Powdery Mildew of Bigleaf Hydrangea: Biology, Control, and Breeding Strategies for Resistance
Abstract
1. Introduction
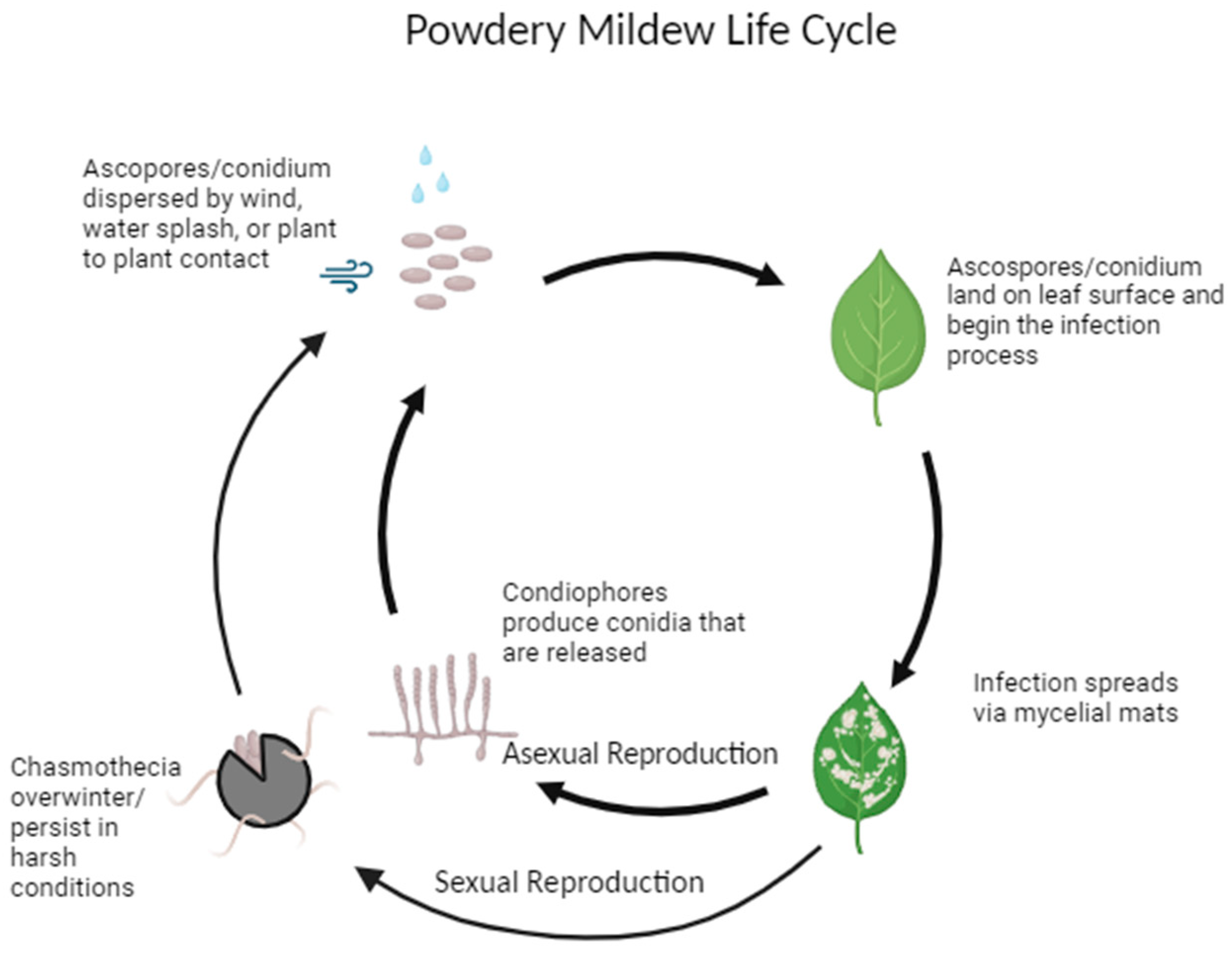
2. Powdery Mildew
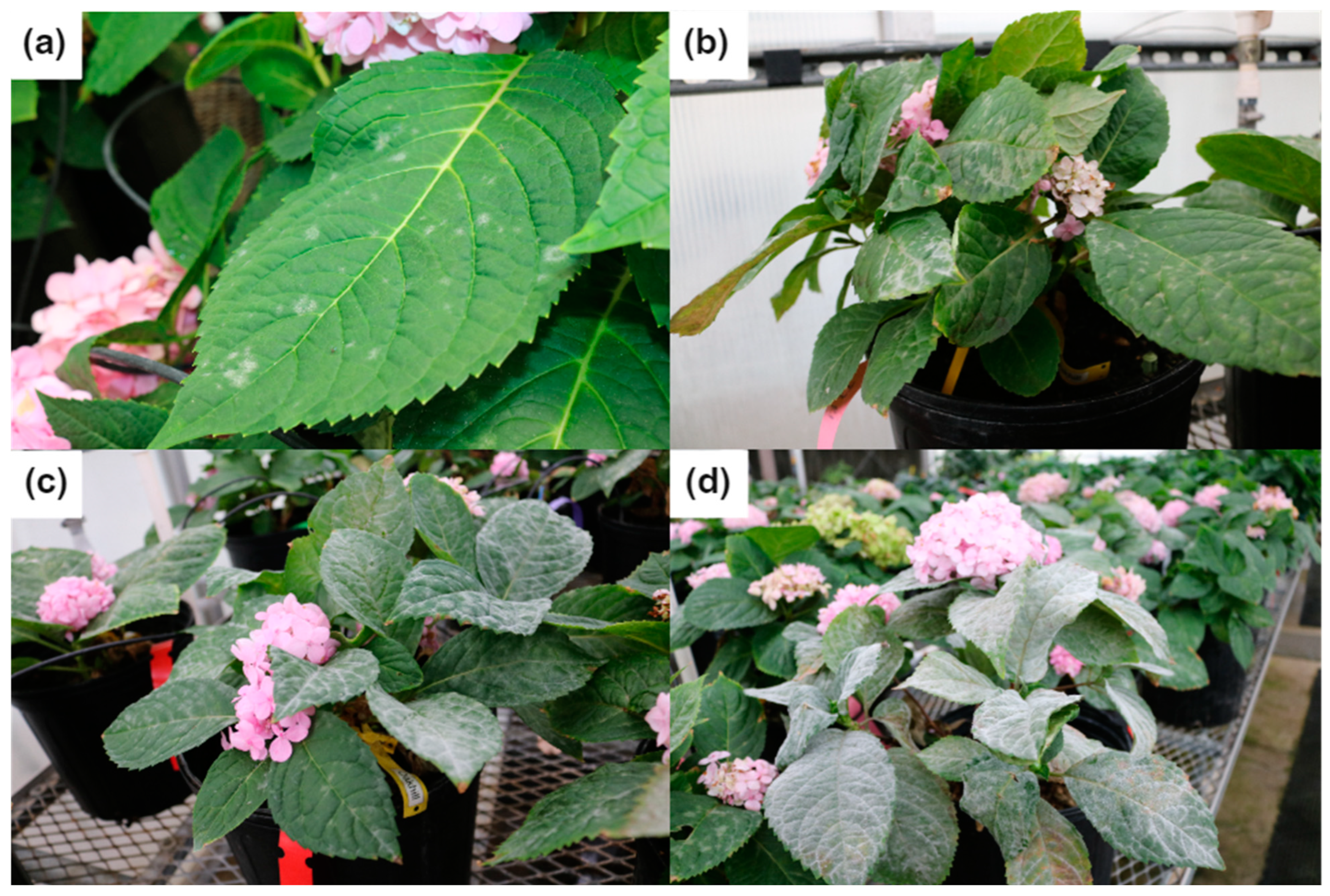
3. Powdery Mildew in Hydrangea
4. Control of Powdery Mildew
4.1. Biological Control
4.2. Chemical Control
4.3. Cultural Control
5. Molecular Mechanisms of Powdery Mildew Resistance
RNA-Seq to Find DEGs in Hydrangea for Future Biotech Use
6. Prospects for Breeding for Powdery Mildew Resistance in Hydrangea
6.1. Evaluating PM Resistance of Cultivars, Species, and Hybrids
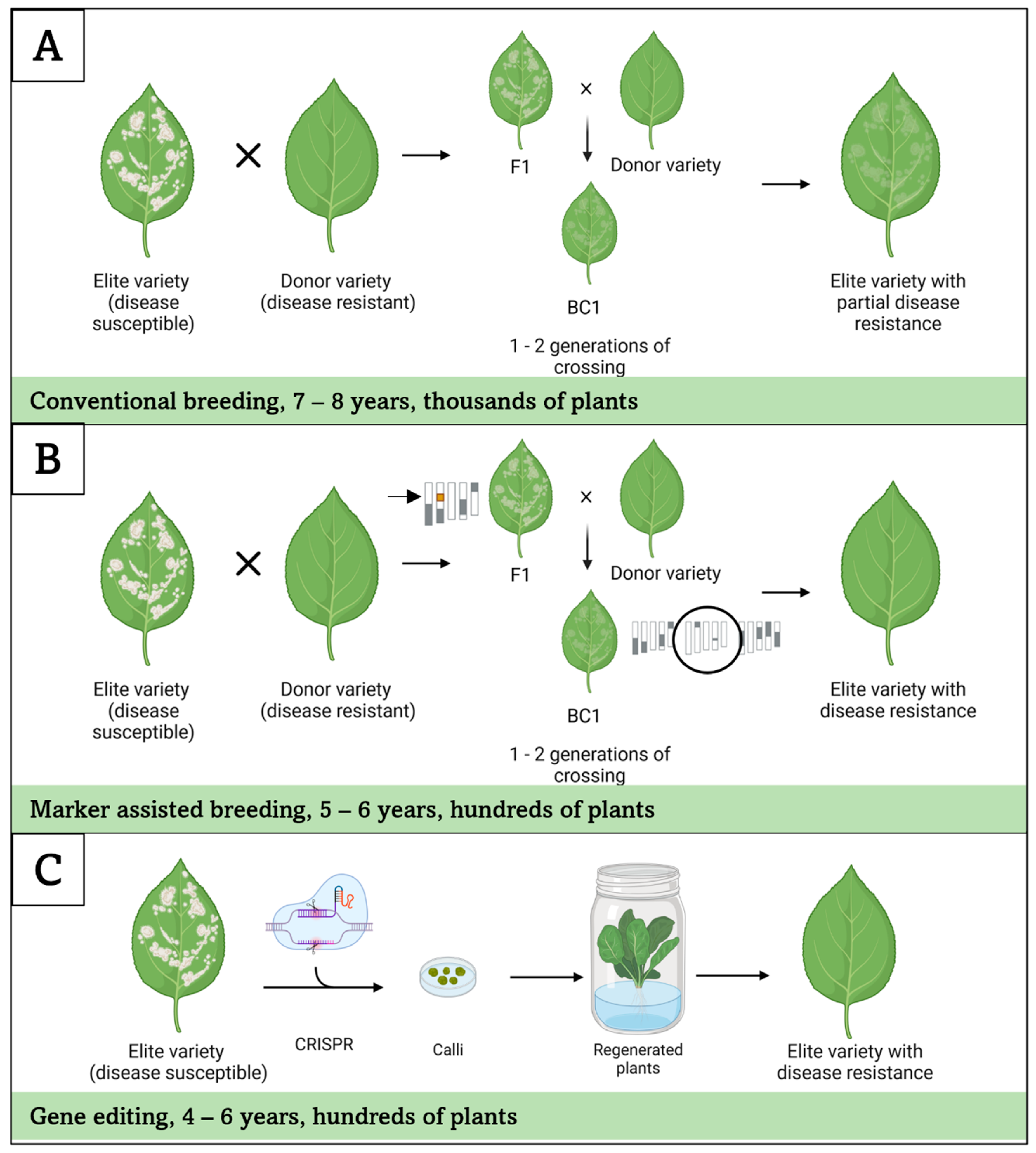
6.2. Breeding for Powdery Mildew Resistance
6.3. Engineering Powdery Mildew Resistance in Bigleaf Hydrangea
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- McClintock, E. A monograph of the genus Hydrangea. Proc. Calif. Acad. Sci. 1957, 29, 147–256. [Google Scholar]
- van Gelderen, C.J.; van Gelderen, D.M. Encyclopedia of Hydrangeas; Timber Press: Portland, OR, USA, 2004. [Google Scholar]
- Reed, S. Compatibility studies in Hydrangea. J. Environ. Hort. 2000, 18, 29–33. [Google Scholar] [CrossRef]
- Dirr, M. Maual of Woody Landscape Plants: Their Identification, Ornamental Characteristics, Culture, Propagation, and Uses, 6th ed.; Stipes Publishing, LLC: Champaign, IL, USA, 2009. [Google Scholar]
- Alexander, L. Production of triploid Hydrangea macrophylla via unreduced gamete breeding. HortScience 2017, 52, 221–224. [Google Scholar] [CrossRef]
- Sherwood, A.; Clark, M.D.; Hokanson, S.C.; Warrington, I. (Eds.) Oakleaf hydrangea (Hydrangea quercifolia Bartr.): Horticulture, Genetics, Breeding, and Conservation. In Horticultural Reviews; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2022; Volume 49, pp. 1–33. [Google Scholar] [CrossRef]
- NASS. 2019 Census of Horticultural Specialties. 2017 Census of Agriculture. 2020. Available online: https://www.nass.usda.gov/Publications/AgCensus/2017/Online_Resources/Census_of_Horticulture_Specialties/HORTIC.pdf (accessed on 30 November 2023).
