Transcriptome Wide Identification and Expression Analysis Revealed BhTALE Gene Family Regulates Wax Gourd (Benincasa hispida) Response to Low Calcium and Magnesium Stress
Abstract
1. Introduction
2. Material and Methods
2.1. Identification of TALE Genes in Wax Gourd
2.2. Phylogenetic Tree, Conserved Motifs, Gene Structure, and Cis-Acting Elements Analysis
2.3. Chromosomal Localization and Protein Interaction
2.4. Plant Material and Nutrient Treatments
2.5. Quantitative RT-PCR (qRT-PCR)
2.6. Statistical Analysis
3. Results
3.1. Identification of TALE Genes in Wax Gourd Genome
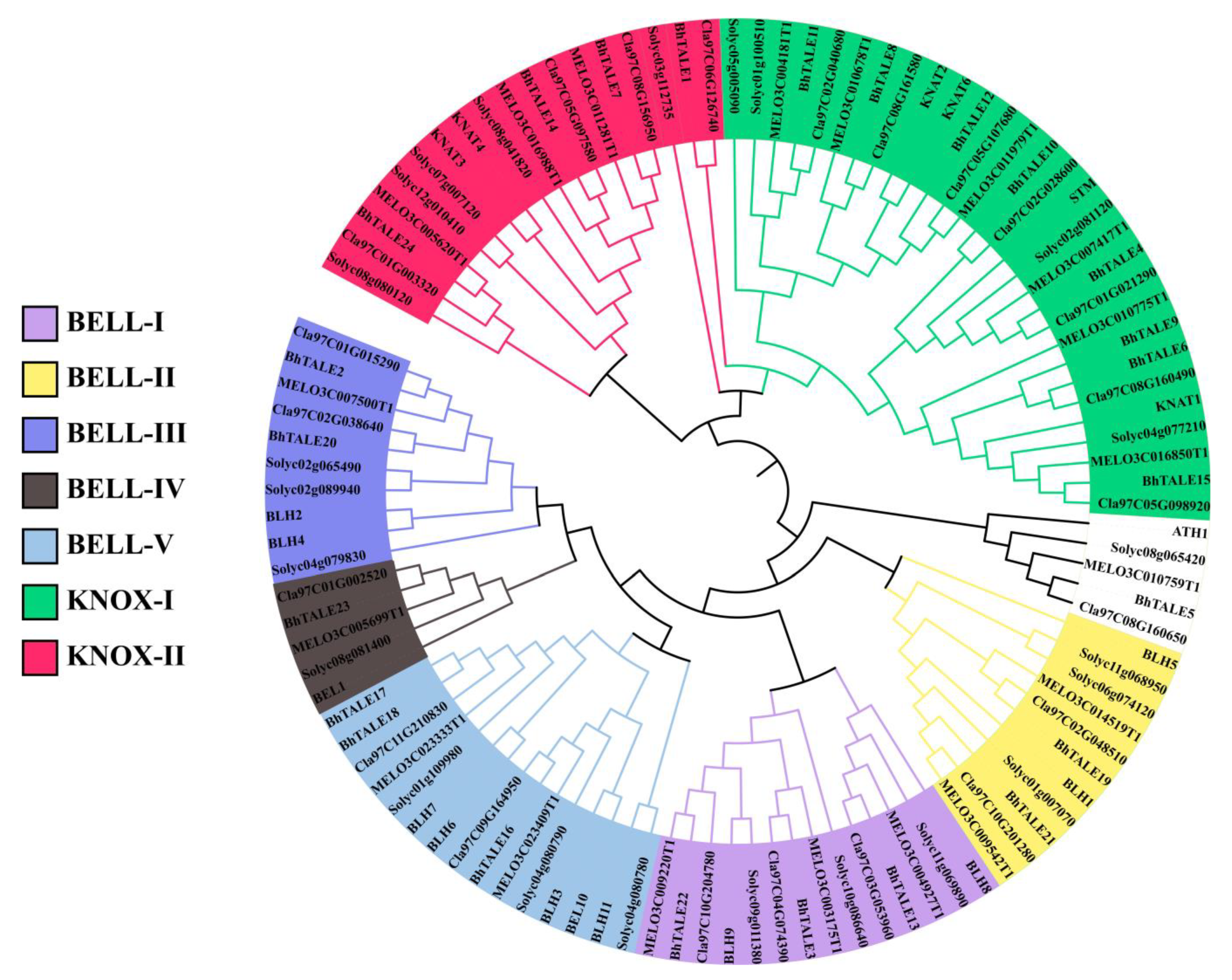
3.2. Evolutionary Analysis of BhTALE Genes
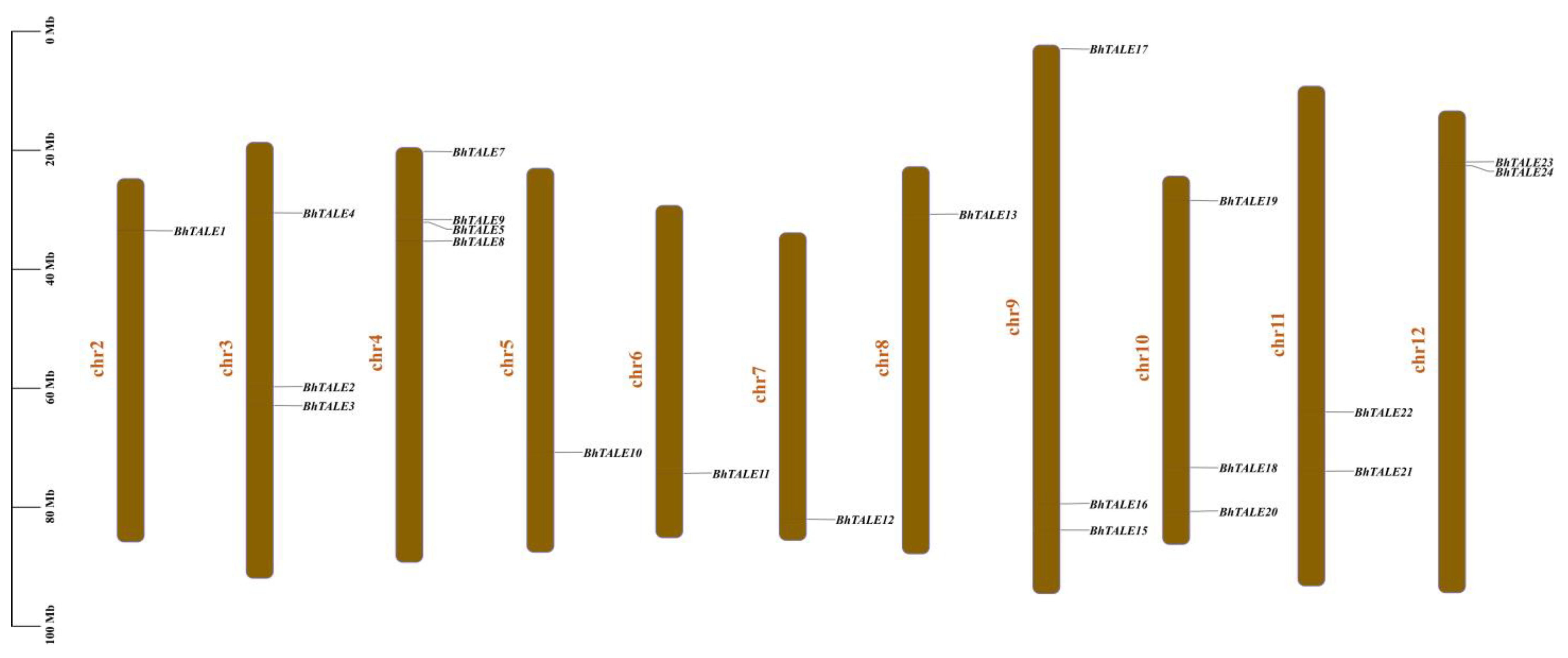
3.3. Chromosomal Localization of BhTALE Genes
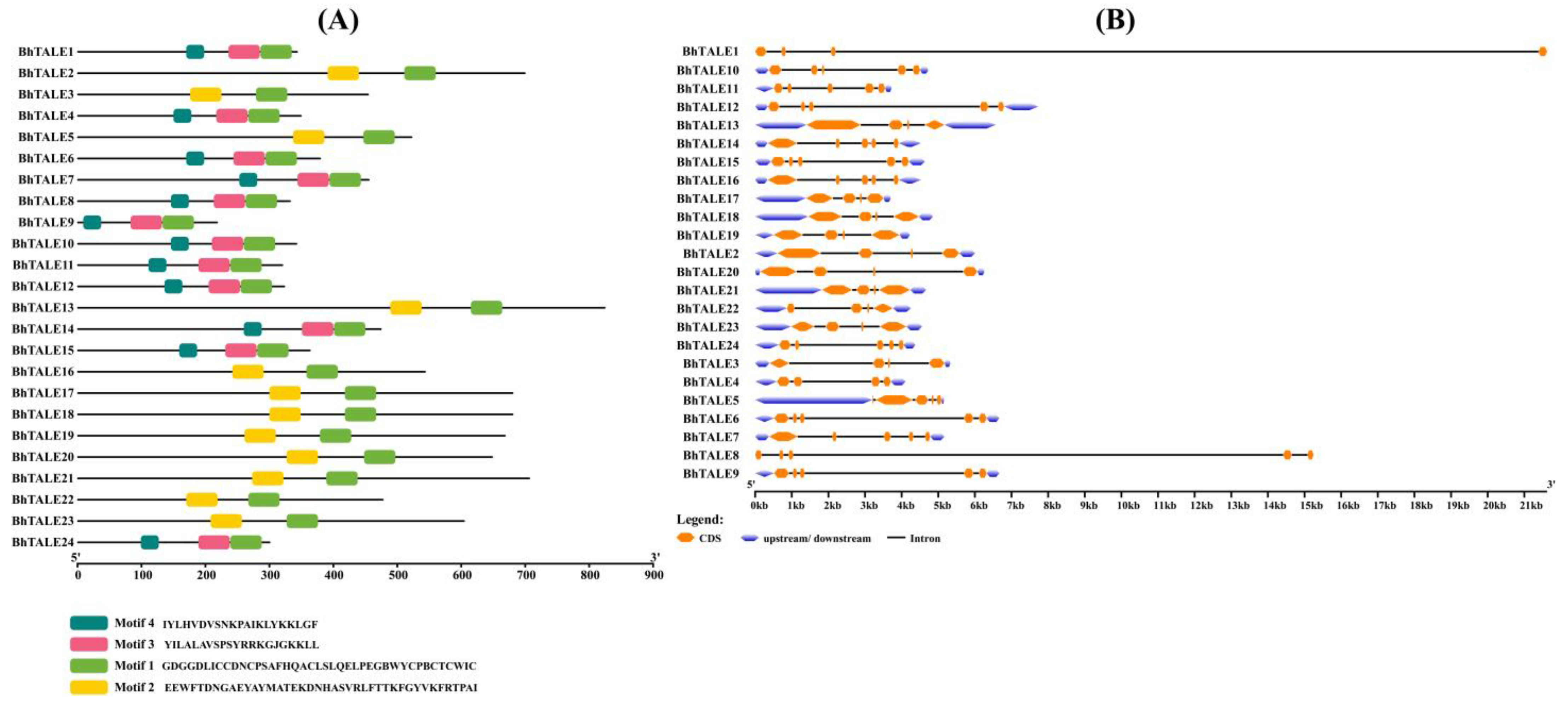
3.4. Gene Structure and Motif Analysis
3.5. Gene Ontology Analysis of BhTALE Genes
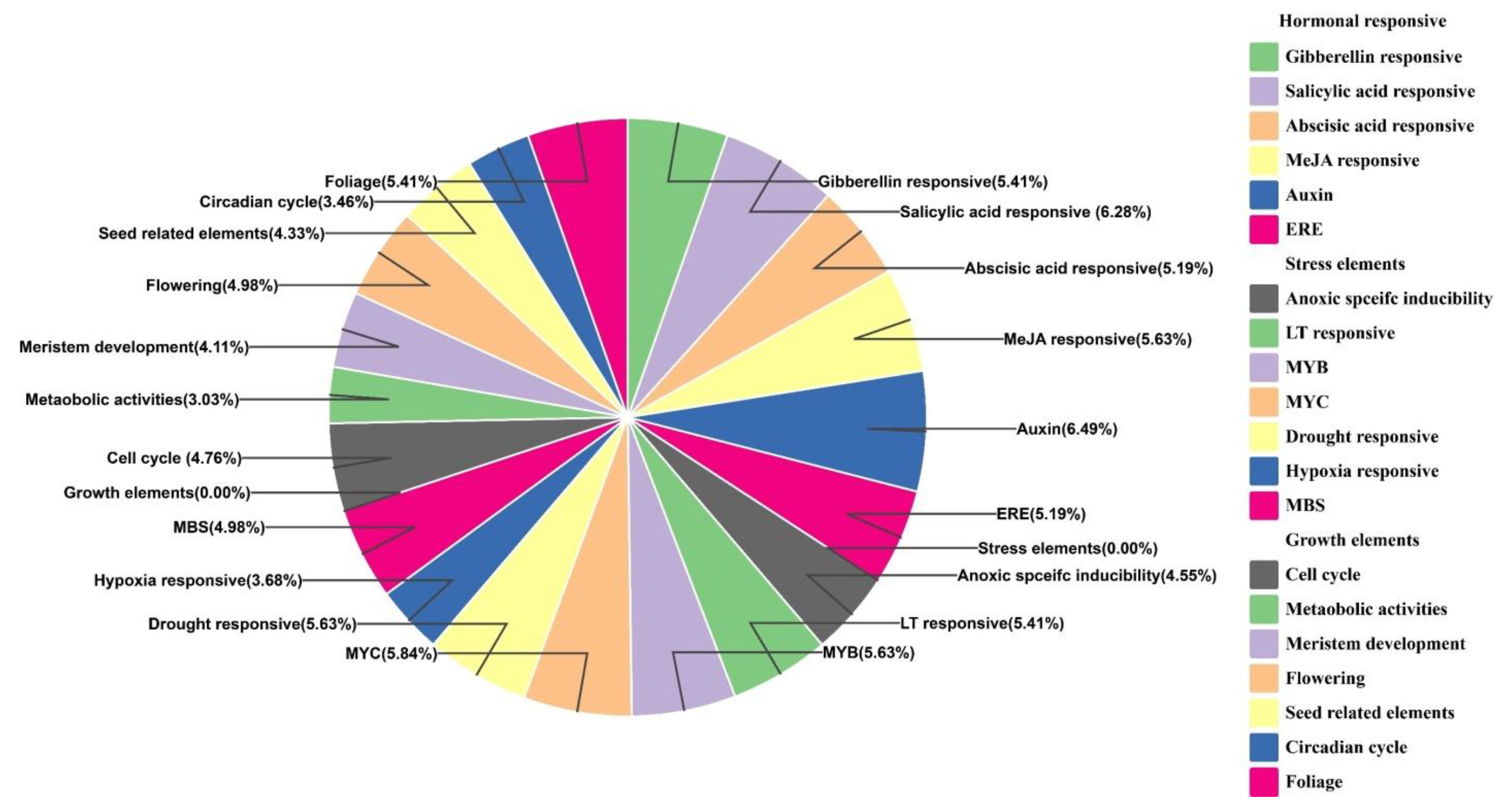
3.6. Promoter Analysis of BhTALE Genes
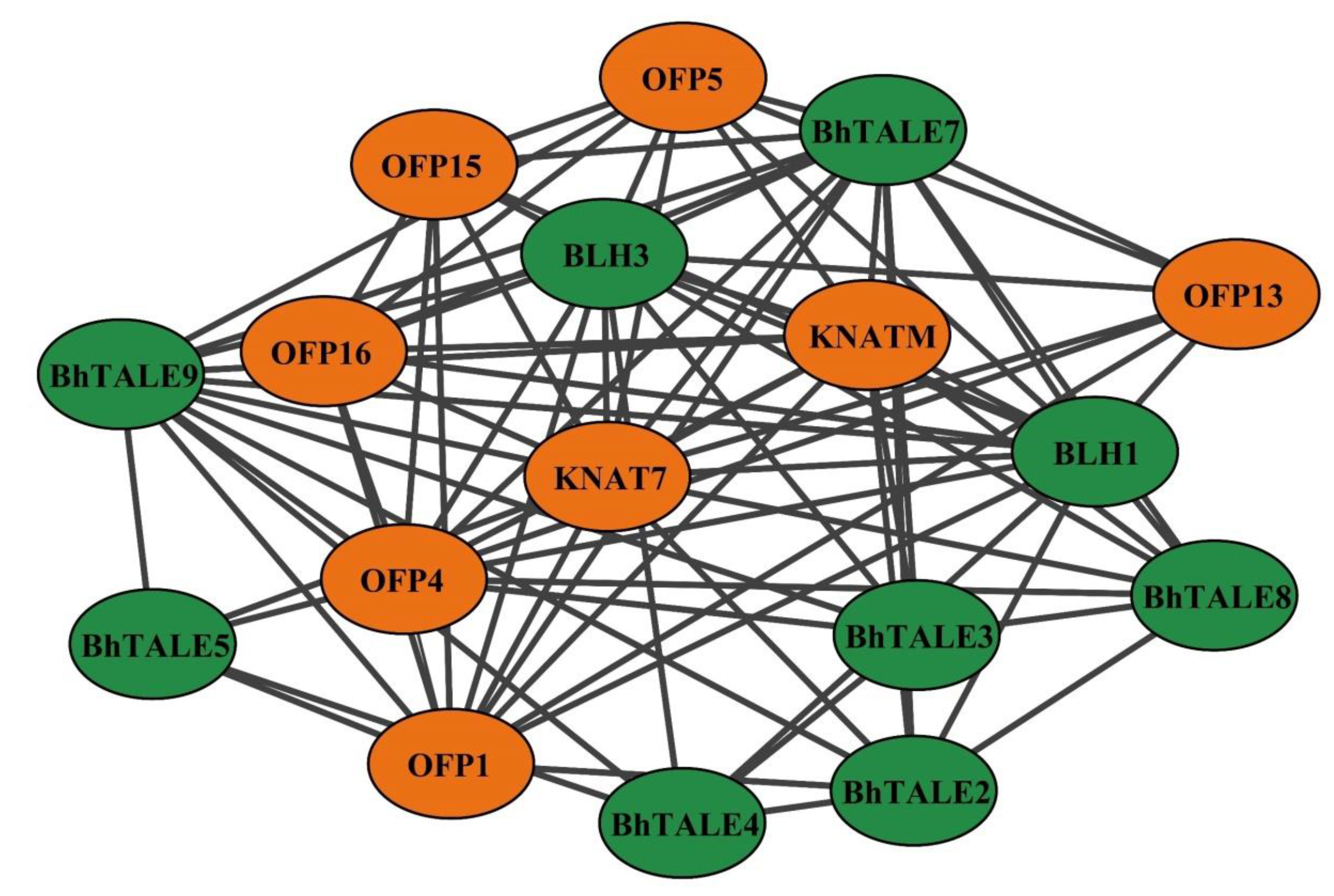
3.7. Interactive Protein Analysis
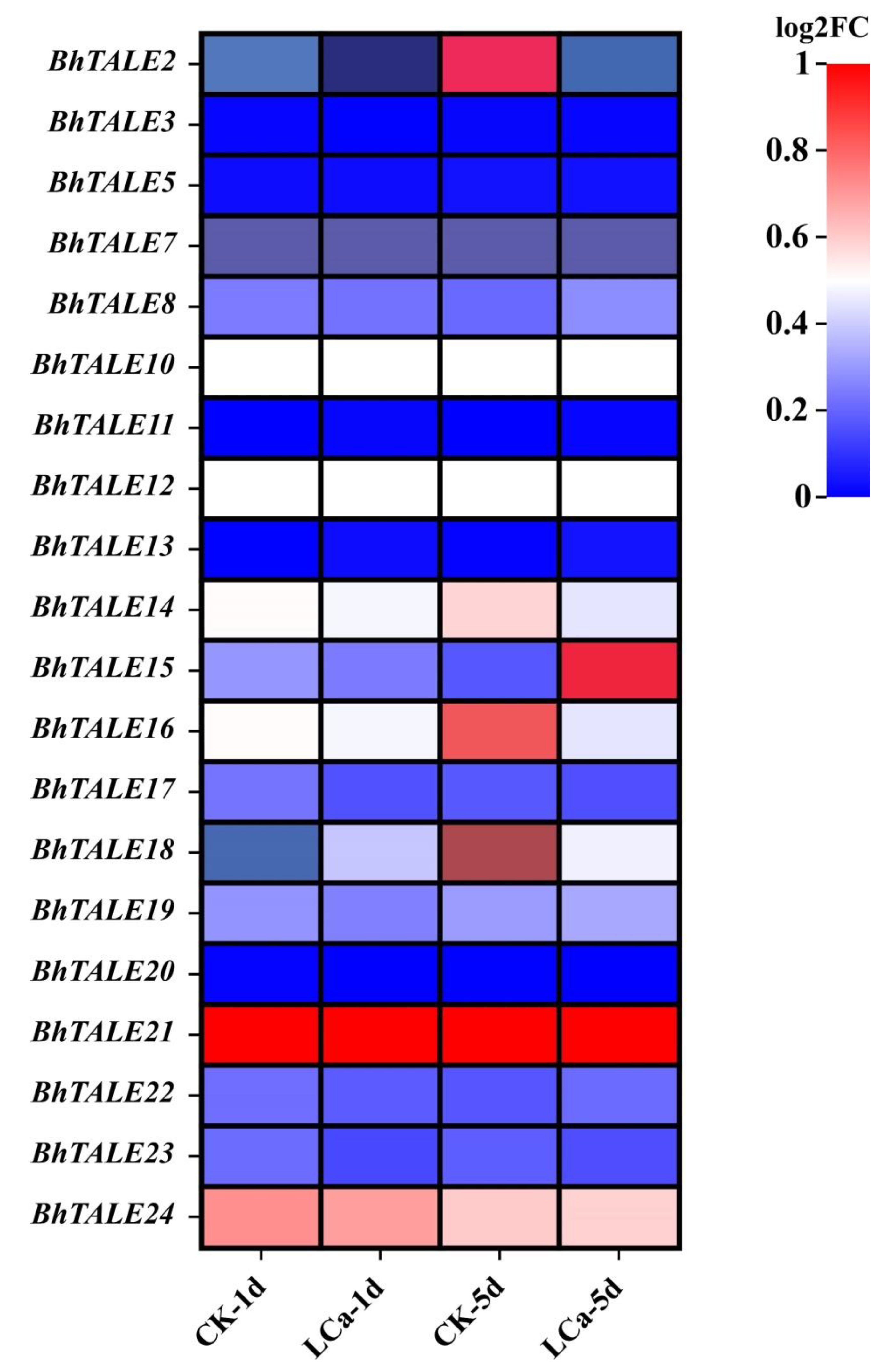
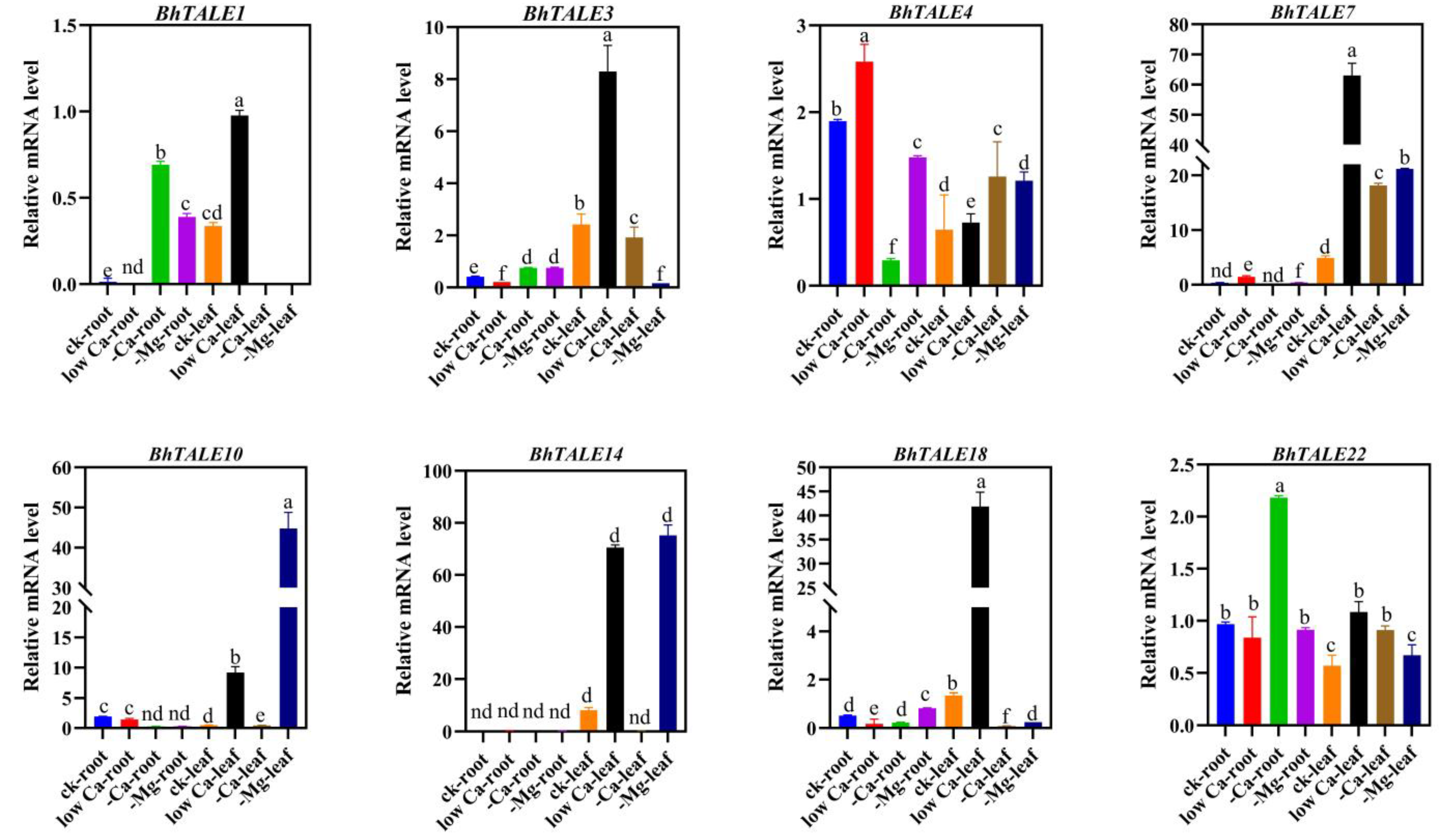
3.8. Expression of BhTALE Genes in Response to Low Calcium and Magnesium
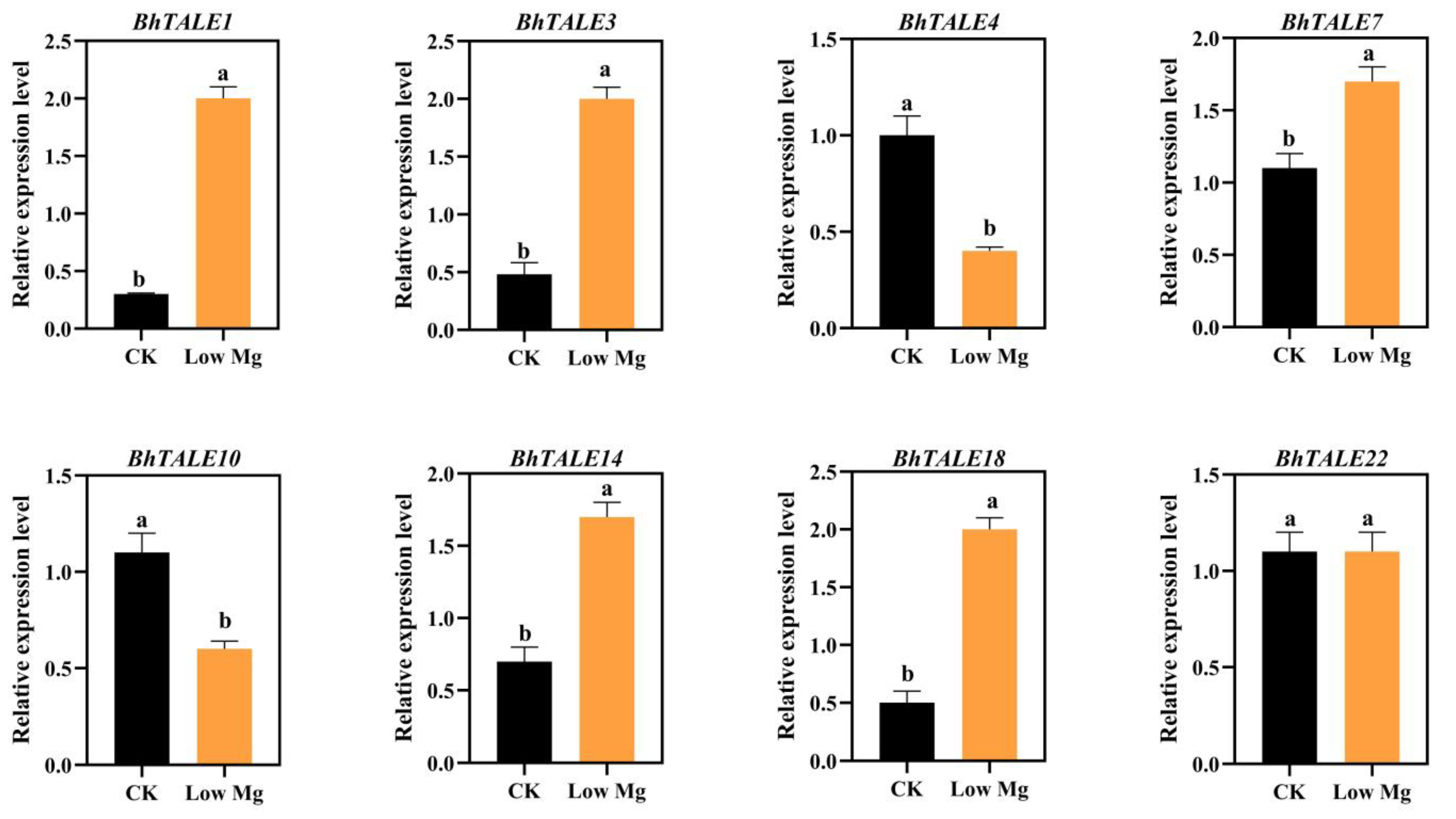
3.9. Expression Analysis under Low Mg Stress
4. Discussion
4.1. Ca and Mg Nutrients
4.2. TALE Genes Are Widely Distributed across the Genome
4.3. In Silico Analysis Shows the Stress-Responsive Nature of BhTALE Genes
4.4. BhTALE Genes Regulate Wax Gourd Response to Low Calcium and Magnesium
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Ahmad, S.; Khan, K.; Saleh, I.A.; Okla, M.K.; Alaraidh, I.A.; AbdElgawad, H.; Naeem, M.; Ahmad, N.; Fahad, S. TALE gene family: Identification, evolutionary and expression analysis under various exogenous hormones and waterlogging stress in Cucumis sativus L. BMC Plant Biol. 2024, 24, 564. [Google Scholar] [CrossRef]
- Bürglin, T.R. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997, 25, 4173–4180. [Google Scholar] [CrossRef]
- Ahmad, S.; Zhu, H.; Chen, Y.; Xi, C.; Shah, A.Z.; Ge, L. Comprehensive Bioinformatics and Expression Analysis of the TLP Gene Family Revealed Its Role in Regulating the Response of Oryza sativa to Nilaparvata lugens, Laodelphax striatellus, and Jinggangmycin. Agronomy 2022, 12, 1297. [Google Scholar] [CrossRef]
