. CYVCD was first detected in the Yunnan Province in China and has been found in the main lemon-producing areas in the Sichuan Province in recent years [2,10,11].
CYVCD is a systemic disease of the lemon caused by CYVCV [2,12,13]. A dangerous abundance of CYVCV can occur throughout the whole growth period of lemons, but mainly in spring and autumn shoots [1]. The side veins of the affected leaves become clear, the leaves near the side veins become yellow, and the leaves subsequently become wrinkled and deformed. In the later stage, ring spots appear on the leaves, leaf vein necrosis occurs, and some young leaves without petioles fall off [7]. The strength of the infected plants decreased, and the yield of the infected plants decreased annually until the garden was destroyed. During latent virus passivation in winter and summer, yellowed leaves turn green, the dorsal veins of the leaves are waterlogged, and the lateral veins are bright when observed by light [1,6]. CYVCV is a member of the genus Mandarivirus of the Alphaflexiviridae family [5]. The 7529-nucleotide positive single-stranded RNA (+ssRNA) genome contains six open reading frames encoding a replication-associated polypolymeric protein (ORF1), a motion-associated protein (ORF2–4), a coat protein (ORF5), and a functionally unknown protein (ORF6). The CYVCV gene structure is stable, and its sequence does not vary significantly due to differences in geographical origin or host variety [6,14].
Phytohormones play a critical role in almost every process of plant development and response. The plant hormones include gibberellins (GAs), salicylic acid (SA), ethylene (ETH), jasmonic acid (JA), abscisic acid (ABA), growth hormone auxin, like Indolacetic acid (IAA), brassinosterol (BR), and strigolactones (SL). These phytohormone regulatory systems have long been known for their roles in tuning abiotic and biotic responses, including the immune response to a viral infection [13,14,15]. Phytohormone-defective mutants have a clear drawback in terms of plant defense against various pathogens, including viruses [15]. Plant defense against viruses is mediated by the small interfering RNA (siRNA) antiviral machinery and is strongly related to the SA signaling pathway [14,16]. The RNA interference (RNAi) system also cooperates with the miRNA system and nucleotide binding site leucine-rich repeat (NBS–LRR) resistance (R) gene in antiviral immunity [17]. Recent studies have further revealed that plant–viral interaction components are also linked to phytohormonal pathways during virus infection [18]. The mechanisms of cross-talk between the siRNA system and phytohormone regulatory systems modulating plant defense against virus diseases have been studied at multiple levels. The primary defense against viruses in plants is triggered by siRNAs derived from the virus, which guide RNA interference and/or DNA methylation to target both RNA and DNA viruses. These viral siRNAs (vsiRNAs) are associated with proteins (Argonaute proteins, Triple Gene Block Protein 1) and phytohormones (BR precursors, PR1, PR2, and PR5) [17,18,19]. Metabolite profiling is a prevalent application of metabolomics in plant and natural product analyses. This method establishes a direct link between changes in metabolite type and content and alterations in biological phenotype. Consequently, metabolomics has emerged as a new research tool, following genomics, transcriptomics, and proteomics, for analyzing complex character systems. Integrating metabolomics with other multiomics approaches allows for a more comprehensive understanding of the genetic mechanisms underlying diverse, complex traits. This integrated analysis enhances our comprehension of the complete biological process encompassing “mutationgene—expression—metabolism—phenotype”, thereby offering a novel means for investigating complex trait mechanisms and introducing innovative ideas for agricultural breeding [20,21,22].
At present, high-throughput RNA sequencing (RNA-seq), metabolomics, and other multiomics techniques have been used to determine the specific transcriptional or metabolic changes in plants in response to viruses [4,12,17]. However, no effective agents have been found to control CYVCD, and no disease-tolerant or disease-resistant lemon varieties have been produced. Viral infection disrupts the physiological environment necessary for plants’ growth and development, affecting plant hormone metabolism and signaling. Conversely, plant hormones play a role in regulating viral replication, assembly, movement, and symptom development in infected plants. The purpose of studying the interplays between host hormones and plant viral infections is to gain a comprehensive understanding of the regulatory mechanisms and cross-talk among different phytohormones in plant–virus interactions. In this study, we performed transcriptomic profiles and targeted metabolomic analyses of plant hormones. The expression of genes involved in phytohormone biosynthesis pathways was analyzed in response to CYVCV infection, revealing changes at both metabolic and transcriptional levels. Understanding the interplays between host hormones and plant viral infections can facilitate innovative approaches for crop protection and improvement.
2. Materials and Methods
2.1. Plant Materials
Samples were collected from ten-year-old Citrus × limon ‘Eureka’ Lemon plants obtained from Anyue, Sichuan Province, at 30.09° N, 105.33° E. The transmission of the virus is facilitated through customary horticultural techniques, including the practice of pruning. Notably, the infection of the lemon with the virus in the context of this study was fortuitous in nature. Our sampling procedures were conducted under authentically natural conditions, leading us to postulate that the involvement of horticultural pruning practices may have been a contributing factor in the dissemination of the viral agent. In order to maintain strict surveillance, afflicted trees are promptly removed upon detection, while the duration of infection with CYVCV spans approximately six months in this study. Tissue specimens of consistent size and color displaying disease symptoms were meticulously harvested from multiple lemon trees affected by the infection. Three biological replicates were obtained for each set. Subsequent to their swift collection, the leaves were promptly immersed in liquid nitrogen, flash frozen, and securely stored at a frigid temperature of −80 °C, being held in readiness for future application.
2.2. Phytohormones Targeting Metabolomics
In this study, the sample’s metabolome was assessed using an LC–MS/MS approach. The raw data files were processed using Compound Discoverer 3.3, which facilitated peak alignment, peak picking, and quantitation for each metabolite. Key parameters, such as peak area correction, mass tolerance, signal intensity tolerance, and minimum intensity, were carefully selected to ensure accurate results. Normalization of peak intensities was performed based on the total spectral intensity. The normalized data were then used to predict the molecular formula and identify metabolites by matching peaks with various databases. To identify, filter, and align peaks, the XCMS package of R (v3.1.3) was employed. This yielded a data matrix containing the mass-to-charge ratio (m/z), retention time, and peak area (intensity). Further, precursor molecules were obtained for both positive and negative ion modes, and the data were exported to Excel for subsequent analysis [23].
2.3. Data Processing and Metabolite Identification
To further understand the biological significance of the identified metabolites, various databases were employed. The KEGG database, HMDB database, and LIPIDMaps database were utilized for annotation purposes. These databases provide valuable information on metabolic pathways, biological functions, and structural details of the metabolites. By leveraging the power of these databases, researchers gain deeper insights into the metabolic profiles under investigation, enabling a more comprehensive understanding of the biological systems being studied. For statistical heatmap clustering analysis of metabolite content, GSEA was performed using the Wekemo BioinCloud platform https://www.bioincloud.tech (accessed on 31 July 2020). R, Python, and Cent OS were employed for statistical analyses involving data standardization and exclusion of compounds with high coefficients of variation (>30%), ultimately enabling the identification and relative quantification of metabolites.
