Prevalence of Lentilacobacillus hilgardii over Lactiplantibacillus plantarum in Low-Temperature Spontaneous Malolactic Fermentation of a Patagonian Pinot Noir
Abstract
:1. Introduction
2. Materials and Methods
2.1. Wine Samples
2.2. Lactiplantibacillus plantarum Strains Collection
2.3. Isolation of Psychrotrophic Lactobacillaceae Strains
2.4. Identification of Psychrotrophic Isolates
2.5. Typification by Random Amplification of Polymorphic DNA-Polymerase Chain Reaction (RAPD-PCR)
2.6. Cell Culture Acclimation
2.7. Fermentation at Low Temperatures in Sterile Wine at a Laboratory Scale
2.8. Fermentation and Implantation at Low Temperatures in Non-Sterile Wine
2.9. L-Malic Acid Consumption (MAC)
2.10. Screening of the hdc Gene
2.11. Reproducibility Assay
3. Results
3.1. Isolation by the Low-Temperature Selection Method
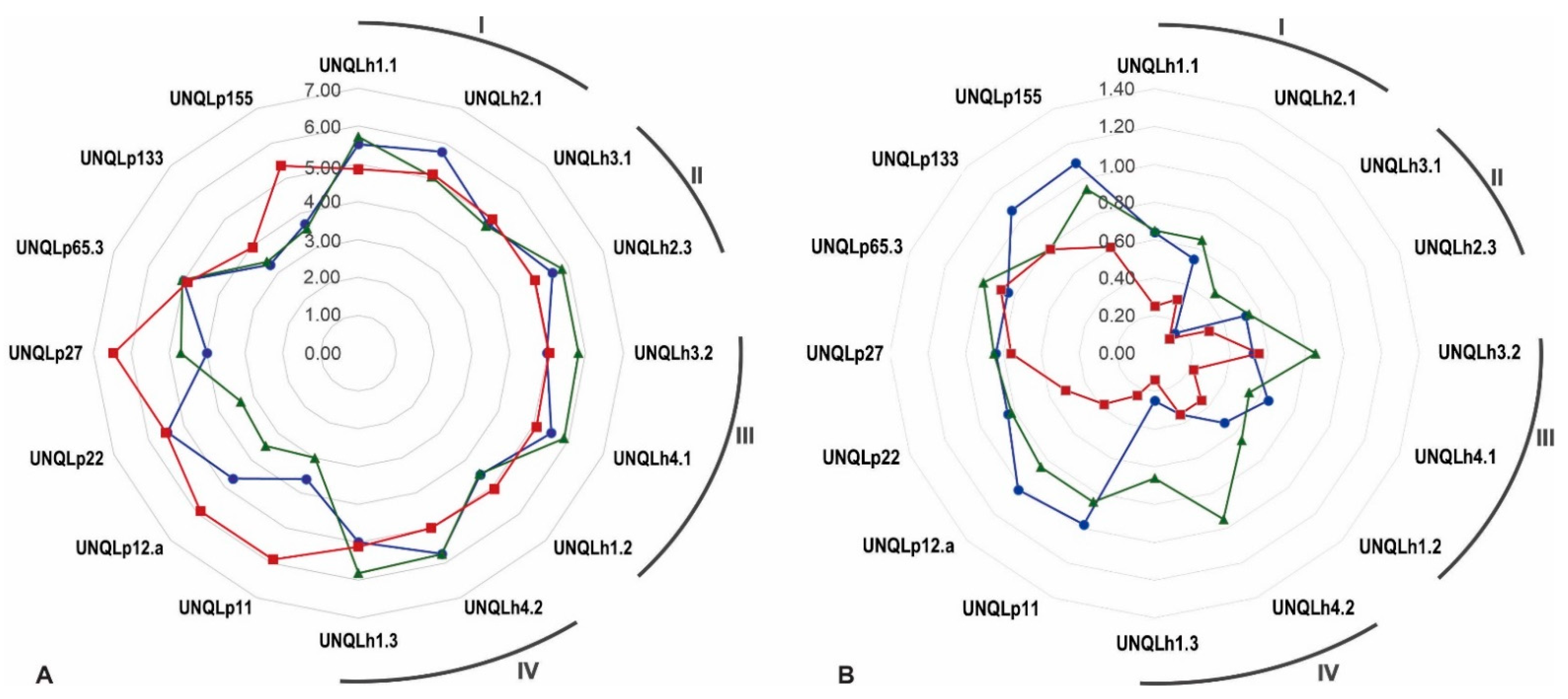
3.2. Identification of Psychrotrophic Isolates
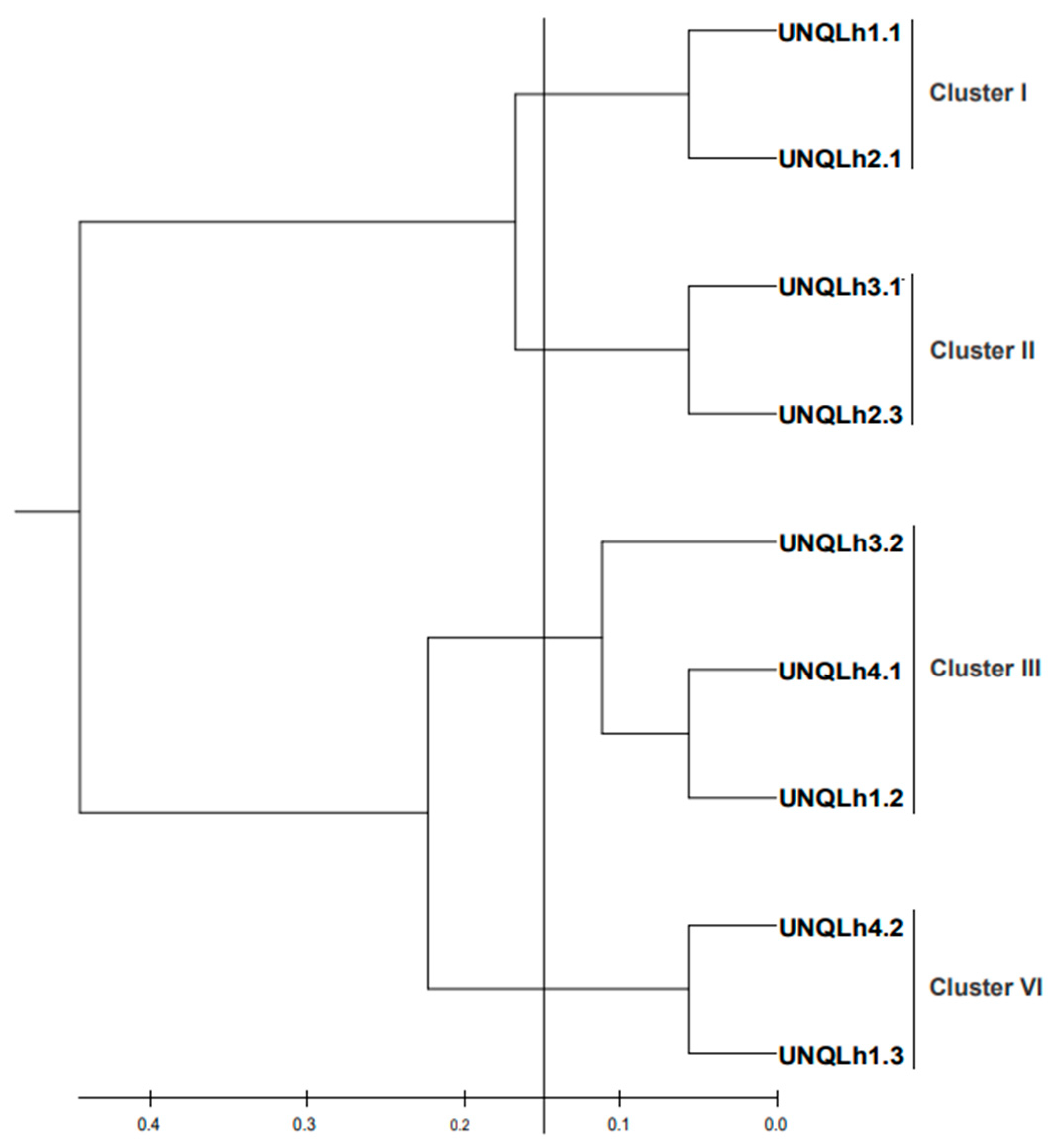
3.3. Typification by RAPD-PCR Analysis
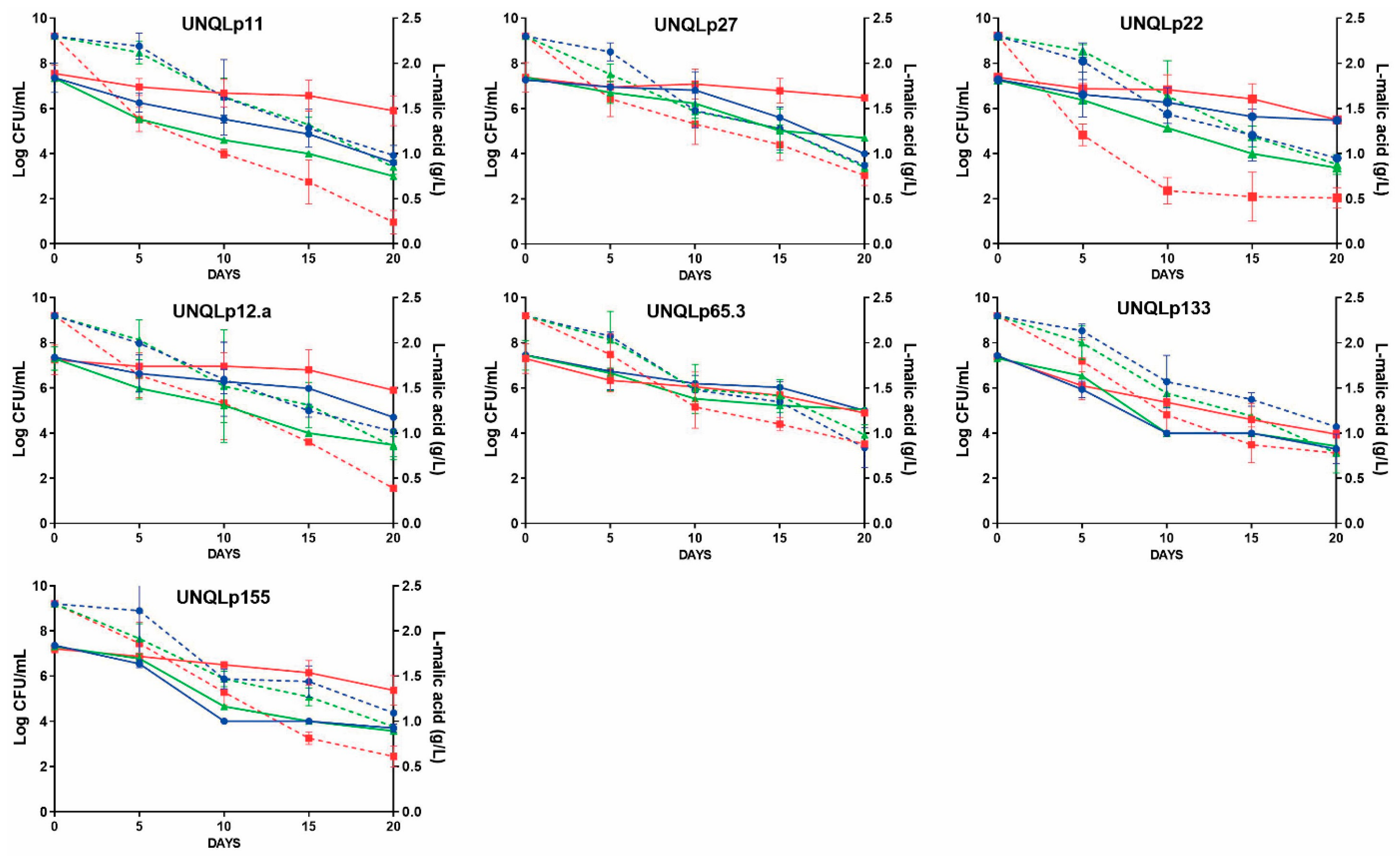
3.4. Fermentation Assays in Sterile Pinot Noir Wine at Laboratory Scale
3.5. Cell Viability and MAC Kinetics Analysis
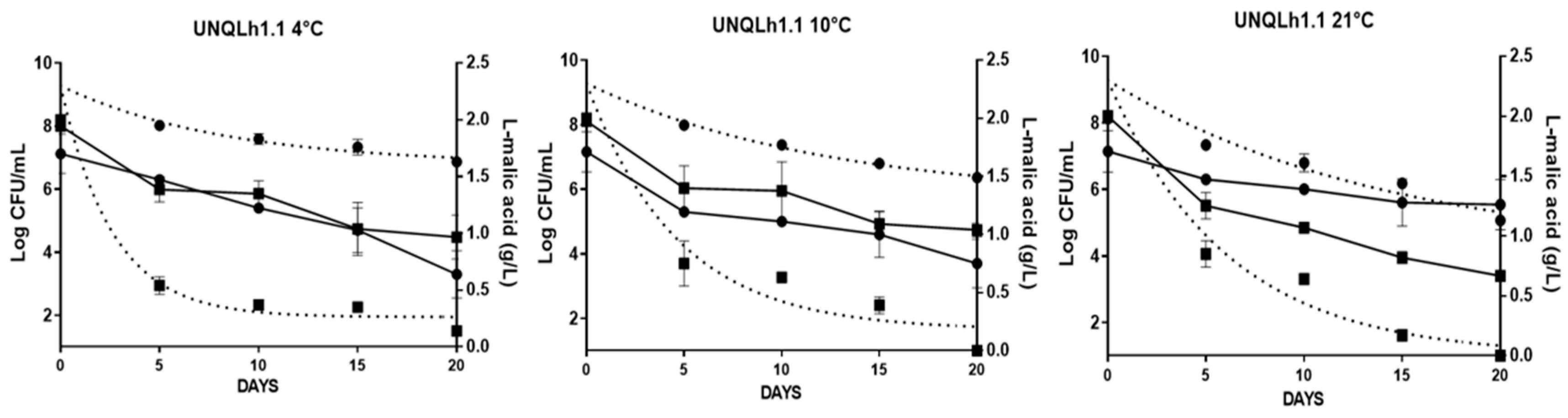
3.6. Implantation Capacity at Low Temperatures in Non-Sterile Wine
3.7. Implantation Capacity by RAPD-PCR
3.8. Screening of the hdc Gene
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bartowsky, E.J.; Borneman, A.R. Genomic variations of Oenococcus oeni strains and the potential to impact on malolactic fermentation and aroma compounds in wine. Appl. Microbiol. Biot. 2011, 92, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Gil-Sánchez, I.; Suáldea, B.B.; Moreno-Arribas, M.V. Malolactic fermentation. In Red Wine Technology, 1st ed.; Morata, A., Ed.; Academic Press: Cambridge, UK, 2019; pp. 85–98. [Google Scholar]
- Capozzi, V.; Tufariello, M.; De Simone, N.; Fragasso, M.; Grieco, F. Biodiversity of oenological lactic acid bacteria: Species-and strain-dependent plus/minus effects on wine quality and safety. Fermentation 2021, 7, 24. [Google Scholar] [CrossRef]
- Nardi, T. Microbial resources as a tool for enhancing sustainability in winemaking. Microorganisms 2020, 8, 507. [Google Scholar] [CrossRef]