- Hagan, A.K.; Oliver, J.W.; Stephenson, J.; Rivas-Davila, M.E. Impact of application rate and interval on the control of powdery mildew and cercospora leaf spot on bigleaf hydrangea with azoxystrobin. J. Environ. Hort. 2004, 22, 58–62. [Google Scholar] [CrossRef]
- Breen, P. Hydrangea macrophylla; Oregon State University: Corvallis, OR, USA, 2023; Available online: https://landscapeplants.oregonstate.edu/plants/hydrangea-macrophylla (accessed on 21 February 2023).
- Zhang, G.; Yuan, S.; Qi, H.; Chu, Z.; Liu, C. Identification of reliable reference genes for the expression of Hydrangea macrophylla ‘Bailmer’ and ‘Duro’ sepal color. Horticulturae 2022, 8, 835. [Google Scholar] [CrossRef]
- Reed, S. Pollination biology of Hydrangea macrophylla. HortScience 2005, 40, 335–338. [Google Scholar] [CrossRef]
- Uemachi, T.; Kato, Y.; Nishio, T. Comparison of decorative and non-decorative flowers in Hydrangea macrophylla (Thunb.) Ser. Sci. Hortic. 2004, 102, 325–334. [Google Scholar] [CrossRef]
- Uemachi, T.; Okumura, A. The inheritance of inflorescence types in Hydrangea macrophylla. J. Jpn. Soc. Hotic. Sci. 2012, 81, 263–268. [Google Scholar] [CrossRef]
- Windham, M.T.; Reed, S.; Mmbaga, M.; Windham, A.; Li, Y.; Rinehart, T.A. Evaluation of powdery mildew resistance in Hydrangea macrophylla. J. Environ. Hort. 2011, 29, 60–64. [Google Scholar] [CrossRef]
- Hacquard, S. The genomics of powdery mildew fungi: Past achievements, present status and future prospects. Adv. Bot. Res. 2014, 70, 109–142. [Google Scholar] [CrossRef]
- Liu, M.; Braun, U. Powdery mildews on crops and ornamentals in Canada: A summary of the phylogeny and taxonomy from 2000–2019. Can. J. Plant Pathol. 2021, 44, 191–218. [Google Scholar] [CrossRef]
- Talgø, V.; Sundheim, L.; Gjærum, H.B.; Herrero, M.L.; Suthaparan, A.; Toppe, B.; Stensvand, A. Powdery mildews on ornamental trees and shrubs in Norway. Eur. J. Pla. Sci. Biotech. 2011, 5, 86–92. [Google Scholar]
- Mekapogu, M.; Jung, J.-A.; Kwon, O.-K.; Ahn, M.-S.; Song, H.-Y.; Jang, S. Recent progress in enhancing fungal disease resistance in ornamental plants. Int. J. Mol. Sci. 2021, 22, 7956. [Google Scholar] [CrossRef]
- Glawe, D. The powdery mildew: A review of the world’s most familiar (yet poorly known) plant pathogens. Annu. Rev. Phytopathol. 2008, 46, 27–51. [Google Scholar] [CrossRef]
- Vielba-Fernández, A.; Polonio, Á.; Ruiz-Jiménez, L.; Vicente, A.; Pérez-García, A.; Fernández-Ortuño, D. Fungicide resistance in powdery mildew fungi. Microorganisms 2020, 8, 1431. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Diaz, D.; Yin, J.; Bloodgood, D.; Sexton, W.; Wei, C.; Xiao, S. An easy and flexible inoculation method for accurately assessing powdery mildew-infection phenotypes of Arabidopsis and other plants. JOVE 2021, 169, e62287. [Google Scholar] [CrossRef]
- Bradshaw, M. Epidemiology and Biology of Powdery Mildews and Their Host Plants. Doctoral Dissertation, University of Washington, Seattle, WA, USA, 2020. Available online: http://hdl.handle.net/1773/46015 (accessed on 9 February 2024).
- Sulima, A.S.; Zhukov, V.A. War and peas: Molecular bases of resistance to powdery mildew in pea (Pisum sativum L.) and other legumes. Plants 2022, 11, 339. [Google Scholar] [CrossRef] [PubMed]
- Gubler, W.D.; Koike, S.T. Powdery Mildew on Ornamentals. UC ANR Pest Notes Publication 7493. 2009. Available online: https://ipm.ucanr.edu/PMG/PESTNOTES/pn7493.html (accessed on 25 January 2024).
- Burgess, C.; Williamson, J. Powdery Mildew. Clemson Cooperative Extension. Home and Garden Information Center. 2021. Available online: https://hgic.clemson.edu/factsheet/powdery-mildew/ (accessed on 30 November 2023).