2.4. RNA Sequencing
RNA was extracted using standard extraction methods [24], followed by quality control using an Agilent2100 bioanalyzer (Palo Alto, Santa Clara, CA, USA). First-strand cDNA synthesis utilized an M–MuLV reverse transcriptase system with fragmented mRNA as the template and random oligonucleotides as primers. RNaseH degraded the RNA strand, and the DNA polymerase I system was used. Second-strand cDNA synthesis employed dNTPs. The purified double-stranded cDNA underwent end repair, adenylation, and connector ligation. T4 DNA polymerase and DNA polymerase I (Klenow-) were used for end repair. In the first round of PCR, the forward primer (f) was designed as 5′-gaacgacatggctacgatccgactt-target-specific primer-3′, and the reverse primer (r) was designed as 5′-ctaagaccgcttggcctccgactt-target-specific primer-3′. In the second round of PCR, the primers used were ad153_pcr2_1: 5′-gaacgacatggctacga-3′ and indexr: 5′-tgtgagccaaggagttgnnnnnnnnnnttgtcttcctaagaccgcttggcct-3′. The 5′ end of the ad153_pcr2_1 primer was phosphorylated, and ‘nnnnnnnnnn’ represents the adapter sequence (index) suitable for the mgiseq-2000rs sequencing platform, with ‘n’ indicating any of the four bases (a, t, g, or c). The universal primer sequence linked to the 5′ ends of multiple PCR forward primers is 5′-gaacgacatggctacgatccgactt-3′ (seqidno:1), and the universal primer sequence linked to the 5′ end of multiple PCR reverse primers is 5′-ctaagaccgcttggcctccgactt-3′ (seqidno:2). The reaction conditions for the multiple PCR were as follows: 95 °C for 15 min; 94 °C for 30 s, 60 °C for 90 s, 72 °C for 60 s for 16 cycles; 60 °C for 30 min. In the second round of PCR, the primers used were ad153_pcr2_1: 5′-gaacgacatggctacga-3′ (seqidno:3) and indexr: 5′-tgtgagccaaggagttgnnnnnnnnnnttgtcttcctaagaccgcttggcct-3′. The 5′ end of the ad153_pcr2_1 primer was phosphorylated, and ‘nnnnnnnnnn’ represents the adapter sequence suitable for the mgiseq-2000rs sequencing platform, with ‘n’ indicating any of the four bases (a, t, g, or c). The reaction conditions for the second round of PCR were as follows: 95 °C for 3 min; 98 °C for 20 s, 60 °C for 15 s, 72 °C for 30 s for 8 cycles; 72 °C for 10 min; 4 °C indefinitely. The reaction was stopped by heating at 94 °C for 10 min in a 25 μL reaction system. After library construction, the initial quantification was performed using a Qubit 2.0 Fluorometer. The libraries were diluted to a concentration of 1.5 ng/μL, and the insert size was analyzed using an Agilent 2100 bioanalyzer. Once the insert size met the expected criteria, qRT–PCR was used to accurately quantify the effective concentration of the libraries, ensuring a library quality with an effective concentration greater than 2 nM. AMPureXP magnetic beads screened 250–300 bp cDNA fragments for PCR amplification, followed by purification. The library was quantified using a Qubit2.0 fluorometer and diluted to 1.5 ng/μL. The library’s insert size was analyzed with an Agilent 2100 bioanalyzer. Libraries meeting the criteria were pooled based on effective concentration and desired Illumina sequencing data volume.
RNA-seq was subsequently performed on the Illumina sequencing platform (HiSeq 2500), and 6 cDNA libraries were collected. An average of 6.44 Gb of high-quality reads, characterized by Q20 values ≥98.06% and Q30 values ≥94.55% (Table S2), and 42,002,012 to 45,198,888 raw reads were detected for the CYVCV-infected group. A total of 39,655,388 to 44,609,390 raw reads were detected for the Mock group (Non-infection). After filtering the raw reads, at least 99.58% of the reads met the quality control requirements. The clean reads were subsequently mapped to the Citrus sinensis genome (GCF_022201045.2) by HISAT2 [4,20,25] software. A total of 20585 expressed genes and 681 novel transcripts were annotated (Table S3) and enriched in the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. New transcripts were identified as unannotated transcripts with less than 100 bp and fewer than 2 exons.
Determining differential gene expression across different samples is a key aspect of the analysis. The selection criteria for differentially expressed genes (DEGs) are of utmost importance. For this study, we proposed selecting DEGs based on the criteria of |log2(fold change)| > 1 and padj < 0.05. Genes meeting these criteria were considered as DEGs [26].
Pathway-based analysis plays a crucial role in gaining insights into the biological functions of genes. The KEGG database is a widely used public database for pathway-related information (https://www.kegg.jp/kegg/, accessed on 31 July 2020). To conduct KEGG pathway enrichment analysis, gene IDs from the reference genome to the corresponding IDs used in the KEGG database were constructed. The same applies to GO (Gene Ontology) enrichment analysis. Prior to performing GO enrichment analysis, the associated GO entries for each gene were determined. The Emapper software (v2.1.12) was employed for identifying matching GO entries through sequence alignment. For gene set enrichment analysis (GSEA), GSEA software (v4.1.0) and MSigDB were utilized to explore the associations between gene sets and specific GO terms and KEGG pathways [27].
2.5. Statistical Analyses
The statistical analysis of the metabolite content was completed using Wekemo BioinCloud https://www.bioincloud.tech (accessed on 31 July 2020). Differences between means were analyzed at different confidence levels with Microsoft Excel. A statistical analysis of the transcript levels of genes in CYVCV-infected groups by log2 (fold change) was performed to determine significant differences compared with the Mock group using Student’s t-test at p values < 0.05.
3. Results
3.1. Transcriptome and Metabolomics Profiling Changes in Lemon Trees Infected by CYVCV
Phytohormones play a central role in plant defense against viral diseases. To investigate the dynamic changes in the metabolic level of plant hormones under natural conditions, lemon trees infected by CYVCV in the field, which clearly exhibit morphological characteristics (Figure 1A), were selected and subsequently verified via PCR. After the identification of amplified products, the plant hormone-targeting metabolomes of these samples were studied by using LC–MS/MS (Figure S1). Before standardized correction, the medians and upper and lower quartiles of metabolite content were uneven and different, but after standardized correction, they were basically at the same level. Unsupervised principal component analysis (PCA) was used to determine the differences in the composition and structure of the metabolites between the Mock group and the CYVCV group (Figure S2A). As shown in Figure 1B and Figure S2B, the percentage of each metabolite in each sample was visualized with an accumulation histogram. Compared with those in healthy leaves, the relative abundances of typhasterol and ABA were lower, while the levels of cis-OPDA, SA, and IAA were greater after CYVCV infection. Clustering was carried out to determine the aggregation rule of the metabolite or sample according to the distribution of metabolite content in each sample. The metabolite content in each sample was clustered and is shown in Figure 1C. Different groups were clustered at different positions, which indicated that the composition of these metabolites was quite different among the groups (Table S1).
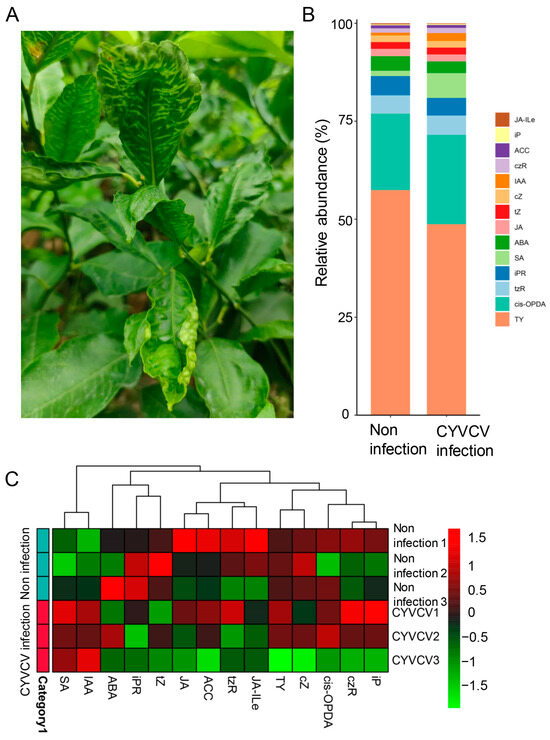
Figure 1.
Changes in the phenotype and phytohormone levels of Citrus limon infected with Citrus yellow vein clearing virus in the field. (A) Phenotype of Citrus limon infected by CYVCV in the field. (B) The relative abundance of plant hormones in the targeted metabolome. (C) The distribution of metabolite content in each sample.