- Franquès, J.; Araque, I.; Palahí, E.; Portillo, M.d.C.; Reguant, C.; Bordons, A. Presence of Oenococcus oeni and Other Lactic Acid Bacteria in Grapes and Wines from Priorat (Catalonia, Spain). LWT Food Sci. Technol. 2017, 81, 326–334. [Google Scholar] [CrossRef]
- Lerm, E.; Engelbrecht, L.; du Toit, M. Selection and Characterisation of Oenococcus oeni and Lactobacillus plantarum South African Wine Isolates for Use as Malolactic Fermentation Starter Cultures. S. Afr. J. Enol. Vitic. 2011, 32, 280–295. [Google Scholar] [CrossRef]
- Lorentzen, M.P.G.; Lucas, P.M. Distribution of Oenococcus oeni Populations in Natural Habitats. Appl. Microbiol. Biotechnol. 2019, 103, 2937–2945. [Google Scholar] [CrossRef]
- Coucheney, F.; Desroche, N.; Bou, M.; Tourdot-Maréchal, R.; Dulau, L.; Guzzo, J. A new approach for selection of Oenococcus oeni strains in order to produce malolactic starters. Int. J. Food Microbiol. 2005, 105, 463–470. [Google Scholar] [CrossRef]
- Lonvaud-Funel, A. Microbiology of the malolactic fermentation: Molecular aspects. FEMS Microbiol. Lett. 1995, 126, 209–214. [Google Scholar] [CrossRef]
- Lonvaud-Funel, A. Malolactic fermentation and its effects on wine quality and safety. In Managing Wine Quality; Woodhead Publishing: Cambridge, UK, 2022; pp. 105–139. [Google Scholar]
- Bokulich, N.A.; Bamforth, C.W.; Mills, D.A. A review of molecular methods for microbial community profiling of beer and wine. J. Am. Soc. Brew. Chem. 2012, 70, 150–162. [Google Scholar] [CrossRef]
- Berbegal, C.; Borruso, L.; Fragasso, M.; Tufariello, M.; Russo, P.; Brusetti, L.; Spano, G.; Capozzi, V. A Metagenomic-Based Approach for the Characterization of Bacterial Diversity Associated with Spontaneous Malolactic Fermentations in Wine. Int. J. Mol. Sci. 2019, 20, 3980. [Google Scholar] [CrossRef]
- Morgan, H.H.; Du Toit, M.; Setati, M.E. The grapevine and wine microbiome: Insights from high-throughput amplicon sequencing. Front. Microbiol. 2017, 8, 820. [Google Scholar] [CrossRef] [PubMed]
- Rivas, G.A.; Semorile, L.; Delfederico, L. Microbial diversity of the soil, rhizosphere and wine from an emerging wine-producing region of Argentina. LWT 2022, 153, 112429. [Google Scholar] [CrossRef]