- Ahmad, S.; Jeridi, M.; Siddiqui, S.; Ali, S.; Shah, A.Z. Genome-wide identification, characterization, and expression analysis of the Chalcone Synthase gene family in Oryza sativa under Abiotic Stresses. Plant Stress 2023, 9, 100201. [Google Scholar] [CrossRef]
- Gehring, W.J. Homeo boxes in the study of development. Science 1987, 236, 1245–1252. [Google Scholar] [CrossRef]
- Hamant, O.; Pautot, V. Plant development: A TALE story. Comptes Rendus Biol. 2010, 333, 371–381. [Google Scholar] [CrossRef]
- Jia, P.; Sharif, R.; Li, Y.; Sun, T.; Li, S.; Zhang, X.; Dong, Q.; Luan, H.; Guo, S.; Ren, X.; et al. The BELL1-like homeobox gene MdBLH14 from apple controls flowering and plant height via repression of MdGA20ox3. Int. J. Biol. Macromol. 2023, 242, 124790. [Google Scholar] [CrossRef]
- Mukherjee, K.; Brocchieri, L.; Bürglin, T.R. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 2009, 26, 2775–2794. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2015, 44, D279–D285. [Google Scholar] [CrossRef]
- Vollbrecht, E.; Veit, B.; Sinha, N.; Hake, S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 1991, 350, 241–243. [Google Scholar] [CrossRef]
- Ma, Q.; Wang, N.; Hao, P.; Sun, H.; Wang, C.; Ma, L.; Wang, H.; Zhang, X.; Wei, H.; Yu, S. Genome-wide identification and characterization of TALE superfamily genes in cotton reveals their functions in regulating secondary cell wall biosynthesis. BMC Plant Biol. 2019, 19, 432. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Quan, S.; Zhang, Z.; Kang, C.; Liu, J.; Niu, J. Genome-wide Identification, Characterization and Expression profile of TALE gene family in (Juglans regia L.). Sci. Hortic. 2022, 297, 110945. [Google Scholar] [CrossRef]
- Jia, P.; Wang, Y.; Sharif, R.; Dong, Q.-L.; Liu, Y.; Luan, H.-A.; Zhang, X.-M.; Guo, S.-P.; Qi, G.-H. KNOTTED1-like homeobox (KNOX) transcription factors—Hubs in a plethora of networks: A review. Int. J. Biol. Macromol. 2023, 253, 126878. [Google Scholar] [CrossRef]