OPLS–DA was used to select features that exhibited the greatest difference between groups. As shown in Figure 2A, the points in the two groups were obviously distributed in different regions, indicating that the metabolite composition and structure of the two groups were quite different between leaves infected by CYVCV and those not infected by CYVCV. An OPLS–DA metabolite importance map showed that SA and IAA were the most regulated metabolites (Figure 2B). The random forest method was used to select the features that contributed the most to the accuracy of sample grouping prediction (Figure 2C, Table S2). The support vector machine (SVM) learning algorithm showed that SA and IAA were induced 3.35- and 2.24-fold, respectively. JA–Ile, ABA, isopentenyladenine riboside (iPR), and JA were reduced 0.48-, 0.71-, 0.76- and 0.82-fold, respectively, in the CYVCV-infected leaves (Table S2). Cis-OPDA, isopentenyl adenine (iP), and cis-zeatin riboside (czR) were slightly induced in the CYVCV-infected leaves. However, trans-zeatin (tZ) was reduced in the CYVCV-infected leaves. Our results suggested that CYVCV infection significantly disrupts plant hormone concentrations and that SA and IAA are induced in CYVCV-infected leaves.
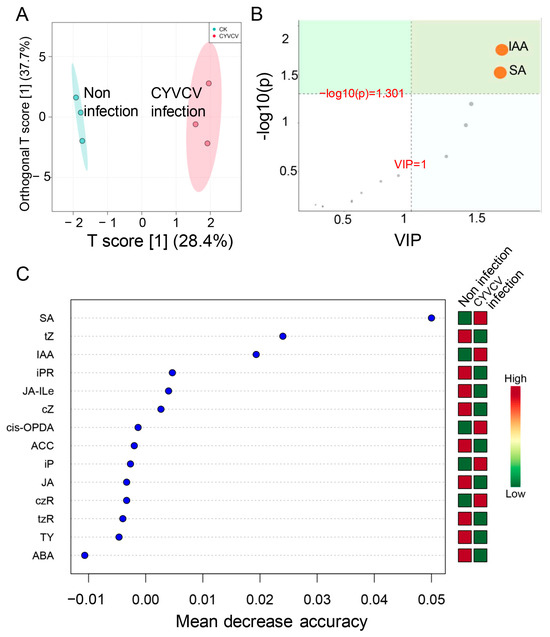
Figure 2.
Differentially abundant metabolite identification. (A) Orthogonal partial least squares discriminant analysis. (B) OPLSDA features the importance of differential hormones in different groups. (C) Detection of differential hormones in Citrus limon infected or not infected by CYVCV using a support vector machine.
Viral infections can disrupt hormonal pathways, causing simultaneous changes in hormone production and triggering defense responses. We investigated transcriptional changes in phytohormone-regulated pathways in lemon plants infected with CYVCV to identify key genes affected by the virus. Box plots were drawn to show the distribution of gene expression in the samples (Figure S3A). To evaluate the repeatability of the biological experiments within the sample group, we used the FPKM data of the samples to calculate the correlation coefficient between the two samples and observed the size of the correlation coefficient, as shown in Figure S3B. The correlation coefficients of the Mock group were 0.94–0.99, and the correlation coefficients of the CYVCV infection groups were 0.98–0.99. A total of 936 differentially expressed genes (DEGs) were identified by screening criteria (padj >= 0.05 and |log2FC| < 1). Among these DEGs, 773 DEGs were downregulated, and only 163 DEGs were upregulated. A volcano map of the differential gene analysis and heatmaps of the gene clusters with the most significant differences are shown in Figure S3C,D. CYVCV infection significantly decreased the expression of the genes in the lemon, and the functions of these genes need to be further explored.
A functional enrichment analysis of the differentially expressed genes (DEGs) was conducted to investigate their association with specific biological processes. Gene Ontology (GO) enrichment revealed that the DEGs were enriched in processes related to the cell wall, external encapsulating structure, and ATPase activity (Figure S4A). The cluster tree diagram also showed enrichment in processes such as the cell cycle, cytoskeleton, and carbohydrate metabolism (Figure S4B). GSEA analysis further demonstrated the downregulation of DEGs associated with cell walls, chemical homeostasis, cytoskeleton, and nucleoside–triphosphatase activity during CYVCV infection (Figure S4C). Protein interaction network analysis revealed the involvement of key genes/proteins in cell wall biogenesis, microtubule-associated complexes, and ATPase activity (Figure S4D). Additionally, KEGG pathway enrichment showed that DEGs were enriched in the plant–pathogen interaction pathway and oxidative phosphorylation (Figure S5A,B). GSEA analysis highlighted the downregulation of DEGs in the cell cycle, oxidative phosphorylation, photosynthesis, and plant–pathogen interaction pathways during CYVCV infection (Figure S5C). Protein interaction network analysis further supported the involvement of key nodes in oxidative phosphorylation, photosynthesis, cell cycle, and plant–pathogen interactions (Figure S5D). These findings suggest that disruption of microtubule-associated processes and hormone signal transduction contribute to CYVCV infection.
3.2. Network Analysis of Genes Regulating Hormone Signaling
Due to the changes in phytohormones at the metabolic and transcriptional levels, we analyzed the expression of genes involved in phytohormone biosynthesis pathways. As shown in Figure 3 and Table S6, CYVCV infection increased SA levels, 11 genes were associated with SA biosynthesis, and 21 genes were associated with SA signal transduction. In the SA biosynthesis process, the majority of these DEGs were downregulated. Isochorismate synthase (ICS, LOC102630235) was significantly upregulated, and enhanced disease susceptibility 1 (EDS1, LOC102618041) was significantly downregulated in the CYVCV infection groups. During the SA signal transduction process, the BTB/POZ domain and ankyrin repeat-containing protein (NPR1, LOC112498891) were induced in the CYVCV infection groups. We also analyzed the expression of genes involved in auxin biosynthesis pathways. As shown in Figure 4 and Table S6, CYVCV infection induced IAA levels, and a total of 154 genes were associated with IAA biosynthesis, inactivation, and signal transduction. The expression of indole-3-pyruvate monooxygenase (YUCCA, LOC102618240, and LOC102616113) was upregulated. Amidase (amiE, LOC112495464) was suppressed significantly during the auxin biosynthesis process. Auxin-responsive GH3 (GH3, LOC102616398) and UDP-glycosyltransferase (UGT74B1, LOC102613517, LOC102612931, LOC102630144, LOC102621570…) genes involved in the auxin inactivation process were differentially expressed in the CYVCV infection groups. Our results explored the key genes related to SA and IAA biosynthesis inactivation and signal transduction processes and elucidated the phytohormone changes after CYVCV infection. The infection led to increased SA levels and altered expression of genes associated with SA biosynthesis and signal transduction. Additionally, CYVCV infection induced IAA levels and affected the expression of genes involved in IAA biosynthesis, inactivation, and signal transduction.
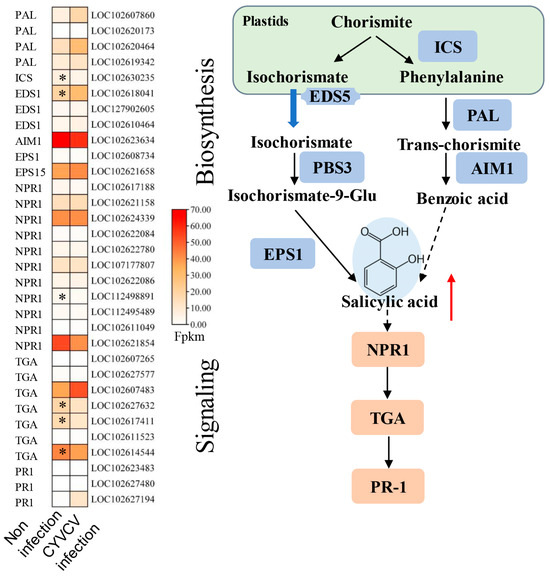
Figure 3.