- Kang, Y.; Zhou, M.; Merry, A.; Barry, K. Mechanisms of powdery mildew resistance of wheat—A review of molecular breeding. Plant Pathol. 2020, 69, 601–617. [Google Scholar] [CrossRef]
- Ale-Agha, N.; Boyle, H.; Braun, U.; Butin, H.; Jage, H.; Kummer, V.; Shin, H.-D. Taxonomy, host range and distribution of some powdery mildew fungi (Erysiphales). Schlechtendalia 2008, 17, 39–54. [Google Scholar]
- Koike, S.T.; Tjosvold, S.A.; Mathews, D.M. Powdery Mildew. UC IPM Management Guidelines: Floriculture and Ornamental Nurseries UC ANR Publication 3392. 2020. Available online: https://ipm.ucanr.edu/agriculture/floriculture-and-ornamental-nurseries/powdery-mildew/ (accessed on 16 October 2023).
- Menzel, C.M. A review of powdery mildew in strawberries: The resistance of species, hybrids and cultivars to the pathogen is highly variable within and across studies with o standard method for assessing the disease. J. Hortic. Sci. Biotechnol. 2022, 97, 273–397. [Google Scholar] [CrossRef]
- Bradshaw, M.; Braun, U.; Pfister, D.H. Powdery mildews on Quercus: A worldwide distribution and rediscovered holotype provide insights into the spread of these ecologically important pathogens. Forest Pathol. 2022, 52, 1. [Google Scholar] [CrossRef]
- Bradshaw, M.; Goolsby, E.; Mason, C.; Tobin, P.C. Evolution of disease severity and susceptibility in the Asteraceae to the powdery mildew Golovinomyces latisporus: Major phylogenetic structure coupled with highly variable disease severity at fine scales. Plant Dis. 2021, 105, 268–275. [Google Scholar] [CrossRef]
- Li, Y.; Mmbaga, M.T.; Windham, A.S.; Windham, M.T.; Trigiano, R.N. Powdery mildew of dogwoods: Current status and future prospects. Plant Dis. 2009, 93, 11. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.W. Morphology and surface characteristics of the anamorphic stage of powdery mildew Erysiphe australiana on crape myrtle leaves. Micron 2021, 143, 103013. [Google Scholar] [CrossRef] [PubMed]
- Boddy, L. Pathogens of autotrophs. In The Fungi, 3rd ed.; Watkinson, S.C., Boddy, L., Money, N.P., Eds.; Academic Press: Cambridge, MA, USA, 2016; Chapter 8; pp. 245–292. [Google Scholar] [CrossRef]
- Hückelhoven, R.; Panstruga, R. Cell biology of the plant-powdery mildew interaction. Curr. Opin. Plant Biol. 2011, 14, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, W.R.; Gubler, W.G.; Grove, G.G. Epidemiology of powdery mildews in agricultural pathosystems. In The Powdery Mildews: A Comprehensive Treatise; Bélanger, R.R., Bushnell, W.R., Dik, A.J., Carver, T.L.W., Eds.; American Phytopathological Society: St. Paul, MN, USA, 2002; pp. 169–199. [Google Scholar]
- Washington State University. Hydrangea: Powdery Mildew. Washington State University CAHNRS and Extension. 2022. Available online: https://hortsense.cahnrs.wsu.edu/fact-sheet/hydrangea-powdery-mildew/#:~:text=The%20cultivar%20’Veitchii’%20is%20resistant,’Holstein’%20show%20intermediate%20susceptibility (accessed on 25 January 2024).
- Baysal-Gurel, F.; Kabir, N.; Blalock, A. Foliar Diseases of Hydrangea; Tennessee State University Extension: Nashville, TN, USA, 2016; Available online: https://www.tnstate.edu/extension/documents/Foliar%20diseases%20of%20Hydrangea%20FBG%20022916%20G1.pdf (accessed on 2 December 2023).
- Thurn, M.; Lamb, E.; Eshenaur, B. Disease and Insect Resistant Ornamental Plants; New York State Integrated Pest Management Program: Geneva, NY, USA, 2019; Available online: https://ecommons.cornell.edu/server/api/core/bitstreams/a3bdfe68-8a30-4aeb-a871-931d597e25cb/content (accessed on 26 January 2024).
- Jennings, C.; Simmons, T.; Parajuli, M.; Oksel, C.; Liyanapathiranage, P.; Hikkaduwa Epa Liyanage, K.; Baysal-Gurel, F. Chemical control of powdery mildew of bigleaf hydrangea. HortScience 2024, 59, 173–178. [Google Scholar] [CrossRef]
- Lebeda, A.; Mieslerová, B. Taxonomy, distribution and biology of lettuce powdery mildew (Golovinomyces cichoracearum sensu stricto). Plant Pathol. 2010, 60, 400–415. [Google Scholar] [CrossRef]
- Li, Y.; Windham, M.T.; Trigiano, R.N.; Reed, S.M.; Spiers, J.M.; Rinehart, T.A. Bright-field and fluorescence microscopic study of development of Erysiphe polygoni in susceptible and resistant bigleaf hydrangea. Plant Dis. 2009, 93, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.; Shang, W.; Bai, L.; Ya, N.; Duo, J.; Ma, Y. First report of Golovinomyces orontii causing Hydrangea macrophylla powdery mildew in Qinghai-Tibet Plateau, China. Dis. Note 2021, 105, 2730. [Google Scholar] [CrossRef] [PubMed]