- Bellaoui, M.; Pidkowich, M.S.; Samach, A.; Kushalappa, K.; Kohalmi, S.E.; Modrusan, Z.; Crosby, W.L.; Haughn, G.W. The Arabidopsis BELL1 and KNOX TALE Homeodomain Proteins Interact through a Domain Conserved between Plants and Animals. Plant Cell 2001, 13, 2455–2470. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, X.; Ju, H.; Chen, J.; Wang, S.; Wang, H.; Zhao, Y.; Chang, Y. Ovate family protein1 interaction with BLH3 regulates transition timing from vegetative to reproductive phase in Arabidopsis. Biochem. Biophys. Res. Commun. 2016, 470, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.M.S.; Hake, S. The Interaction of Two Homeobox Genes, BREVIPEDICELLUS and PENNYWISE, Regulates Internode Patterning in the Arabidopsis Inflorescence. Plant Cell 2003, 15, 1717–1727. [Google Scholar] [CrossRef]
- Kim, D.; Cho, Y.H.; Ryu, H.; Kim, Y.; Kim, T.H.; Hwang, I. BLH1 and KNAT3 modulate ABA responses during germination and early seedling development in Arabidopsis. Plant J. Cell Mol. Biol. 2013, 75, 755–766. [Google Scholar] [CrossRef]
- Sharif, R.; Su, L.; Chen, X.; Qi, X. Hormonal interactions underlying parthenocarpic fruit formation in horticultural crops. Hortic. Res. 2022, 9, uhab024. [Google Scholar] [CrossRef] [PubMed]
- Sharif, R.; Su, L.; Chen, X.; Qi, X. Involvement of auxin in growth and stress response of cucumber. Veg. Res. 2022, 2, 1–9. [Google Scholar] [CrossRef]
- Sharif, R.; Zhu, Y.; Huang, Y.; Sohail, H.; Li, S.; Chen, X.; Qi, X. microRNA regulates cytokinin induced parthenocarpy in Cucumber (Cucumis sativus L.). Plant Physiol. Biochem. 2024, 212, 108681. [Google Scholar] [CrossRef]
- Pan, J.; Sohail, H.; Sharif, R.; Hu, Q.; Song, J.; Qi, X.; Chen, X.; Xu, X. Cucumber JASMONATE ZIM-DOMAIN 8 interaction with transcription factor MYB6 impairs waterlogging-triggered adventitious rooting. Plant Physiol. 2024. ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Cakmak, I.; Feng, J.; Yu, C.; Chen, X.; Xie, D.; Wu, L.; Song, Z.; Cao, J.; He, Y. Magnesium Deficiency Reduced the Yield and Seed Germination in Wax Gourd by Affecting the Carbohydrate Translocation. Front. Plant Sci. 2020, 11, 797. [Google Scholar] [CrossRef]
- Chang, J.; Liao, D.; Li, J.; Li, J.; Li, Z.; Chen, X.; Song, Z.; Zhang, B. Calcium deficiency leads to fruit blackheart formation by disrupting glycometabolism and phenylpropanoid metabolism in wax gourd. Postharvest Biol. Technol. 2024, 211, 112851. [Google Scholar] [CrossRef]