Expression of genes involved in SA biosynthesis and signaling pathways. Diagram showing the phytohormone signaling pathways. Heatmap analysis of log2-fold changes; the asterisks in the heatmaps represent the DEGs. The list of DEGs is shown in Table S6. ICS: Isochorismate Synthase, NPR1: BTB/POZ domain and ankyrin repeat-containing protein, TGA: transcription factor TGA, PR1: pathogenesis-related protein 1, PAL: phenylalanine ammonia-lyase, EDS1: enhanced disease susceptibility 1 protein, EPS1: epidermal growth factor receptor substrate 1. The red arrow indicates upward adjustment, * indicated p values ≤ 0.05.
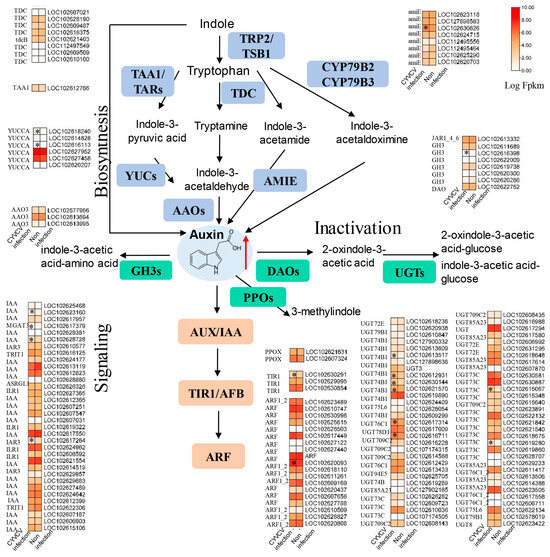
Figure 4.
Expression of genes involved in IAA biosynthesis and signaling pathways. Diagram showing the phytohormone signaling pathways. Heatmap analysis of log2-fold changes; the asterisks in the heatmaps represent the DEGs. The list of DEGs is shown in Table S6. amiE: amidase, TDC: L-tryptophan decarboxylase, TAA1: L-tryptophan-pyruvate aminotransferase. YUCs: indole 3 pyruvate monooxygenase, AAO3: abscisic-aldehyde oxidase, GH3: auxin responsive GH3, DAO: 2-oxoglutarate-dependent dioxygenase, PPOs: protoporphyrinogen oxidase, UGTs: coniferyl-alcohol glucosyltransferase, IAA: auxin-responsive protein IAA, ARF; ADP-ribosylation factor, TIR1: transport inhibitor response 1, CYP79, cytochrome P450 79. The red arrow indicates upward adjustment, * indicated p values ≤ 0.05.
Because ABA was reduced to a 0.71-fold change in the CYVCV-infected group, JA was reduced to a 0.82-fold change in the CYVCV-infected group, and JA–Ile was reduced to a 0.48-fold change in the CYVCV-infected group (Table S2); therefore, we also analyzed the expression of genes involved in the ABA and JA biosynthesis pathways. As shown in Figure 5 and Table S6, the expression of JA biosynthesis-related genes, including nine lipoxygenase (LOX) genes, was mostly downregulated, LOXS2 (LOC102620930, LOC102621545) was significantly downregulated, and allene oxide cyclase (AOC, LOC102625302, LOC112499525, LOC102623979)/amine oxidase (AOS) was upregulated in the CYVCV-infected group. 12-Oxophytodienoic acid reductase (OPR, LOC102608507) was significantly downregulated. Jasmonate ZIM domain-containing protein (JAZ, LOC102629841) and jasmonic acid amino synthetase (JAR1_4_6, LOC102611440) in the JA signaling pathway were significantly downregulated. As shown in Figure 6 and Table S6, the expression of the ABA biosynthesis-related genes zeaxanthin epoxidase (ZEP, LOC102619293), 9 cis-epoxycarotenoid dioxygenase (NCED, LOC102578070), and neoxanthin synthase (NSY, LOC102625209) were significantly downregulated. The expression of ABA signaling transduction-related genes, and the abscisic acid receptor PYR/PYL family (PYL, LOC102613559) was induced in the CYVCV infection groups. 3 oxoacyl [acyl carrier protein] synthase II (LOC102621824) and ABA responsive element binding factor (ABF, LOC102608145, LOC102613575, LOC102618173) were the most strongly downregulated genes. Our results illustrated the dynamic changes in ABA and JA hormones at the transcriptome level.
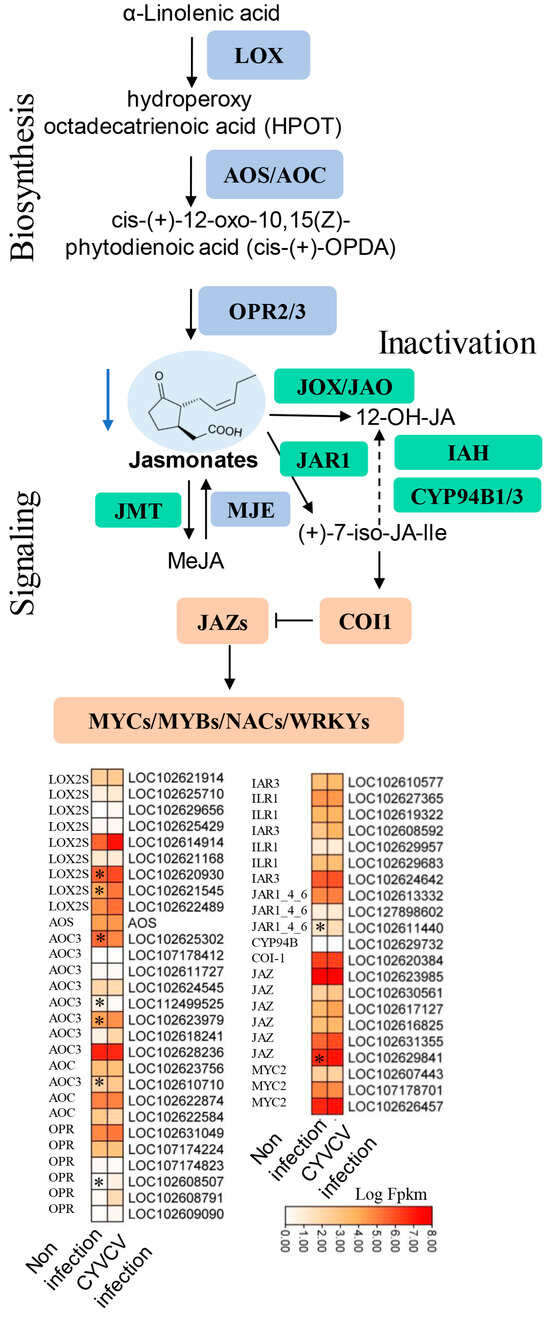
Figure 5.
Expression of genes involved in JA biosynthesis and signaling pathways. Diagram showing the phytohormone signaling pathways. Heatmap analysis of log2-fold changes; the asterisks in the heatmaps represent the DEGs. The list of DEGs is shown in Table S6. LOX: lipoxygenase, AOS: allene oxide synthase, AOC: allene oxide cyclase, OPR: 12-oxophytodienoic acid reductase, JAR: jasmonic acid amino synthetase, CYP94B, cytochrome P450 94B, COI-1: coronatine-insensitive protein 1, JAZ: jasmonate ZIM domain-containing protein, MYC: transcription factor MYC, NAC (NAM, ATAF1/2, CUC1/2) transcription factors, The blue arrow indicates down adjustment, * indicated p values ≤ 0.05.