- Braun, U.; Shin, H.D.; Takamatsu, S.; Meeboon, J.; Kiss, L.; Lebeda, A.; Kitner, M.; Götz, M. Phylogeny and taxonomy of Golovinomyces orontii revisited. Mycol. Prog. 2019, 18, 335–537. [Google Scholar] [CrossRef]
- Appiano, M.; Pavan, S.; Catalano, D.; Zheng, Z.; Bracuto, V.; Lotti, C.; Visser, R.G.F.; Ricciardi, L.; Bai, Y. Identification of candidate MLO powdery mildew susceptibility genes in cultivated Solanaceae and functional characterization of tobacco NtMLO1. Transgenic Res. 2015, 24, 847–858. [Google Scholar] [CrossRef]
- Jennings, C.; Alexander, L.W.; Baysal-Gurel, F. Comparison of powdery mildew susceptibility in Hydrangea cultivars and hybrids. In Abstracts of Presentations from American Society of Horticultural Science 2023 Annual Conference, Proceedings of the American Society of Horticultural Science 2023 Annual Conference, Orlando, FL, USA, 31 July–4 August 2023; HortScience; American Society for Horticultural Science: Alexandria, VA, USA, 2023; Volume 58, (Suppl. S3). [Google Scholar] [CrossRef]
- He, D.C.; He, M.-H.; Amalin, D.M.; Liu, W.; Alvindia, D.G.; Zhan, J. Biological control of plant diseases: An evolutionary and eco-economic consideration. Pathogens 2021, 10, 1311. [Google Scholar] [CrossRef]
- Hegazi, M.A.; El-Kot, G.A. Biological control of powdery mildew on zinnia (Zinnia elegans, L.) using some biocontrol agents and plant extracts. J. Agric. Sci. 2010, 2, 221–230. [Google Scholar] [CrossRef]
- Bika, R.; Baysal-Gurel, F.; Jennings, C. Botrytis cinerea management in ornamental production: A continuous battle. Can. J. Plant Pathol. 2020, 43, 345–365. [Google Scholar] [CrossRef]
- Bika, R.; Palmer, C.; Alexander, L.; Baysal-Gurel, F. Comparative performance of reduced-risk fungicides and biorational products in management of postharvest botrytis blight on bigleaf hydrangea cut flowers. HortTechnology 2020, 30, 659–669. [Google Scholar] [CrossRef]
- Sawant, I.; Wadkar, P.; Ghule, S.; Rajguru, Y.; Salunkhe, V.; Sawant, S. Enhanced biological control of powdery mildew in vineyards by integrating a strain of Trichoderma afroharzianum with sulphur. Biol. Control 2017, 114, 133–143. [Google Scholar] [CrossRef]
- English-Loeb, G.; Norton, A.P. Biological control of grape powdery mildew using mycophagous mites. Plant Dis. 2006, 91, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Pandit, M.A.; Kumar, J.; Gulati, S.; Bhandari, N.; Mehta, P.; Katyal, R.; Rawat, C.D.; Mishra, V.; Kaur, J. Major biological control strategies for plant pathogens. Pathogens 2022, 11, 273. [Google Scholar] [CrossRef] [PubMed]
- Matocha, M.A. Pesticide Mode of Action and Resistance Management. AgriLife Extension. 2010. Available online: https://www-aes.tamu.edu/files/2010/10/Pesticide-MOA3.pdf (accessed on 20 December 2023).
- Burrows, M.; Rupp, J. Fungicide Use in Field Crops: Benefits and Risks. Montana IPM Bulletin. 2017. Available online: https://www.montana.edu/extension/plantpath/resources/fungicide_use_in_field_crops.html#:~:text=Fungicides%20work%2C%20in%20general%2C%20by,of%20action%E2%80%9D%20(MOA) (accessed on 20 December 2023).
- Hagan, A.K.; Olive, J.W.; Stephenson, J.; Rivas-Davila, M.E. Control of Powdery Mildew and Cercospora Leaf Spot on Bigleaf Hydrangea with Heritage and Milstop Fungicides; Bulletin 658; The Alabama Agricultural Experiment Station: Auburn, AL, USA, 2005; Available online: https://aurora.auburn.edu/bitstream/handle/11200/4098/BULL0658.pdf?sequence=1 (accessed on 27 January 2024).
- Newman, S.; Pottorff, L.P. Powdery Mildews; Fact Sheet No. 2.902; Colorado State University Extension: Fort Collins, CO, USA, 2013; Available online: https://extension.colostate.edu/topic-areas/yard-garden/powdery-mildews-2-902/ (accessed on 26 January 2024).
- Harmon, P.F.; Bledsoe, S.D. Professional Disease Management Guide for Ornamental Plants; #PP-202; University of Florida IFAS Extension Publication: Gainesville, FL, USA, 2019; Available online: https://edis.ifas.ufl.edu/publication/PP123 (accessed on 13 December 2023).
- Fondevilla, R. Powdery mildew control in pea. A review. Agron. Sustain. Dev. 2012, 32, 401–409. [Google Scholar] [CrossRef]
- NC State. Cultural Control. Available online: https://genent.cals.ncsu.edu/bug-bytes/tactics/cultural/ (accessed on 25 January 2024).
- NC State Extension. Ornamentals. Available online: https://plantpathology.ces.ncsu.edu/pp-ornamentals/ (accessed on 27 January 2024).
- Hill, S.B. Cultural Methods of Pest, Primarily Insect, Control; Publication 58; Ecological Agriculture Projects; McGill University (Macdonald Campus): Sainte-Anne-de-Bellevue, QC, Canada, 1989; Available online: https://www.eap.mcgill.ca/publications/eap58.htm (accessed on 25 January 2024).