- Xie, D.; Xu, Y.; Wang, J.; Liu, W.; Zhou, Q.; Luo, S.; Huang, W.; He, X.; Li, Q.; Peng, Q.; et al. The wax gourd genomes offer insights into the genetic diversity and ancestral cucurbit karyotype. Nat. Commun. 2019, 10, 5158. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Ali, S.; Shah, A.Z.; Khan, A.; Faria, S. Chalcone synthase (CHS) family genes regulate the growth and response of cucumber (Cucumis sativus L.) to Botrytis cinerea and abiotic stresses. Plant Stress 2023, 8, 100159. [Google Scholar] [CrossRef]
- Ahmad, S.; Zhang, J.; Wang, H.; Zhu, H.; Dong, Q.; Zong, S.; Wang, T.; Chen, Y.; Ge, L. The Phosphoserine Phosphatase Alters the Free Amino Acid Compositions and Fecundity in Cyrtorhinus lividipennis Reuter. Int. J. Mol. Sci. 2022, 23, 15283. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Li, J.; Hu, X.; Zhang, R.; Li, Q.; Xu, W.; Zhang, L.; Guo, F.; Zhao, H.; Wang, P.; Wang, Y.; et al. The plasma membrane magnesium transporter CsMGT5 mediates magnesium uptake and translocation under magnesium limitation in tea plants (Camellia sinensis L.). Sci. Hortic. 2023, 310, 111711. [Google Scholar] [CrossRef]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef]
- Pan, J.; Tu, J.; Sharif, R.; Qi, X.; Xu, X.; Chen, X. Study of JASMONATE ZIM-Domain gene family to waterlogging stress in Cucumis sativus L. Veg. Res. 2021, 1, 1–12. [Google Scholar] [CrossRef]
- Yang, Q.; Sharif, Y.; Zhuang, Y.; Cai, T.; Wang, L.; Fu, H.; Lu, W.; Ma, M.; Yang, H.; Li, H.; et al. Comprehensive in-silico characterization and expression analysis of UbiA prenyltransferase genes in peanut (Arachis hypogaea L.) against abiotic stresses. Plant Stress 2023, 10, 100229. [Google Scholar] [CrossRef]
- Zaman, Q.U.; Hussain, M.A.; Khan, L.U.; Hui, L.; Khan, D.; Khokhar, A.A.; Cui, J.; Raza, A.; Wang, H.-F. Genome-wide identification and expression profiling of APX gene family under multifactorial stress combinations and melatonin-mediated tolerance in pitaya. Sci. Hortic. 2023, 321, 112312. [Google Scholar] [CrossRef]
- Sharif, R.; Liu, P.; Wang, D.; Jin, Z.; Uzair, U.; Yadav, V.; Mujtaba, M.; Chen, P.; Li, Y. Genome-wide characterisation and expression analysis of cellulose synthase genes superfamily under various environmental stresses in Cucumis sativus L. N. Z. J. Crop Hortic. Sci. 2021, 49, 127–150. [Google Scholar] [CrossRef]