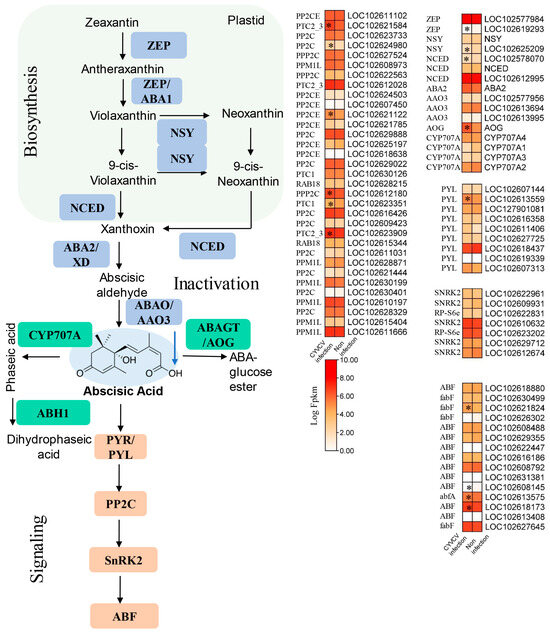
Figure 6.
Expression of genes involved in ABA biosynthesis and signaling pathways. Diagram showing the phytohormone signaling pathways. Heatmap analysis of log2-fold changes; the asterisks in the heatmaps represent the DEGs. The list of DEGs is shown in Table S6. ZEP: zeaxanthin epoxidase, NSY: neoxanthin synthase, NCED: 9-cis-epoxycarotenoid dioxygenase, ABA2: xanthoxin dehydrogenase, AAO3: abscisic-aldehyde oxidase, AOG: abscisate beta-glucosyltransferase, CYP707A: (+)-abscisic acid 8′-hydroxylase, PYL: abscisic acid receptor PYR/PYL family, PP2CE: protein phosphatase 1L, SNRK2: serine/threonine protein kinase SRK2, ABF: ABA-responsive element binding factor. The blue arrow indicates down adjustment, * indicated p values ≤ 0.05.
4. Discussion
CYVCD, also known as yellow vein clearing disease, is a viral disease of most citrus species. Although the host range of CYVCV is wide, lemons are the most sensitive to the virus; CYVCV causes yellow vein clearing disease in Citrus × limon ‘Eureka’, which is a significant and devastating disease. How is the virus disease controlled? How to avoid the spread of CYVCV and its adverse effects on the yield and quality of lemons is a critical issue in current research. Previous studies have focused on the molecular characterization and rapid identification of CYVCV, and only a few studies have investigated the pathogenesis of CYVCV. Multiomics analysis provides another effective strategy for revealing the dynamic changes in citrus-CYVCV interactions. In this study, phytohormone targeting metabolomics and transcriptomic analysis were performed, and networks of genes regulating SA, IAA, ABA, and JA hormone metabolism at the transcriptome level were explored. This study provides some reference for the subsequent research on the interaction between plants and viruses.
Transcriptomic and metabolomic data have been widely used to explore plant defenses against viral diseases. Our study was performed under natural conditions at the late infection stage, during which time a change in phytohormone levels could occur during CYVCV infection. A total of 936 DEGs were identified in this study, most of which were downregulated. Phytohormones are disrupted by viral infection and participate in plant defense responses [14,28,29]. GSEA showed that the DEGs were enriched in the microtubule-related complex, ATPase activity, hormone signal transduction pathway, and plant-pathogen interaction pathway. Microtubules and associated complexes play important roles in viruses subverting host cell mechanisms, replication, and transmission [30,31]. Many DEGs in the lemon’s response to CYVCV are enriched in functions related to cytoskeletal protein binding and microtubule motor activity. This suggests that CYVCV utilizes the host cytoskeleton for transport, which may be linked to the distinctive phenotype observed in the new leaves during the spring and autumn growth stages. Some coat proteins (CPs) of potato viruses X (PVX) exhibit enzyme activities, such as ATPase activities [32], and host ATPase activities also respond to virus infection [33,34]. The coat protein (CP) of CYVCV serves as a potent RNA silencing suppressor linked to symptom severity in citrus, interacting with the 40S ribosomal subunit protein S9-2 (ClRPS9-2) through its N-terminal 8–108 amino acid sequence, while it would be valuable to investigate whether the coat proteins of CYVCV exhibit ATPase activity or modulate the ATPase activities of their host cells. Virus-mediated phytohormone disruption and alteration are common ways to facilitate virus invasion [14,16,29]. RNAi, in combination with salicylic acid, mediates host antiviral immunity in stem cells [35], and the virus defense response is cross-linked with the phytohormone regulation system [36,37]. A previous study suggests that phytohormone biosynthesis, signaling, and photosynthesis-related genes play a role in CYVCV infection and symptom occurrence in lemon plants [4]. So, we further analyzed phytohormone regulation gene expression patterns in lemon plants infected with CYVCV.
Plant hormones play a crucial role in viral infection. They can affect virus replication and accumulation, symptom development, virus movement, host resistance, and the relationship with insect vectors [38,39,40]. Viral infection can also disrupt hormonal pathways, triggering defense responses and inducing synergistic or antagonistic hormones [39,41]. In our study on CYVCV infection, we observed increased levels of SA and IAA. Genes involved in SA biosynthesis, such as isochorismate synthase (ICS) and BTB/POZ domain protein NPR1, were upregulated. SA treatment provides broad-spectrum defense against many viruses [42,43]. NUCLEAR INCLUSION B (NIb) of the Turnip mosaic virus can bind NPR1 to prevent sumoylation and phosphorylation of NPR1 [42]. The increased resistance of lemon plants to CYVCV could be linked to the up-regulation of salicylic acid-associated pathways in our and other studies [4,6]. CYVCV infection affected IAA levels and the expression of genes related to IAA biosynthesis, inactivation, and signal transduction. Higher production of SA enhances resistance against tobacco mosaic virus (TMV) and induces PR gene expressing [43]. The TMV disrupts IAA26′s localization and regulates auxin response during disease development; The Rice dwarf virus P2 protein interacts with OsIAA10 and OsARF17 to disrupt auxin signaling to facilitate infection [44]. The analysis of gene expression in ABA and JA biosynthesis pathways revealed downregulation of JA-related genes (LOX, LOXS2, OPR, JAZ, JAR1_4_6) and upregulation of AOC/AOS genes. In rice, increased OsJAZ4 levels and decreased JA signaling resulted in susceptibility to Rice black-streaked dwarf virus (RBSDV) [45]. JA and GA activate MYC2 and induce sesquiterpene biosynthesis, while ABA promotes interaction between ABA receptor PYL6 and MYC2, hindering MYC2’s binding ability to target promoters. In the Tomato yellow leaf curl China virus, βC1 interacts with MYC2, suppressing MYC2-regulated terpene synthase and accelerating viral spread [46]. CYVCV infection induces ABA signaling-related genes (PYL) but downregulates 3-oxoacyl [acyl carrier protein] synthase II and ABF. This leads to reduced levels of ABA and JA hormones, potentially affecting plant defense responses against viral infections. ABA negatively regulates rice defense against RBSDV infection by suppressing the JA pathway and ROS accumulation [39,47]. A comprehension of hormone-mediated functions in plant–virus interactions can facilitate the development of strategies for disease control.
In plants, the cell wall not only provides rigid support for the plant but is also the first barrier against the invasion of diseases and pests [48]. Cellulose synthase A, trichome birefringence [49], and chitinase-like protein 2 were found to be downregulated during CYVCV infection. Lectin can bind to viral glycoproteins [50,51] and is significantly reduced after CYVCV infection (Figure S6). This may contribute to CYVCV infection and transmission. Surprisingly, we found that the expression of MLO-like protein 12 was significantly induced (Figure S6). Mlo is a susceptibility gene that barely protects against powdery mildew and loss-of-function. Mlo confers broad-spectrum resistance against all known isolates of powdery mildew [52,53]. Another unpublished study has found that cli-miRn6 plays a role in the lemon’s response to viruses by targeting and cleaving the Mlo gene. So, whether Mlo can be a pathogenic factor in CYVCV infection is worth exploring further. Disease-resistance proteins are also involved in the CYVCV-lemon interaction [54,55,56]. CYVCV can be transmitted by a wide array of pathways, such as mechanical transmission and insect transmission pathways. Insect transmission is also an important pathway, and thaumatin-like proteins, tetrahydroberberine oxidase-like proteins, eugenol synthase, and laccase 4 [57,58] may contribute to flavor changes and promote insect feeding. These DEGs also respond to biotic stress [59,60,61]. Through the analysis of differentially expressed genes in lemons infected by CYVCV and combined with changes in hormone levels, marker genes, and quantitative methods for plant hormones, future research on the interaction between plants and viruses can be identified. By genetically reducing or even eliminating the impact of viruses on yield, resistant and tolerant varieties can be developed.