- Du Toit, M.; Engelbrecht, L.; Lerm, E.; Krieger-Weber, S. Lactobacillus: The Next Generation of Malolactic Fermentation Starter Cultures—An Overview. Food Bioprocess Technol. 2011, 4, 876–906. [Google Scholar] [CrossRef]
- Ribéreau-Gayon, P.; Dubourdieu, D.; Donèche, B.; Lonvaud, A. Handbook of Enology: Volume 1: The Microbiology of Wine and Vinifications, 2nd ed.; John Wiley & Sons: New York, NY, USA, 2006; pp. 149–151. [Google Scholar]
- Reynolds, A.G. Managing Wine Quality: Volume 2: Oenology and Wine Quality, 2nd ed.; Woodhead Publishing: Sawston, UK, 2021. [Google Scholar]
- Berbegal, C.; Peña, N.; Russo, P.; Grieco, F.; Pardo, I.; Ferrer, S.; Spano, G.; Capozzi, V. Technological properties of Lactobacillus plantarum strains isolated from grape must fermentation. Food Microbiol. 2016, 57, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Ferrada, B.M.; Hollmann, A.; Delfederico, L.; Valdés la Hens, D.; Caballero, A.; Semorile, L. Patagonian red wines: Selection of Lactobacillus plantarum isolates as potential starter cultures for malolactic fermentation. World J. Microbiol. Biotechnol. 2013, 29, 1537–1549. [Google Scholar] [CrossRef] [PubMed]
- Brizuela, N.; Tymczyszyn, E.E.; Semorile, L.C.; La Hens, D.V.; Delfederico, L.; Hollmann, A.; Bravo-Ferrada, B. Lactobacillus plantarum as a malolactic starter culture in winemaking: A new (old) player? Electron. J. Biotech. 2019, 38, 10–18. [Google Scholar] [CrossRef]
- Lucio, O.; Pardo, I.; Krieger-Weber, S.; Heras, H.M.; Ferrer, S. Selection of Lactobacillus strains to induce biological acidification in low acidity wines. LWT Food Sci. Technol. 2016, 73, 334–341. [Google Scholar] [CrossRef]
- Krieger-Weber, S.; Heras, J.M.; Suarez, C. Lactobacillus plantarum, a new biological tool to control malolactic fermentation: A review and an outlook. Beverages 2020, 6, 23. [Google Scholar] [CrossRef]
- Wang, L.; Huang, G.; Ma, W.; Jin, G. Preparation and Application of Directed Vat Set Indigenous Freeze-Drying Lentilactobacillus hilgardii Q19 Starter in Winemaking. Foods 2023, 12, 1053. [Google Scholar] [CrossRef]
- López-Seijas, J.; García-Fraga, B.; Da Silva, A.F.; Zas-García, X.; Lois, L.C.; Gago-Martínes, A.; Leão-Martins, J.M.; Sieiro, C. Evaluation of malolactic bacteria associated with wines from Albariño variety as potential; starter cultures for quality and safety. Foods 2020, 9, 99. [Google Scholar] [CrossRef]