- Li, Y.; Windham, M.; Trigiano, R.; Reed, S.; Rinehart, T.; Spiers, J. Assessment of resistance components of bigleaf hydrangeas (Hydrangea macrophylla) to Erysiphe polygoni in vitro. Can. J. Plant Path. 2009, 31, 348–355. [Google Scholar] [CrossRef]
- Berg, J.A.; Appiano, M.; Martínez, M.S.; Hermans, F.W.K.; Vrizen, W.H.; Visser, R.G.F.; Bai, Y.; Schouten, H.J. A transposable element insertion in the susceptibility gene CsaMLO8 results in hypocotyl resistance to powdery mildew in cucumber. BMC Plant Biol. 2015, 15, 243. [Google Scholar] [CrossRef]
- Xiang, G.; Zhang, H.; Jian, H.; Yan, H.; Wang, Q.; Zhou, N.; Li, S.; Tang, K.; Qiu, X. De novo assembly and characterization of the transcriptome of susceptible and resistant rose species in response to powdery mildew. Sci. Hortic. 2019, 257, 108653. [Google Scholar] [CrossRef]
- Kusch, S.; Panstruga, R. mlo-Based resistance: An apparently universal “weapon” to defeat powdery mildew disease. MPMI 2017, 30, 179–189. [Google Scholar] [CrossRef]
- Uemachi, T.; Mizuhara, Y.; Deguchi, K.; Shinjo, Y.; Kajino, E.; Ohba, H. Phylogenetic relationship of Hydrangea macrophyla (Thunb.) Ser. and H. serrata (Thunb.) Ser. evaluated using RAPD markers and plastid DNA sequences. J. Japan. Soc. Hort. Sci. 2014, 83, 163–171. [Google Scholar] [CrossRef]
- Chandran, N.K.; Sriram, S.; Prakash, T.; Budhwar, R. Transcriptome changes in resistant and susceptible rose in response to powdery mildew. J. Phytopathol. 2021, 169, 556–569. [Google Scholar] [CrossRef]
- Gill, U.S.; Lee, S.; Mysore, K.S. Host versus nonhost resistance: Distinct wars with similar arsenals. Phytopathology 2015, 105, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Lipka, U.; Fuchs, R.; Lipka, V. Arabidopsis non-host resistance to powdery mildews. Curr. Opin. Plant Biol. 2008, 11, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Martínez, M.I.S.; Bracuto, V.; Koseoglou, E.; Appiano, M.; Jacobsen, E.; Visser, R.G.F.; Wolters, A.A.; Bai, Y. CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol. 2020, 20, 284. [Google Scholar] [CrossRef]
- Rodriquez-Esteban, R.; Jiang, X. Differential gene expression in disease: A comparison between high-throughput studies and the literature. BMC Med. Genom. 2017, 10, 59. [Google Scholar] [CrossRef] [PubMed]
- de Sá, P.H.C.G.; Guimarães, L.C.; das Graças, D.A.; de Oliveira Veras, A.A.; Barh, D.; Azevedo, V.; da Costa da Silva, A.L.; Ramos, R.T.J. Next-generation sequencing and data analysis: Strategies, tools, pipelines and protocols. In Omics Technologies and Bio-Engineering; Barh, D., Azevedo, V., Eds.; Academic Press: Cambridge MA, USA, 2018; pp. 191–207. [Google Scholar]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, R. RNA-Seq: Basics, Applications and Protocol; Technology Networks: Sudbury, UK, 2018; Available online: https://www.technologynetworks.com/genomics/articles/rna-seq-basics-applications-and-protocol-299461 (accessed on 18 December 2023).
- Fang, Z.; Martin, J.; Wang, Z. Statistical methods for identifying differentially expressed genes in RNA-Seq experiments. Cell Biosci. 2012, 2, 26. [Google Scholar] [CrossRef] [PubMed]
- Anjum, A.; Jaggi, S.; Varghese, E.; Lall, S.; Bhowmik, A.; Rai, A. Identification of differentially expressed genes in RNA-seq data of Arabidopsis thaliana: A compound distribution approach. J. Comput. Biol. 2016, 23, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Adam, L.; Ellwood, S.; Wilson, I.; Saenz, G.; Xiao, S.; Oliver, R.P.; Turner, J.G.; Somerville, S. Comparison of Erysiphe cichioracearum and E. cruciferarum and a survey of 360 Arabidopsis thaliana accessions for resistance to these two powdery mildew pathogens. MPMI 1999, 12, 1031–1043. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Qiu, X.; Wang, Q.; Zhang, H.; Jian, H.; Zhou, N.; Ji, C.; Yan, H.; Bao, M.; Tang, K. Antisense RhMLO1 gene transformation enhances resistance to the powdery mildew pathogen in Rosa multiflora. Plant Mol. Biol. Rep. 2015, 33, 1659–1665. [Google Scholar] [CrossRef]
- Pessina, S.; Angeli, D.; Martens, S.; Visser, R.G.F.; Bai, Y.; Salamini, F.; Velasco, R.; Schouten, H.J.; Malnoy, M. The knock-down of the expression of MdMLO19 reduces susceptbility to powdery mildew (Podosphaera leucotricha) in apple (Malus domestica). Plant Biotechnol. J. 2016, 14, 2033–2044. [Google Scholar] [CrossRef]
- Huibers, R.P.; Loonen, A.E.H.M.; Gao, D.; Van den Ackerveken, G.; Visser, R.G.F.; Bai, Y. Powdery mildew resistance in tomato by impairment of SIPMR4 and SIDMR1. PLoS ONE 2013, 8, e67467. [Google Scholar] [CrossRef]
- Consonni, C.; Humphry, M.E.; Hartmann, H.A.; Livaja, M.; Durner, J.; Westphal, L.; Vogel, J.; Lipka, V.; Kemmerling, B.; Schulze-Lefert, P.; et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 2006, 38, 716–720. [Google Scholar] [CrossRef]
- Adam, L.; Somerville, S.C. Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana. Plant J. 1996, 9, 341–356. [Google Scholar] [CrossRef]
- Yang, I.S.; Kim, S. Analysis of whole transcriptome sequencing data: Workflow and software. Genom. Inform. 2015, 13, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wan, H.; Cheng, T.; Wang, J.; Yang, W.; Pan, H.; Zhang, Q. Comparative RNA-seq analysis of transcriptome dynamics during petal development in Rosa chinensis. Sci. Rep. 2017, 7, 43882. [Google Scholar] [CrossRef] [PubMed]
- NC Cooperative Extension. Hydrangea febrifuga. North Carolina Extension Gardener Plant Toolbox. Available online: https://plants.ces.ncsu.edu/plants/hydrangea-febrifuga/ (accessed on 9 December 2023).