5. Conclusions
Viruses often exploit the components of the host’s phytohormone pathways as a strategy for their own pathogenesis. In this study, we revealed the changes in phytohormone levels at the metabolic and transcriptional levels in lemon plants infected with CYVCV. CYVCV infection significantly depressed lemon plant gene expression. In response to CYVCV infection, they upregulated ICS and downregulated EDS1 in an increased SA regulation pathway, with SA signal transduction involving NPR1. Moreover, IAA biosynthesis is enhanced by upregulated YUCCA genes and suppressed amidase genes, while GH3 and UGT74B1 genes are involved in IAA inactivation during CYVCV infection. Additionally, CYVCV infection results in decreased levels of ABA, JA, and JA-Ile, impacting the expression of genes, such as upregulation of AOC/AOS and downregulation of genes involved in JA signaling like JAZ and JAR1_4_6. Genes associated with ABA biosyntheses, such as ZEP, NCED, and NSY, are significantly downregulated, while ABA signaling-related genes like PYL and ABF are induced in response to CYVCV infection. Notably, the involvement of pathogenic factors such as Mlo, lectin, microtubules, associated complexes, cell wall-related enzymes, and disease-resistance proteins among the DEGs needs to be further explored. The changes in the levels of endogenous phytohormones are a direct result of viral infection and are closely coordinated with the movement and replication of the virus, the development of symptoms, and the activation of defense responses. Further exploration is needed to understand the involvement of these genes in the CYVCV-lemon interaction.
Supplementary Materials
The following supporting information can be downloaded from https://www.mdpi.com/article/10.3390/horticulturae10030231/s1. Figure S1: Normalization of hormone targeting metabolomics data; Figure S2: (A) Principal component analysis of the biological replicates from each treatment. (B) Percentage content bar chart of different hormones in CYVCV-infected lemon plants and non-infected (Mock) plants; Figure S3: (A) Box plots of the gene expression distribution in the samples. (B) Spearman correlation heatmap of TPM expression among samples. (C) Differential analysis volcano map. (D) Heatmaps of the gene clusters with the most significant differences; Figure S4: GSEA enrichment in GO terms of differentially expressed genes. (A) Bubble map of the enrichment analysis of the DEGs. (B) The clustering tree of the items in the enrichment analysis. Tree graph for clustering according to the Jaccard correlation coefficient between GO entries. (C) GSEA enrichment analysis. (D) Enrichment analysis of the gene concept network. At the center of the nodes of each cluster are the GO entries, and these peripheral nodes are the corresponding genes; Figure S5: GSEA enrichment in the KEGG pathway of differentially expressed genes. (A) Bubble map of KEEG enrichment analysis of DEGs. (B) The clustering tree of the items in the KEEG enrichment analysis. Tree graph for clustering according to the Jaccard correlation coefficient between KEGG pathways. (C) GSEA enrichment analysis. (D) Enrichment analysis of the gene concept network. The KEEG pathway is located at the center of the nodes of each cluster, and these peripheral nodes are the corresponding genes. * indicate p values ≤ 0.05; Figure S6: Gene expression (Fpkm) of lectin and Mlo in different samples; Table S1: Relative abundance of phytohormones in different samples; Table S2: SVM feature importance of phytohormones in different samples; Table S3: Transcriptome quality control data; Table S4: Novel transcript identification in the transcriptome; Table S5: DEGs identified in this study; Table S6: Network analysis of genes regulating phytohormone metabolism.
Author Contributions
T.Q. and Q.W.: conceptualization and data curation. X.H.: writing—original draft. L.L., X.C. and L.D.: visualization, investigation. L.Z., X.H. and M.D.: supervision, funding acquisition. T.Q. and Q.W.: management and coordination responsibility for the research; X.H.: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (No. 32001851, No. 32301838) and the Natural Science Foundation of Sichuan Province (2023NSFSC0149, 2022NSFSC1715). The Scientific Research Initiation Project of Mianyang Normal University (QD2023A01, QD2023A01). The Mainyang Normal University Innovation Team Project (CXTD2023PY02, CXTD2023LX01).
Data Availability Statement
The data are available in the Supplemental Materials, and further information can be provided if needed.
Acknowledgments
We thank Microeco Tech Co., Ltd. We thank Shenzhen, China, for sequencing services and metabolomic profiling services.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Zhou, Y.; Chen, H.M.; Cao, M.J.; Wang, X.F.; Jin, X.; Liu, K.H.; Zhou, C.Y. Occurrence, Distribution, and Molecular Characterization of Citrus Yellow Vein Clearing Virus in China. Plant Dis. 2017, 101, 137–143. [Google Scholar] [CrossRef]
- Zhen, S.; Kurth, E.G.; Peremyslov, V.V.; Changyong, Z.; Dolja, V.V. Molecular characterization of a citrus yellow vein clearing virus strain from China. Arch. Virol. 2015, 160, 1811–1813. [Google Scholar] [CrossRef]
- Bin, Y.; Xu, J.; Duan, Y.; Ma, Z.; Zhang, Q.; Wang, C.; Su, Y.; Jiang, Q.; Song, Z.; Zhou, C. The Titer of Citrus Yellow Vein Clearing Virus Is Positively Associated with the Severity of Symptoms in Infected Citrus Seedlings. Plant Dis. 2022, 106, 828–834. [Google Scholar] [CrossRef]
- Bin, Y.; Zhang, Q.; Su, Y.; Wang, C.; Jiang, Q.; Song, Z.; Zhou, C. Transcriptome Analysis of Citrus limon Infected with Citrus Yellow Vein Clearing Virus. BMC Genom. 2023, 24, 65. [Google Scholar] [CrossRef]
- Loconsole, G.; Önelge, N.; Potere, O.; Giampetruzzi, A.; Bozan, O.; Satar, S.; De Stradis, A.; Savino, V.; Yokomi, R.K.; Saponari, M. Identification and Characterization of Citrus Yellow Vein Clearing Virus, A Putative New Member of the Genus Mandarivirus. Phytopathology 2012, 102, 1168–1175. [Google Scholar] [CrossRef]