- Bravo-Ferrada, B.M.; Delfederico, L.; Hollmann, A.; La Hens, V.D.; Curilén, Y.; Caballero, A.; Semorile, L. Oenococcus oeni from Patagonian red wines: Isolation, characterization and technological properties. Int. J. Microbiol. Res. 2011, 3, 48. [Google Scholar]
- Brizuela, N.S.; Bravo-Ferrada, B.M.; La Hens, D.V.; Hollmann, A.; Delfederico, L.; Caballero, A.; Tymczyszyn, E.E.; Semorile, L. Comparative vinification assays with selected Patagonian strains of Oenococcus oeni and Lactobacillus plantarum. LWT Food Sci. Technol. 2017, 77, 348–355. [Google Scholar] [CrossRef]
- Valdes la Hens, D.; Bravo-Ferrada, B.M.; Delfederico, L.; Caballero, A.C.; Semorile, L. Prevalence of Lactobacillus plantarum and Oenococcus oeni during spontaneous malolactic fermentation in Patagonian red wines revealed by polymerase chain reaction-denaturing gradient gel electrophoresis with two targeted genes. Aust. J. Grape Wine Res. 2015, 21, 49–56. [Google Scholar] [CrossRef]
- Brizuela, N.S.; Bravo-Ferrada, B.M.; Pozo-Bayón, M.Á.; Semorile, L.; Tymczyszyn, E.E. Changes in the volatile profile of Pinot noir wines caused by Patagonian Lactobacillus plantarum and Oenococcus oeni strains. Food Res. Int. 2018, 106, 22–28. [Google Scholar] [CrossRef]
- Lasik, M. The application of malolactic fermentation process to create good-quality grape wine produced in cool-climate countries: A review. Eur. Food Res. Technol. 2013, 237, 843–850. [Google Scholar] [CrossRef]
- Manera, C.; Olguin, N.T.; Bravo-Ferrada, B.M.; Tymczyszyn, E.E.; Delfederico, L.; Bibiloni, H.; Caballero, A.; Semorile, C.; La Hens, D.V. Survival and implantation of indigenous psychrotrophic Oenococcus oeni strains during malolactic fermentation in a Patagonian Pinot noir wine. LWT 2019, 108, 353–360. [Google Scholar] [CrossRef]
- Olguín, N.T.; Valdés La Hens, D.; Delfederico, L.; Semorile, L. Relative expression of stress-related genes during acclimation at low temperature of psychrotrophic Oenococcus oeni strains from Patagonian wine. World J. Microbiol. Biotechnol. 2019, 35, 5. [Google Scholar] [CrossRef]