- Bin, M.; Yi, G.; Zhang, X. Discovery and characterization of magnesium transporter (MGT) gene family in Citrus sinensis and their role in magnesium deficiency stress. Plant Growth Regul. 2023, 100, 733–746. [Google Scholar] [CrossRef]
- Li, J.; Muneer, M.A.; Sun, A.; Guo, Q.; Wang, Y.; Huang, Z.; Li, W.; Zheng, C. Magnesium application improves the morphology, nutrients uptake, photosynthetic traits, and quality of tobacco (Nicotiana tabacum L.) under cold stress. Front. Plant Sci. 2023, 14, 1078128. [Google Scholar]
- Kobayashi, N.I.; Tanoi, K. Critical issues in the study of magnesium transport systems and magnesium deficiency symptoms in plants. Int. J. Mol. Sci. 2015, 16, 23076–23093. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.-F.; Qian, X.-X.; Wang, K.-Q.; Yu, Y.-H.; Guo, Y.-Y.; Zhao, X.; Wang, B.; Yang, N.-Y.; Huang, J.-R.; Yang, Z.-N. Slowing development facilitates Arabidopsis mgt mutants to accumulate enough magnesium for pollen formation and fertility restoration. Front. Plant Sci. 2021, 11, 621338. [Google Scholar] [CrossRef]
- Hou, J.; Riaz, M.; Yan, L.; Lu, K.; Jiang, C. Effect of exogenous l-aspartate nano-calcium on root growth, calcium forms and cell wall metabolism of Brassica napus L. NanoImpact 2022, 27, 100415. [Google Scholar] [CrossRef]
- Winkler, A.; Knoche, M. Calcium and the physiology of sweet cherries: A review. Sci. Hortic. 2019, 245, 107–115. [Google Scholar] [CrossRef]
- Jing, T.; Li, J.; He, Y.; Shankar, A.; Saxena, A.; Tiwari, A.; Maturi, K.C.; Solanki, M.K.; Singh, V.; Eissa, M.A.; et al. Role of calcium nutrition in plant Physiology: Advances in research and insights into acidic soil conditions—A comprehensive review. Plant Physiol. Biochem. 2024, 210, 108602. [Google Scholar] [CrossRef] [PubMed]
- Lenz, H.; Dombinov, V.; Dreistein, J.; Reinhard, M.R.; Gebert, M.; Knoop, V. Magnesium deficiency phenotypes upon multiple knockout of Arabidopsis thaliana MRS2 clade B genes can be ameliorated by concomitantly reduced calcium supply. Plant Cell Physiol. 2013, 54, 1118–1131. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.-J.; Luan, S. Regulation of calcium and magnesium homeostasis in plants: From transporters to signaling network. Curr. Opin. Plant Biol. 2017, 39, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Chen, Y.; Shah, A.Z.; Wang, H.; Xi, C.; Zhu, H.; Ge, L. The Homeodomain-Leucine Zipper Genes Family Regulates the Jinggangmycin Mediated Immune Response of Oryza sativa to Nilaparvata lugens, and Laodelphax striatellus. Bioengineering 2022, 9, 398. [Google Scholar] [CrossRef]