- Liu, C.; Liu, H.; Hurst, J.; Timko, M.P.; Zhou, C. Recent Advances on Citrus Yellow Vein Clearing Virus in Citrus. Hortic. Plant J. 2020, 6, 216–222. [Google Scholar] [CrossRef]
- Chen, H.M.; Zhou, Y.; Wang, X.F.; Zhou, C.Y.; Li, Z.A. Detection of Citrus Yellow Vein Clearing Virus Based on a Real-Time RT-PCR Approach. Acta Hortic. Sin. 2016, 2, 188–192. [Google Scholar]
- Catara, A.; Azzaro, A.; Davino, M.; Polizzi, G. Yellow Vein Clearing of Lemon in Pakistan. In Proceedings of the International Organization of Citrus Virologists Conference Proceedings, India, New Delhi, 23 November 1992. [Google Scholar] [CrossRef]
- Hashemian, S.M.B.; Aghajanzadeh, S. Occurrence of Citrus Yellow Vein Clearing Virus In Citrus Species In Iran. J. Plant Pathol. 2017, 99, 290. [Google Scholar]
- Yang, X.; Xu, Q.; Liu, Z.; Zhou, C.; Cao, M. First Report of Citrus Virus A Infecting Citrus (Citrus reticulata) in China. Plant Dis. 2023, 107, 2269. [Google Scholar] [CrossRef]
- Chen, H.M.; Li, Z.A.; Wang, X.F.; Zhou, Y.; Tang, K.Z.; Zhou, C.Y.; Zhao, X.Y.; Yue, J.Q. First Report of Citrus Yellow Vein Clearing Virus on Lemon in Yunnan, China. Plant Dis. 2014, 98, 1747. [Google Scholar] [CrossRef]
- Meena, R.P.; Prabha, K.; Baranwal, V.K. Genome Characterization of Citrus Yellow Vein-Clearing Virus: Limited Heterogeneity of Viral Genomes in Mandarivirus-Infecting Different Citrus Species. 3 Biotech 2019, 9, 348. [Google Scholar] [CrossRef]
- Denancé, N.; Sánchez-Vallet, A.; Goffner, D.; Molina, A. Disease Resistance or Growth: The Role of Plant Hormones in Balancing Immune Responses and Fitness Costs. Front. Plant Sci. 2013, 4, 155. [Google Scholar] [CrossRef]
- Islam, W.; Naveed, H.; Zaynab, M.; Huang, Z.; Chen, H.Y.H. Plant Defense against Virus Diseases; Growth Hormones in Highlights. Plant Signal. Behav. 2019, 14, 1596719. [Google Scholar] [CrossRef]
- Ma, K.-W.; Ma, W. Phytohormone Pathways as Targets of Pathogens to Facilitate Infection. Plant Mol. Biol. 2016, 91, 713–725. [Google Scholar] [CrossRef]
- Alazem, M.; Lin, N. Roles of Plant Hormones in the Regulation of Host–Virus Interactions. Mol. Plant Pathol. 2015, 16, 529–540. [Google Scholar] [CrossRef]
- Jin, Y.; Zhao, J.-H.; Guo, H.-S. Recent Advances in Understanding Plant Antiviral RNAi and Viral Suppressors of RNAi. Curr. Opin. Virol. 2021, 46, 65–72. [Google Scholar] [CrossRef]
- Nafisi, M.; Fimognari, L.; Sakuragi, Y. Interplays between the Cell Wall and Phytohormones in Interaction between Plants and Necrotrophic Pathogens. Phytochemistry 2015, 112, 63–71. [Google Scholar] [CrossRef]
- Senshu, H.; Ozeki, J.; Komatsu, K.; Hashimoto, M.; Hatada, K.; Aoyama, M.; Kagiwada, S.; Yamaji, Y.; Namba, S. Variability in the Level of RNA Silencing Suppression Caused by Triple Gene Block Protein 1 (TGBp1) from Various Potexviruses during Infection. J. Gen. Virol. 2009, 90, 1014–1024. [Google Scholar] [CrossRef]
- Pandohee, J.; Kyereh, E.; Kulshrestha, S.; Xu, B.; Mahomoodally, M.F. Review of the Recent Developments in Metabolomics-Based Phytochemical Research. Crit. Rev. Food Sci. Nutr. 2023, 63, 3734–3749. [Google Scholar] [CrossRef]
- Dalio, R.J.D.; Litholdo, C.G.; Arena, G.; Magalhães, D.; Machado, M.A. Contribution of Omics and Systems Biology to Plant Biotechnology. In Advances in Plant Omics and Systems Biology Approaches; Springer: Cham, Switzerland, 2021; pp. 171–188. [Google Scholar]
- Aghdam, S.A.; Brown, A.M.V. Deep Learning Approaches for Natural Product Discovery from Plant Endophytic Microbiomes. Environ. Microbiome 2021, 16, 6. [Google Scholar] [CrossRef]
- Chong, J.; Xia, J. MetaboAnalystR: An R Package for Flexible and Reproducible Analysis of Metabolomics Data. Bioinformatics 2018, 34, 4313–4314. [Google Scholar] [CrossRef]
- Rio, D.C.; Ares, M.; Hannon, G.J.; Nilsen, T.W. Purification of RNA Using TRIzol (TRI Reagent); Cold Spring Harbor: New York, NY, USA, 2010. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Ghosh, D.; Chakraborty, S. Molecular Interplay between Phytohormones and Geminiviruses: A Saga of a Never-Ending Arms Race. J. Exp. Bot. 2021, 72, 2903–2917. [Google Scholar] [CrossRef]
- Collum, T.D.; Culver, J.N. The Impact of Phytohormones on Virus Infection and Disease. Curr. Opin. Virol. 2016, 17, 25–31. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Y.; Luo, L.; Hao, J.; Li, J. Characterization of Cmcp Gene as a Pathogenicity Factor of Ceratocystis Manginecans. Front. Microbiol. 2020, 11, 1824. [Google Scholar] [CrossRef]
- Wang, Z.; Xia, Y.; Lin, S.; Wang, Y.; Guo, B.; Song, X.; Ding, S.; Zheng, L.; Feng, R.; Chen, S.; et al. Osa-MiR164a Targets OsNAC60 and Negatively Regulates Rice Immunity against the Blast Fungus Magnaporthe Oryzae. Plant J. 2018, 95, 584–597. [Google Scholar] [CrossRef]
- Rakitina, D.V.; Kantidze, O.L.; Leshchiner, A.D.; Solovyev, A.G.; Novikov, V.K.; Morozov, S.Y.; Kalinina, N.O. Coat Proteins of Two Filamentous Plant Viruses Display NTPase Activity in Vitro. FEBS Lett. 2005, 579, 4955–4960. [Google Scholar] [CrossRef]
- Cheng, Z.; Yang, J.; Xia, H.; Qiu, Y.; Wang, Z.; Han, Y.; Xia, X.; Qin, C.-F.; Hu, Y.; Zhou, X. The Nonstructural Protein 2C of a Picorna-Like Virus Displays Nucleic Acid Helix Destabilizing Activity that Can Be Functionally Separated from Its ATPase Activity. J. Virol. 2013, 87, 5205–5218. [Google Scholar] [CrossRef][Green Version]
- Tu, Y.; Zhang, Z.; Li, D.; Li, H.; Dong, J.; Wang, T. Potato Virus Y HC-Pro Reduces the ATPase Activity of NtMinD, Which Results in Enlarged Chloroplasts in HC-Pro Transgenic Tobacco. PLoS ONE 2015, 10, e0136210. [Google Scholar] [CrossRef]
- Incarbone, M.; Bradamante, G.; Pruckner, F.; Wegscheider, T.; Rozhon, W.; Nguyen, V.; Gutzat, R.; Mérai, Z.; Lendl, T.; MacFarlane, S.; et al. Salicylic Acid and RNA Interference Mediate Antiviral Immunity of Plant Stem Cells. Proc. Natl. Acad. Sci. USA 2023, 120, e2302069120. [Google Scholar] [CrossRef]
- Silva-Martins, G.; Roussin-Léveillée, C.; Bolaji, A.; Veerapen, V.P.; Moffett, P. A Jasmonic Acid–Related Mechanism Affects ARGONAUTE5 Expression and Antiviral Defense against Potato Virus X in Arabidopsis Thaliana. Mol. Plant-Microbe Interact. 2023, 36, 425–433. [Google Scholar] [CrossRef]