- Bravo-Ferrada, B.M.; Hollmann, A.; Brizuela, N.; La Hens, D.V.; Tymczyszyn, E.; Semorile, L. Growth and consumption of L-malic acid in wine-like medium by acclimated and non-acclimated cultures of Patagonian Oenococcus oeni strains. Folia Microbiol. 2016, 61, 365–373. [Google Scholar] [CrossRef]
- Brizuela, N.S.; Bravo-Ferrada, B.M.; Curilén, Y.; Delfederico, L.; Caballero, A.; Semorile, L.; Pozo-Bayón, M.A.; Tymczyszyn, E.E. Advantages of using blend cultures of native L. plantarum and O. oeni strains to induce malolactic fermentation of Patagonian Malbec wine. Front. Microbiol. 2018, 9, 2109. [Google Scholar] [CrossRef]
- Iglesias, N.G.; Brizuela, N.S.; Tymczyszyn, E.E.; Hollmann, A.; Valdes la Hens, D.; Semorile, L.; Bravo-Ferrada, B.M. Complete genome sequencing of Lactobacillus plantarum UNQLp 11 isolated from a Patagonian pinot noir wine. S. Afr. J. Enol. Vitic. 2020, 41, 197–209. [Google Scholar] [CrossRef]
- De Man, J.C.; Rogosa, D.; Sharpe, M.E. A medium for the cultivation of Lactobacilli. J. Appl. Bacteriol. 1960, 23, 130–135. [Google Scholar] [CrossRef]
- Rodas, A.M.; Ferrer, S.; Pardo, I. 16S-ARDRA, a tool for identification of lactic acid bacteria isolated from grape must and wine. Syst. Appl. Microbiol. 2003, 26, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Stenlid, J.; Karlsson, J.O.; Högberg, N. Intraspecific genetic variation in Heterobasidion annosum revealed by amplification of minisatellite DNA. Mycol. Res. 1994, 98, 57–63. [Google Scholar] [CrossRef]
- Apostol, B.L.; Black, W.; Miller, B.R.; Reiter, P.; Beaty, B.J. Estimation of the number of full sibling families at an oviposition site using RAPD-PCR markers: Applications to the mosquito Aedes aegypti. Theoret. Appl. Genet. 1993, 86, 991–1000. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Ferrada, B.M.; Tymczyszyn, E.E.; Gómez-Zavaglia, A.; Semorile, L. Effect of acclimation medium on cell viability, membrane integrity and ability to consume malic acid in synthetic wine by oenological Lactobacillus plantarum strains. J. Appl. Microbiol. 2014, 116, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Le Jeune, C.; Lonvaud-Funel, A.; Ten Brink, B.; Hofstra, H.; Van der Vossen, J.M.B.M. Development of a detection system for histidine decarboxylating lactic acid bacteria based on DNA probes, PCR and activity test. J. Appl. Bacteriol. 1995, 78, 316–326. [Google Scholar] [CrossRef]