- Zheng, Y.; Wu, S.; Bai, Y.; Sun, H.; Jiao, C.; Guo, S.; Zhao, K.; Blanca, J.; Zhang, Z.; Huang, S. Cucurbit Genomics Database (CuGenDB): A central portal for comparative and functional genomics of cucurbit crops. Nucleic Acids Res. 2018, 47, D1128–D1136. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, L.; Yan, L.; Xiong, X.; Wang, W.; Zhang, X.-H.; Min, D.-H. Genome-wide analysis of TALE superfamily in Triticum aestivum reveals TaKNOX11-A is involved in abiotic stress response. BMC Genom. 2022, 23, 89. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, P.; Cheng, B.; Zhang, Y.; Shen, Y.; Wang, X.; Zhang, Q.; Lou, Q.; Zhang, S.; Wang, B.; et al. Identification of TALE Transcription Factor Family and Expression Patterns Related to Fruit Chloroplast Development in Tomato (Solanum lycopersicum L.). Int. J. Mol. Sci. 2022, 23, 4507. [Google Scholar] [CrossRef]
- Ullah, U.; Shalmani, A.; Ilyas, M.; Raza, A.; Ahmad, S.; Shah, A.Z.; Khan, F.U.; AzizUd, D.; Bibi, A.; Rehman, S.U.; et al. BZR proteins: Identification, evolutionary and expression analysis under various exogenous growth regulators in plants. Mol. Biol. Rep. 2022, 49, 12039–12053. [Google Scholar] [CrossRef]
- Zhao, K.; Zhang, X.; Cheng, Z.; Yao, W.; Li, R.; Jiang, T.; Zhou, B. Comprehensive analysis of the three-amino-acid-loop-extension gene family and its tissue-differential expression in response to salt stress in poplar. Plant Physiol. Biochem. 2019, 136, 1–12. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Yan, M.; Zhao, H.; Zhang, X.; Yuan, Z. Genome-wide Identification and Expression Analysis of TALE Gene Family in Pomegranate (Punica granatum L.). Agronomy 2020, 10, 829. [Google Scholar] [CrossRef]
- Muhammad, I.; Jing, X.-Q.; Shalmani, A.; Ali, M.; Yi, S.; Gan, P.-F.; Li, W.-Q.; Liu, W.-T.; Chen, K.-M. Comparative in Silico Analysis of Ferric Reduction Oxidase (FRO) Genes Expression Patterns in Response to Abiotic Stresses, Metal and Hormone Applications. Molecules 2018, 23, 1163. [Google Scholar] [CrossRef] [PubMed]