- Farinas, C.; Jourdan, P.; Paul, P.A.; Hand, F.P. Development and evaluation of laboratory bioassays to study powdery mildew pathogens of Phlox in vitro. Plant Disease 2019, 103, 1536–1543. [Google Scholar] [CrossRef]
- Yan, Z.; Dolstra, O.; Prins, T.; Stam, P.; Visser, P. Assessment of partial resistance to powdery mildew (Podosphaera pannosa) in a tetraploid rose population using a spore-suspension inoculation method. European J. Plant Pathol. 2006, 114, 301–308. [Google Scholar] [CrossRef]
- Reed, S.M.; Riedel, G.L.; Pooler, M.R. Verification and establishment of Hydrangea macrophylla ‘Kardinal’ x H. paniculata ‘Brussels Lace’ interspecific hybrids. J. Environ. Hort. 2001, 19, 85–88. [Google Scholar] [CrossRef]
- Reed, S.M. Self-incompatibility and time of stigma receptivity in two species of Hydrangea. HortScience 2004, 39, 312–315. [Google Scholar] [CrossRef]
- Reed, S.M.; Jones, K.D.; Rinehart, A. Production and characterization of intergeneric hybrids between Dichroa febrifuga and Hydrangea macrophylla. J. Am. Soc. Hort. Sci. 2008, 133, 84–91. [Google Scholar] [CrossRef]
- Rinehart, T.A.; Scheffler, B.E.; Reed, S.M. Ploidy variation and genetic diversity in Dichroa. HortScience 2010, 45, 208–213. [Google Scholar] [CrossRef]
- Mendoza, C.; Wanke, S.; Goetghebeur, P.; Samain, M.-S. Facilitating wide hybridization in Hydrangea s. l. cultivars: A phylogenetic and marker-assisted breeding approach. Mol. Breed. 2013, 32, 233–239. [Google Scholar] [CrossRef]
- Cai, M.; Wang, K.; Luo, L.; Pan, H.-t.; Zhang, Q.-x.; Yang, Y.-y. Production of interspecific hybrids between Hydrangea macrophylla and Hydrangea arborescens via ovary culture. HortScience 2015, 50, 1765–1769. [Google Scholar] [CrossRef]
- Nesi, B.; Lazzereschi, S.; Pecchioli, S.; Pacifici, S.; Venturieri, G.A.; Burchi, G. Development of complementary techniques applied to Hydrangea hybridizations. Acta Hortic. 2015, 1104, 341–348. [Google Scholar] [CrossRef]
- Ramalho, M.A.P.; Santos, H.G.; Ferreira, G.C.; Gonçalves, F.M.A. Viability of the use of inbred progenies in recurrent selection in perennial plants. Crop Breed. Appl. Biotechnol. 2023, 23, e469523415. [Google Scholar] [CrossRef]
- Wu, X.; Hulse-Kemp, A.M.; Wadl, P.A.; Smith, Z.; Mockaitis, K.; Staton, M.E.; Rinehart, T.A.; Alexander, L.W. Genomic resource development for hydrangea (Hydrangea macrophylla (Thunb. Ser.)—A transcriptome assembly and a high-density genetic linkage map. Horticulturae 2021, 7, 25. [Google Scholar] [CrossRef]
- Wu, X.; Alexander, L. Genome-wide association studies for inflorescence type and remontancy in Hydrangea macrophylla. Hortic. Res. 2020, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Simpson, S.A.; Youngblood, R.C.; Liu, X.F.; Scheffler, B.E.; Rinehart, T.; Alexander, L.; Hulse-Kemp, A.M. Two haplotype-resolved genomes reveal important flower traits in bigleaf hydrangea (Hydrangea macrophylla) and insights into Asterid Evolution. Hortic. Res. 2023, 10, uhad217. [Google Scholar] [CrossRef]
- Tränkner, C.; Krüger, J.; Wanke, S.; Naumann, J.; Wenke, T.; Engel, F. Rapid identification of inflorescence type markers by genotyping-by-sequencing of diploid and triploid F1 plants of Hydrangea macrophylla. BMC Genetics 2019, 20, 60. [Google Scholar] [CrossRef]
- Waki, T.; Kodama, M.; Akutsu, M.; Namai, K.; Iigo, M.; Kurokura, T.; Yamamoto, T.; Nashima, K.; Nakayama, M.; Yagi, M. Development of DNA markers linked to double-flower and hortensia traits in Hydrangea macrophyla (Thunb.) Ser. Hortic. J. 2018, 87, 264–273. [Google Scholar] [CrossRef]
- Nashima, K.; Shirasawa, K.; Ghelfi, A.; Hirakawa, H.; Isobe, S.; Suyama, T.; Wada, T.; Kurokura, T.; Uemachi, T.; Azuma, M.; et al. Genome sequence of Hydrangea macrophylla and its application in analysis of the double flower phenotype. DNA Res. 2020, 28, dsaa026. [Google Scholar] [CrossRef]
- Rinehart, T.A.; Scheffler, B.E.; Reed, S.M. Genetic diversity estimates for the genus Hydrangea and development of a molecular key based on SSR. J. Americ. Soc. Hortic. Sci. 2006, 131, 787–797. [Google Scholar] [CrossRef]
- Reed, S.M.; Rinehart, T.A. Simple sequence repeat marker analysis of genetic relationships within Hydrangea macrophylla. J. Americ. Soc. Hortic. Sci. 2007, 132, 341–351. [Google Scholar] [CrossRef]
- Wu, X.; Alexander, L.W. Genetic diversity and population structure analysis of bigleaf hydrangea using genotyping-by-sequencing. J. AMeric. Soc. Hortic. Sci. 2019, 144, 257–263. [Google Scholar] [CrossRef]
- Negishi, T.; Oshima, K.; Hattori, M.; Kanai, M.; Mano, S.; Nishimura, M.; Yoshida, K. Tonoplast- and plasma membrane-localized aquaporin-family transporters in blue hydrangea sepals of aluminum hyperaccumuating plant. PLoS ONE 2012, 7, e43189. [Google Scholar] [CrossRef]
- Chen, H.; Lu, C.; Jiang, H.; Peng, J. Global transcriptome analysis reveals distinct aluminnum-tolerance pathways in the Al-accumulating species Hydrangea macrophylla and marker identification. PLoS ONE 2015, 10, e0144927. [Google Scholar] [CrossRef]
- Rahmati, R.; Hamid, R.; Ghorbanzadeh, Z.; Jacob, F.; Azadi, P.; Zeinalabedini, M.; Farsad, L.K.; Kazemi, M.; Ebrahimi, M.A.; Shahinnia, F.; et al. Comparative transcriptome analysis unveils the molecular mechanism underlying sepal colour changes under acidic pH substratum in Hydrangea macrophylla. Int. J. Mol. Sci. 2022, 23, 15428. [Google Scholar] [CrossRef]