- Iorizzo, M.; Testa, B.; Lombardi, S.J.; García-Ruiz, A.; Muñoz-González, C.; Bartolomé, B.; Moreno-Arribas, M.V. Selection and technological potential of Lactobacillus plantarum bacteria suitable for wine malolactic fermentation and grape aroma release. LWT 2016, 73, 557–566. [Google Scholar] [CrossRef]
- Sun, J.; Ge, Y.; Gu, X.; Li, R.; Ma, W.; Jin, G. Identification and Characterization of Malolactic Bacteria Isolated from the Eastern Foothills of Helan Mountain in China. Foods 2022, 11, 2455. [Google Scholar] [CrossRef]
- Mtshali, P.S.; Divol, B.; Van Rensburg, P.; Du Toit, M. Genetic screening of wine-related enzymes in Lactobacillus species isolated from South African wines. J. Appl. Microbiol. 2010, 108, 1389–1397. [Google Scholar] [CrossRef]
- Ruiz, P.; Izquierdo, P.M.; Seseña, S.; Palop, M.L. Analysis of lactic acid bacteria populations during spontaneous malolactic fermentation of Tempranillo wines at five wineries during two consecutive vintages. Food Control 2010, 21, 70–75. [Google Scholar] [CrossRef]
- Zhuravleva, D.E.; Iskhakova, Z.I.; Ozhegov, G.D.; Gogoleva, N.E.; Khusnutdinova, D.R.; Shagimardanova, E.I.; Forchhammer, K.; Kayumov, A.R. Complete Genome Sequence of Lactobacillus hilgardii LMG 7934, Carrying the Gene Encoding for the Novel PII-Like Protein PotN. Curr. Microbiol. 2020, 77, 3538–3545. [Google Scholar] [CrossRef]
- Iglesias, N.G.; Navarro, M.E.; Brizuela, N.S.; Valdés La Hens, D.; Semorile, L.C.; Tymczyszyn, E.E.; Bravo Ferrada, B.M. Analysis of the Genome Architecture of Lacticaseibacillus paracasei UNQLpc 10, a Strain with Oenological Potential as a Malolactic Starter. Fermentation 2022, 8, 726. [Google Scholar] [CrossRef]
- Iglesias, N.G.; Valdés La Hens, D.; Olguin, N.T.; Bravo-Ferrada, B.M.; Brizuela, N.S.; Tymczyszyn, E.E.; Bibiloni, H.; Caballero, A.C.; Delfederico, L.; Semorile, L. Genome sequence of Oenococcus oeni UNQOe19, the first fully assembled genome sequence of a Patagonian psychrotrophic oenological strain. Microbiol. Resour. Announc. 2018, 7, e00889-18. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, L.M.; Merin, M.G.; Morata, V.I.; Farias, M.E. Characterization of wines produced by mixed culture of autochthonous yeasts and Oenococcus oeni from the northwest region of Argentina. J. Ind. Microbiol. Biotechnol. 2011, 38, 1777–1785. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. USA 2014, 111, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Bartowsky, E.J.; Costello, P.J.; Chambers, P.J. Emerging trends in the application of malolactic fermentation. Aust. J. Grape Wine Res. 2015, 21, 663–669. [Google Scholar] [CrossRef]
- Jin, G.; Jiranek, V.; Hayes, A.M.; Grbin, P.R. Isolation and Characterization of High-Ethanol-Tolerance Lactic Acid Bacteria from Australian Wine. Foods 2022, 11, 1231. [Google Scholar] [CrossRef] [PubMed]
- Virdis, C.; Sumby, K.; Bartowsky, E.; Jiranek, V. Lactic acid bacteria in wine: Technological advances and evaluation of their functional role. Front. Microbiol. 2021, 11, 612118. [Google Scholar] [CrossRef]
- Emer, C.D.; Marques, S.; Colla, L.M.; Reinehr, C.O. Biogenic amines and the importance of starter cultures for malolactic fermentation. Aust. J. Grape Wine Res. 2021, 27, 26–33. [Google Scholar] [CrossRef]
- Costantini, A.; Vaudano, E.; Pulcini, L.; Carafa, T.; Garcia-Moruno, E. An overview on biogenic amines in wine. Beverages 2019, 5, 19. [Google Scholar] [CrossRef]
- Li, B.; Lu, S. The importance of amine-degrading enzymes on the biogenic amine degradation in fermented foods: A review. Process Biochem. 2020, 99, 331–339. [Google Scholar] [CrossRef]



| UNQLh1.1 | UNQLh1.2 | UNQLh1.3 | |||||||
| 4 °C | 10 °C | 21 °C | 4 °C | 10 °C | 21 °C | 4 °C | 10 °C | 21 °C | |
| N0 (CFU/mL) | 2.8 × 107 | 2.8 × 107 | 3.0 × 107 | 2.4 × 107 | 2.6 × 107 | 2.8 × 107 | 2.6 × 107 | 2.0 × 107 | 2.7 × 107 |
| MAC (%) | 72.17 ± 1.30 c | 71.30 ± 5.21 b | 89.13 ± 5.22 cb | 79.17 ± 5.65 | 71.30 ± 5.22 d | 87.13 ± 1.30 d | 89.13 ± 6.09 | 71.30 ± 6.09 a | 93.91 ± 0.87 a |
| K | 0.15 ± 0.06 | 0.15 ± 0.06 | 0.29 ± 0.09 | 0.15 ± 0.06 | 0.15 ± 0.07 | 0.29 ± 0.09 | 0.08 ± 0.02 | 0.08 ± 0.03 | 0.19 ± 0.06 |
| R2 | 0.937 | 0.9364 | 0.9682 | 0.937 | 0.9364 | 0.9682 | 0.9693 | 0.9724 | 0.9666 |
| [MAi] | 0.73 | 0.76 | 0.45 | 0.73 | 0.76 | 0.45 | 0.00 | 0.41 | 0.28 |