- Shalmani, A.; Fan, S.; Jia, P.; Li, G.; Muhammad, I.; Li, Y.; Sharif, R.; Dong, F.; Zuo, X.; Li, K. Genome Identification of B-BOX Gene Family Members in Seven Rosaceae Species and Their Expression Analysis in Response to Flower Induction in Malus domestica. Molecules 2018, 23, 1763. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.-Q.; Shalmani, A.; Zhou, M.-R.; Shi, P.-T.; Muhammad, I.; Shi, Y.; Sharif, R.; Li, W.-Q.; Liu, W.-T.; Chen, K.-M. Genome-wide identification of malectin/malectin-like domain containing protein family genes in rice and their expression regulation under various hormones, abiotic stresses, and heavy metal treatments. J. Plant Growth Regul. 2019, 39, 492–506. [Google Scholar] [CrossRef]
- Gupta, S.; Kaur, N.; Kant, K.; Jindal, P.; Ali, A.; Naeem, M. Calcium: A master regulator of stress tolerance in plants. S. Afr. J. Bot. 2023, 163, 580–594. [Google Scholar] [CrossRef]
- Franz, S.; Ehlert, B.; Liese, A.; Kurth, J.; Cazalé, A.-C.; Romeis, T. Calcium-Dependent Protein Kinase CPK21 Functions in Abiotic Stress Response in Arabidopsis thaliana. Mol. Plant 2011, 4, 83–96. [Google Scholar] [CrossRef]
- Shi, X.; Bao, J.; Lu, X.; Ma, L.; Zhao, Y.; Lan, S.; Cao, J.; Ma, S.; Li, S. The mechanism of Ca2+ signal transduction in plants responding to abiotic stresses. Environ. Exp. Bot. 2023, 216, 105514. [Google Scholar] [CrossRef]

| Name | Gene ID | Chromosomal Location | Subcellular |
|---|---|---|---|
| BhTALE1 | Bhi02G000349 | Chr2: 8718933 … 8740554 (−) | Nucleus |
| BhTALE2 | Bhi03G001512 | Chr 3: 41081772 … 41087777 (−) | Nucleus |
| BhTALE3 | Bhi03G001611 | Chr 3: 44246973 … 44252304 (−) | Nucleus |
| BhTALE4 | Bhi03G000473 | Chr 3: 11851118 … 11855225 (−) | Nucleus |
| BhTALE5 | Bhi04G000450 | Chr 4: 12570212 … 12575367 (−) | Nucleus |
| BhTALE6 | Bhi04G000431 | Chr 4: 12094162 … 12100830 (−) | Nucleus |
| BhTALE7 | Bhi04G000020 | Chr 4: 649004 … 654299 (−) | Nucleus |
| BhTALE8 | Bhi04G000553 | Chr 4: 15727426 … 15742670 (+) | Nucleus |
| BhTALE9 | Bhi04G000431 | Chr 4: 12094162 … 12100830 (−) | Nucleus |
| BhTALE10 | Bhi05G001210 | Chr 5: 47747978 … 47752706 (−) | Nucleus |
| BhTALE11 | Bhi06G001339 | Chr 5: 47747978 … 47752706 (−) | Nucleus |