- Jay, F.; Wang, Y.; Yu, A.; Taconnat, L.; Pelletier, S.; Colot, V.; Renou, J.-P.; Voinnet, O. Misregulation of AUXIN RESPONSE FACTOR 8 Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis. PLoS Pathog. 2011, 7, e1002035. [Google Scholar] [CrossRef]
- Collum, T.D.; Padmanabhan, M.S.; Hsieh, Y.-C.; Culver, J.N. Tobacco Mosaic Virus-Directed Reprogramming of Auxin/Indole Acetic Acid Protein Transcriptional Responses Enhances Virus Phloem Loading. Proc. Natl. Acad. Sci. USA 2016, 113, E2740–E2749. [Google Scholar] [CrossRef]
- Zhao, S.; Li, Y. Current Understanding of the Interplays between Host Hormones and Plant Viral Infections. PLoS Pathog. 2021, 17, e1009242. [Google Scholar] [CrossRef]
- Pan, L.; Miao, H.; Wang, Q.; Walling, L.L.; Liu, S. Virus-induced Phytohormone Dynamics and Their Effects on Plant–Insect Interactions. New Phytol. 2021, 230, 1305–1320. [Google Scholar] [CrossRef]
- Zhu, S.; Gao, F.; Cao, X.; Chen, M.; Ye, G.; Wei, C.; Li, Y. The Rice Dwarf Virus P2 Protein Interacts with Ent -Kaurene Oxidases in Vivo, Leading to Reduced Biosynthesis of Gibberellins and Rice Dwarf Symptoms. Plant Physiol. 2005, 139, 1935–1945. [Google Scholar] [CrossRef]
- Liu, J.; Wu, X.; Fang, Y.; Liu, Y.; Bello, E.O.; Li, Y.; Xiong, R.; Li, Y.; Fu, Z.Q.; Wang, A.; et al. A Plant RNA Virus Inhibits NPR1 Sumoylation and Subverts NPR1-Mediated Plant Immunity. Nat. Commun. 2023, 14, 3580. [Google Scholar] [CrossRef]
- Yalpani, N.; Leon, J.; Lawton, M.A.; Raskin, I. Pathway of Salicylic Acid Biosynthesis in Healthy and Virus-Inoculated Tobacco. Plant Physiol. 1993, 103, 315–321. [Google Scholar] [CrossRef]
- He, X.; Jiang, J.; Wang, C.; Dehesh, K. ORA59 and EIN3 Interaction Couples Jasmonate-ethylene Synergistic Action to Antagonistic Salicylic Acid Regulation of PDF Expression. J. Integr. Plant Biol. 2017, 59, 275–287. [Google Scholar] [CrossRef]
- He, Y.; Hong, G.; Zhang, H.; Tan, X.; Li, L.; Kong, Y.; Sang, T.; Xie, K.; Wei, J.; Li, J.; et al. The OsGSK2 Kinase Integrates Brassinosteroid and Jasmonic Acid Signaling by Interacting with OsJAZ4. Plant Cell 2020, 32, 2806–2822. [Google Scholar] [CrossRef]
- Li, R.; Weldegergis, B.T.; Li, J.; Jung, C.; Qu, J.; Sun, Y.; Qian, H.; Tee, C.; van Loon, J.J.A.; Dicke, M.; et al. Virulence Factors of Geminivirus Interact with MYC2 to Subvert Plant Resistance and Promote Vector Performance. Plant Cell 2014, 26, 4991–5008. [Google Scholar] [CrossRef]
- Xie, K.; Li, L.; Zhang, H.; Wang, R.; Tan, X.; He, Y.; Hong, G.; Li, J.; Ming, F.; Yao, X.; et al. Abscisic Acid Negatively Modulates Plant Defence against Rice Black-streaked Dwarf Virus Infection by Suppressing the Jasmonate Pathway and Regulating Reactive Oxygen Species Levels in Rice. Plant Cell Environ. 2018, 41, 2504–2514. [Google Scholar] [CrossRef]
- Kozieł, E.; Otulak-Kozieł, K.; Bujarski, J.J. Plant Cell Wall as a Key Player During Resistant and Susceptible Plant-Virus Interactions. Front. Microbiol. 2021, 12, 656809. [Google Scholar] [CrossRef]
- Sun, A.; Yu, B.; Zhang, Q.; Peng, Y.; Yang, J.; Sun, Y.; Qin, P.; Jia, T.; Smeekens, S.; Teng, S. MYC2-Activated TRICHOME BIREFRINGENCE-LIKE37 Acetylates Cell Walls and Enhances Herbivore Resistance. Plant Physiol. 2020, 184, 1083–1096. [Google Scholar] [CrossRef]
- Francis, F.; Chen, J.; Yong, L.; Bosquee, E. Aphid Feeding on Plant Lectins Falling Virus Transmission Rates: A Multicase Study. J. Econ. Entomol. 2020, 113, 1635–1639. [Google Scholar] [CrossRef]
- Konozy, E.H.E.; Osman, M.E.M.; Dirar, A.I.; Ghartey-Kwansah, G. Plant Lectins: A New Antimicrobial Frontier. Biomed. Pharmacother. 2022, 155, 113735. [Google Scholar] [CrossRef]
- Acevedo-Garcia, J.; Kusch, S.; Panstruga, R. Magical Mystery Tour: MLO Proteins in Plant Immunity and Beyond. New Phytol. 2014, 204, 273–281. [Google Scholar] [CrossRef]
- Jacott, C.N.; Ridout, C.J.; Murray, J.D. Unmasking Mildew Resistance Locus O. Trends Plant Sci. 2021, 26, 1006–1013. [Google Scholar] [CrossRef]
- Sett, S.; Prasad, A.; Prasad, M. Resistance Genes on the Verge of Plant–Virus Interaction. Trends Plant Sci. 2022, 27, 1242–1252. [Google Scholar] [CrossRef]
- Poque, S.; Pagny, G.; Ouibrahim, L.; Chague, A.; Eyquard, J.-P.; Caballero, M.; Candresse, T.; Caranta, C.; Mariette, S.; Decroocq, V. Allelic Variation at the Rpv1 Locus Controls Partial Resistance to Plum Pox Virus Infection in Arabidopsis Thaliana. BMC Plant Biol. 2015, 15, 159. [Google Scholar] [CrossRef]
- Helderman, T.A.; Deurhof, L.; Bertran, A.; Richard, M.M.S.; Kormelink, R.; Prins, M.; Joosten, M.H.A.J.; van den Burg, H.A. Members of the Ribosomal Protein S6 (RPS6) Family Act as Pro-viral Factor for Tomato Spotted Wilt Orthotospovirus Infectivity in Nicotiana Benthamiana. Mol. Plant Pathol. 2022, 23, 431–446. [Google Scholar] [CrossRef]
- Zippilli, C.; Botta, L.; Bizzarri, B.M.; Nencioni, L.; De Angelis, M.; Protto, V.; Giorgi, G.; Baratto, M.C.; Pogni, R.; Saladino, R. Laccase-Catalyzed 1,4-Dioxane-Mediated Synthesis of Belladine N-Oxides with Anti-Influenza A Virus Activity. Int. J. Mol. Sci. 2021, 22, 1337. [Google Scholar] [CrossRef]
- Lu, Z.; Deng, J.; Wang, H.; Zhao, X.; Luo, Z.; Yu, C.; Zhang, Y. Multifunctional Role of a Fungal Pathogen-secreted Laccase 2 in Evasion of Insect Immune Defense. Environ. Microbiol. 2021, 23, 1256–1274. [Google Scholar] [CrossRef]
- Green, C. Thaumatin: A Natural Flavour Ingredient. In Low-Calories Sweeteners: Present and Future; KARGER: Basel, Switzerland, 1999; pp. 129–132. [Google Scholar]
- de Jesús-Pires, C.; Ferreira-Neto, J.R.C.; Pacifico Bezerra-Neto, J.; Kido, E.A.; de Oliveira Silva, R.L.; Pandolfi, V.; Wanderley-Nogueira, A.C.; Binneck, E.; da Costa, A.F.; Pio-Ribeiro, G.; et al. Plant Thaumatin-like Proteins: Function, Evolution and Biotechnological Applications. Curr. Protein Pept. Sci. 2020, 21, 36–51. [Google Scholar] [CrossRef]
- Lin, Y.; Qiu, Z.; Lin, X.; Wu, Y.; Niu, X.; Yin, G.; Shao, D.; Xiang, X.; Li, Y.; Yang, C. The Role of MbEGS1 and MbEGS2 in Methyleugenol Biosynthesis by Melaleuca Bracteata. Plants 2023, 12, 1026. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).