| UNQLh2.1 | UNQLh2.3 | UNQLh3.1 | |||||||
| 4 °C | 10 °C | 21 °C | 4 °C | 10 °C | 21 °C | 4 °C | 10 °C | 21 °C | |
| N0 (CFU/mL) | 2.7 × 107 | 2.70 × 107 | 2.6 × 107 | 3.0 × 107 | 2.7 × 107 | 2.8 × 107 | 2.8 × 107 | 2.8 × 107 | 3.0 × 107 |
| MAC (%) | 76.52 ± 5.65 | 71.74 ± 1.74 c | 86.52 ± 0.43 c | 77.82 ± 4.87 | 76.52 ± 5.22 | 86.52 ± 0.87 | 93.48 ± 5.21 | 80.43 ± 1.30 c | 95.22 ± 5.22 c |
| K | 0.13 ± 0.07 | 0.17 ± 0.10 | 0.31 ± 0.08 | 0.07 ± 0.03 | 0.06 ± 0.01 | 0.22 ± 0.07 | 0.08 ± 0.02 | 0.06 ± 0.03 | 0.20 ± 0.07 |
| R2 | 0.9117 | 0.8751 | 0.9814 | 0.9649 | 0.9626 | 0.9606 | 0.9029 | 0.915 | 0.95 |
| [MAi] | 0.63 | 0.85 | 0.46 | 0.11 | 0.00 | 0.48 | 0.00 | 0.00 | 0.31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manera, C.; Rivas, G.A.; Flores, N.E.; Brizuela, N.S.; Caballero, A.C.; Semorile, L.C.; Valdes La Hens, D. Prevalence of Lentilacobacillus hilgardii over Lactiplantibacillus plantarum in Low-Temperature Spontaneous Malolactic Fermentation of a Patagonian Pinot Noir. Fermentation 2023, 9, 809. https://doi.org/10.3390/fermentation9090809
Manera C, Rivas GA, Flores NE, Brizuela NS, Caballero AC, Semorile LC, Valdes La Hens D. Prevalence of Lentilacobacillus hilgardii over Lactiplantibacillus plantarum in Low-Temperature Spontaneous Malolactic Fermentation of a Patagonian Pinot Noir. Fermentation. 2023; 9(9):809. https://doi.org/10.3390/fermentation9090809
Chicago/Turabian StyleManera, Camila, Gabriel Alejandro Rivas, Naiquen Elizabeth Flores, Natalia Soledad Brizuela, Adriana Carmen Caballero, Liliana Carmen Semorile, and Danay Valdes La Hens. 2023. "Prevalence of Lentilacobacillus hilgardii over Lactiplantibacillus plantarum in Low-Temperature Spontaneous Malolactic Fermentation of a Patagonian Pinot Noir" Fermentation 9, no. 9: 809. https://doi.org/10.3390/fermentation9090809
APA StyleManera, C., Rivas, G. A., Flores, N. E., Brizuela, N. S., Caballero, A. C., Semorile, L. C., & Valdes La Hens, D. (2023). Prevalence of Lentilacobacillus hilgardii over Lactiplantibacillus plantarum in Low-Temperature Spontaneous Malolactic Fermentation of a Patagonian Pinot Noir. Fermentation, 9(9), 809. https://doi.org/10.3390/fermentation9090809