- Wang, Q.; Lyu, T.; Lyu, Y. Exploring the molecular mechanism of sepal formation in the decorative flowers of Hydrangea macrophylla ‘Endless Summer’ based on the ABCDE model. Int. J. Mol. Sci. 2022, 23, 14112. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, W.; Hu, D.; Shen, P.; Zhang, G.; Zhu, Y. Comparative analsysi of the metabolome and transcriptome between green and albino zones of variegated leaves from Hydrangea macrophylla ’Maculata’ infected by hydrangea ringspot virus. Plant Physiol. Biochem. 2020, 157, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Chen, S.; Wang, H.; Feng, J.; Chen, H.; Qin, Z.; Deng, Y. Comparative physiology and transcriptome analysis reveals that chloroplast development influences silver-white leaf color formation in Hydrangea macrophylla var. maculata. BMC Plant Biol. 2022, 22, 345. [Google Scholar] [CrossRef]
- Chen, S.; Qi, X.; Feng, J.; Chen, H.; Qin, Z.; Wang, H.; Deng, Y. Biochemistry and transcriptome analyses reveal key genes and pathways involved in high-aluminum stress ressponse and tolerance in hydrangea sepals. Plant Physiol. Biochem. 2022, 185, 268–278. [Google Scholar] [CrossRef]
- Todkar, L.; Krishna, H.; Singh, G.P.; Jain, N.; Singh, P.K.; Prabhu, K. Introgression of drought tolerance QTLs through marker assisted backcross breeding in wheat (Triticum aestivum L.). Indian J. Genet. Plant Breed. 2020, 80, 209–212. [Google Scholar] [CrossRef]
- Lopez Arias, D.C.; Chastellier, A.; Thouroude, T.; Bradeen, J.; Van Eck, L.; De Oliveira, Y.; Paillard, S.; Foucher, F.; Hibrand-Saint Oyant, L.; Soufflet-Freslon, V. Characterization of black spot resistance in diploid roses with QTL detection, meta-analysis and candidate-gene identification. Theor. Appl. Genet. 2020, 133, 3299–3321. [Google Scholar] [CrossRef]
- Zurn, J.D.; Zlesak, D.C.; Holen, M.; Bradeen, J.M.; Hokanson, S.C.; Bassil, N.V. Mapping the black spot resistance locus Rdr3 in the shrub rose ‘George Vancouver’ allows for the development of improved diagnostic markers for DNA-informed breeding. Theor. Appl. Genet. 2020, 133, 2011–2022. [Google Scholar] [CrossRef]
- Giovannini, A.; Laura, M.; Nesi, B.; Savona, M.; Cardi, T. Genes and genome editing tools for breeding desirable phenotypes in ornamentals. Plant Cell Rep. 2021, 40, 461–478. [Google Scholar] [CrossRef]
- Alexander, L. Ploidy level influences pollen tube growth and seed viability in interploidy crosses of Hydrangea macrophylla. Front. Plant Sci. 2020, 11, 100. [Google Scholar] [CrossRef]
- Doudna, J.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 6213. [Google Scholar] [CrossRef]
- Kishi-Kaboshi, M.; Aida, R.; Sasaki, K. Genome engineering in ornamental plants: Current status and future prospects. Plant Physiol. Biochem. 2018, 131, 47–52. [Google Scholar] [CrossRef]
- Knott, G.J.; Douda, J.A. CRISPR-Cas guides the future of genetic engineering. Science 2018, 361, 866–869. [Google Scholar] [CrossRef]
- Liu, H.; Wang, Z.; Liu, Y.; Wei, Y.; Hu, Z.; Wu, X.; Zheng, C.; Wang, C. Applications of CRISPR/Cas9 technology in ornamental plants. Plant Mol. Biol. Rep. 2023. [Google Scholar] [CrossRef]
- Wang, C.; Li, Y.; Wang, N.; Yu, Q.; Li, Y.; Gao, J.; Zhou, X.; Ma, N. An efficient CRISPR/Cas9 platform for targeted genome editing in rose (Rosa hybrida). J. Integr. Plant Biol. 2022, 65, 895–899. [Google Scholar] [CrossRef]
- Boccan-Gibod, J.; Billard, C.; Maltete, S. In vitro regeneration system of Hydrangea macrophylla plantlets from leaves and internodes. Acta Hortic. 2000, 508, 229–232. [Google Scholar] [CrossRef]
- Ledbetter, D.I.; Preece, J.E. Thidiazuron stimulates adventitious shoot production from Hydrangea quercifolia Batr. Leaf explants. Sci. Hortic. 2004, 101, 121–126. [Google Scholar] [CrossRef]
- Doil, A.; Zhang, R.; Schum, A.; Serek, M.; Winkelmann, T. In vitro regeneration and propagation of Hydrangea macrophylla Thunb. ‘Nachtigall’. Propag. Ornom. Plants 2008, 8, 151–153. [Google Scholar]
- Liu, F.; Huang, L.-L.; Li, Y.-L.; Reinhoud, P.; Jongsma, M.A.; Wang, C.-Y. Shoot organogenesis in leaf explants of Hydrangea macrophylla ‘Hyd1’ and assessing genetic stability of regenerants using ISSR markers. Plant Cell Tissue Organ. Cult. 2011, 104, 111–117. [Google Scholar] [CrossRef]
- Maher, M.F.; Nasti, R.A.; Vollbrecht, M.; Starker, C.G.; Clark, M.D.; Voytas, D.F. Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 2020, 38, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, S.; Ma, H.; Tao, Y.; Feng, S.; Gong, S.; Zhang, J.; Zhou, A. Reliable and efficient Agrobacterium tumefaciens-mediated genetic transformation of Dianthus spiculifolius. Hortic. Plant J. 2020, 6, 199–204. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jennings, C.; Baysal-Gurel, F.; Alexander, L.W. Powdery Mildew of Bigleaf Hydrangea: Biology, Control, and Breeding Strategies for Resistance. Horticulturae 2024, 10, 216. https://doi.org/10.3390/horticulturae10030216
Jennings C, Baysal-Gurel F, Alexander LW. Powdery Mildew of Bigleaf Hydrangea: Biology, Control, and Breeding Strategies for Resistance. Horticulturae. 2024; 10(3):216. https://doi.org/10.3390/horticulturae10030216
Chicago/Turabian StyleJennings, Christina, Fulya Baysal-Gurel, and Lisa W. Alexander. 2024. "Powdery Mildew of Bigleaf Hydrangea: Biology, Control, and Breeding Strategies for Resistance" Horticulturae 10, no. 3: 216. https://doi.org/10.3390/horticulturae10030216
APA StyleJennings, C., Baysal-Gurel, F., & Alexander, L. W. (2024). Powdery Mildew of Bigleaf Hydrangea: Biology, Control, and Breeding Strategies for Resistance. Horticulturae, 10(3), 216. https://doi.org/10.3390/horticulturae10030216