| BhTALE12 | Bhi07G001403 | Chr 7: 48154292 … 48163762 (+) | Nucleus |
| BhTALE13 | Bhi08G000192 | Chr 8: 7980115 … 7986683 (−) | Nucleus |
| BhTALE14 | Bhi09G002444 | Chr 9: 77103490 … 77108059 (−) | Nucleus |
| BhTALE15 | Bhi09G002609 | Chr 9: 81520308 … 81524945 (+) | Nucleus |
| BhTALE16 | Bhi09G002444 | Chr 9: 77103490 … 77108059 (−) | Nucleus |
| BhTALE17 | Bhi09G000031 | Chr 9: 606684 … 610404 (−) | Nucleus |
| BhTALE18 | Bhi10G001574 | Chr 10: 48974608 … 48979463 (+) | Nucleus |
| BhTALE19 | Bhi10G000146 | Chr 10: 4078350 … 4083322 (+) | Nucleus |
| BhTALE20 | Bhi10G001790 | Chr 10: 56344392 … 56350654 (−) | Nucleus |
| BhTALE21 | Bhi11G001983 | Chr 11: 64704085 … 64708775 (−) | Nucleus |
| BhTALE22 | Bhi11G001620 | Chr 11: 54758024 … 54762277 (+) | Nucleus |
| BhTALE23 | Bhi12G000276 | Chr 12: 8560884 … 8566030 (+) | Nucleus |
| BhTALE24 | Bhi12G000297 | Chr 12: 9196808 … 9201184 (−) | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussain, S.; Chang, J.; Li, J.; Chen, X.; Xie, D.; Zhang, B. Transcriptome Wide Identification and Expression Analysis Revealed BhTALE Gene Family Regulates Wax Gourd (Benincasa hispida) Response to Low Calcium and Magnesium Stress. Horticulturae 2024, 10, 1083. https://doi.org/10.3390/horticulturae10101083
Hussain S, Chang J, Li J, Chen X, Xie D, Zhang B. Transcriptome Wide Identification and Expression Analysis Revealed BhTALE Gene Family Regulates Wax Gourd (Benincasa hispida) Response to Low Calcium and Magnesium Stress. Horticulturae. 2024; 10(10):1083. https://doi.org/10.3390/horticulturae10101083
Chicago/Turabian StyleHussain, Shahid, Jingjing Chang, Jing Li, Xiao Chen, Dasen Xie, and Baige Zhang. 2024. "Transcriptome Wide Identification and Expression Analysis Revealed BhTALE Gene Family Regulates Wax Gourd (Benincasa hispida) Response to Low Calcium and Magnesium Stress" Horticulturae 10, no. 10: 1083. https://doi.org/10.3390/horticulturae10101083
APA StyleHussain, S., Chang, J., Li, J., Chen, X., Xie, D., & Zhang, B. (2024). Transcriptome Wide Identification and Expression Analysis Revealed BhTALE Gene Family Regulates Wax Gourd (Benincasa hispida) Response to Low Calcium and Magnesium Stress. Horticulturae, 10(10), 1083. https://doi.org/10.3390/horticulturae10101083






