Abstract
Cassava plays an important role in the life and diet of several communities worldwide. It is used in various processed forms and has become the focus of research and innovation in recent years. Bioprocessing techniques, such as fermentation, have been utilized to create new avenues for its application in food and functional products. The fermentation of cassava can enhance its nutritional value, improve its shelf life, and increase its digestibility. However, the effect of salt-mediated fermentation on microbial diversity and potential benefits has not been widely reported. In this study, the effect of six levels of salt (NaCl), ranging from 0 to 25% after 50 days of spontaneous cassava fermentation at 25–30 °C, was investigated. A total of 30 bacterial isolates were selected for molecular characterization. A proprietary pool of universal 16S rRNA primers and species-specific primers were used to amplify a wide variety of species in clonal samples. The bacteria observed include Lactiplantibacillus, Limosilactobacillus, and Weissella. The population of Lactiplantibacillus in the cassava microflora increased with and without salt treatment, while Weissella, a new genus, was detected in 20 and 25% of salt-treated samples. Lactiplantibacillus and Weissella accounted for 32 and 68% and 60 and 40% of the bacterial populations detected at 20 and 25% salt treatment, respectively. A total of 17 strains and sequences were identified from the 30 isolates screened. Sequencing results classified the 30 isolates into four groups, of which 76.67% were Limosilactobacillus. Phylogenetic analysis showed that all 17 strains were divided into three clusters. These results show that high salt-mediated fermentation of cassava can trigger a shift in dominance in the bacterial community, changing the diversity of the microbial community.
1. Introduction
Cassava (Manihot esculenta Cranz/Manihot utillisima Phol) is an important root tuber native to South America, a major source of carbohydrates, and a food security crop in several countries [1]. It is the third most important crop in tropical regions after rice and maize. In Africa and other parts of the world, cassava production reached 193 and 302 million tons in 2020, respectively [2]. Cassava has been processed into food products such as fufu, gari, tapioca, and attieke in several West African countries [3]. Cassava is also increasingly becoming important due to its potential for novel applications in product development [4]. Traditional methods of processing cassava into food are dependent on root yield, cyanide and nutrient content, and processing capacity [1]. In most African countries, fermentation is important for the preparation of several products, including gari, lafun, and agbelima, using simple equipment at low production costs. Several vegetables, other starchy roots, meat, milk, cereals, legumes, and tea have been fermented to obtain products such as sauerkraut, kimchi, kefir, tempeh, kombucha, and cheese [5,6,7,8].
Solid-state fermentation (SSF) and submerged fermentation (SMF), as well as lactic acid, alkaline, and alcoholic fermentation, have been described [9,10], with varying impacts on the value and quality of the final product. Unlike inoculated fermentation, backslopping and spontaneous fermentation (without starter culture) are mediated by endogenous microbes. These fermentation techniques result in diverse microbial populations with distinct organoleptic and nutritional properties [8,11]. Fermentation has also been used to reduce the content of cyanogenic glucosides in cassava with the aid of linamarase [12]. This results in the release of hydrogen cyanide, detoxifying the cassava. It also improves product stability and texture, bio-concentrate proteins, vitamins, essential amino acids, and fatty acids, and reduces cooking [12,13,14]. Other studies have described the fermentation of food and agricultural wastes to produce value-added and innovative bioproducts [6,7,8,13]. Increasing consumer awareness of healthy living and the role of food is expanding the market, and demand for new foods with health benefits and fermented foods and beverages are among the current trends.
Fermentation is influenced by several factors, including pH, time, oxygen availability, salinity, selection of starter culture, inoculum concentration, microbial community, nutrients, and food/raw materials used [5,8,15]. Salt-mediated fermentation has been used to promote nutrient extraction for use by microbes, reduce excessive softening, inhibit the growth of spoilage bacteria, and extend the food shelf life [16,17,18]. It is a traditional method used in some regions to detoxify, preserve, enhance flavor, and improve the texture of cassava and products [19]. The salt creates an environment that promotes the growth of certain beneficial bacteria, yeasts, and other microorganisms, which contribute to fermentation and the development of unique flavors [20,21], as well as a hostile environment for spoilage-causing bacteria and molds, inhibiting their growth [22].
Cassava fermentation results in dynamic changes in the microbial community strain of bacteria and yeast, such as Bacillus, Leuconostoc, Klebsiella, Corynebacterium, Lactobacillus, and Candida, Aspergillus, have been isolated [6,9,13,23,24], as well as a variety of metabolites and other end products [6,8]. Culture-dependent and culture-independent metagenomic analysis (16S rRNA gene amplicon, whole genome sequencing), phenotypic detection, and RNA polymerase alpha subunit sequencing identification methods have been used in combination with other bioinformatic pipelines as reliable systems to identify and characterize the microbial community present during fermentation [25].
Although fermentation is known to cause significant changes in the physicochemical and functional properties of the raw material, attempts have seldom been made to study the influence of salt mediation on the diversity of the microbial community in fermented cassava. The aim of this work was to identify and characterize the microbiota present during cassava fermentation at different salt content using amplicon and bioinformatic analysis.
2. Materials and Methods
2.1. Sample Preparation
Cassava was purchased from the Produce Junction, Dover, DE. Food-grade sodium chloride (NaCl; all-purpose granulated) was purchased from a local grocer. Cassava was cleaned by washing under running tap water and manually chopped into pieces of approximately 0.1–0.2 cm2. About 400 g portions of chopped cassava were then treated by adding 20 g/5% (FCS20), 40 g/10% (FCS40), 60 g/15% (FCS60), 80 g/20% (FCS80), and 100 g/25% (FCS100) of salt in 64 oz glass mason jars with lids and bands. The samples were covered with equal amounts of water, lids and bands were replaced, and the samples were allowed to ferment naturally/spontaneously (without the addition of starter culture) for 50 days at ambient temperature (25–30 °C). Chopped cassava samples without salt treatment (FCS0) served as fermentation control. Unfermented fresh cassava samples without salt treatment served as process control. There was no control of environmental conditions such as temperature or relative humidity. After 50 days of fermentation, samples were taken and analyzed for microbial growth, and the remaining samples were frozen for subsequent analysis. The viable counts of LAB were determined using plate count technique on MRS and M17 agar (Oxoid, Basingstoke, UK). Yeast was enumerated on yeast chloramphenicol glucose agar (Difco Laboratories, Detroit). The acidity (pH and titratable acidity) of fermented cassava was determined as described [26].
2.2. Enumeration and Isolation of Microorganisms during Cassava Fermentation
Ten milliliters of fermented cassava sample each were homogenized in 90 mL sterile phosphate-buffered peptone water and serially diluted (10−1 to 10−9). For aerobic plate count, 10−9 dilutions were spread directly onto the surface of plate count agar and incubated at 35 °C for 48 h. LAB were enumerated by pour plating on MRS and M17 agar. Inoculated plates were then incubated anaerobically (AnaeroPack-Anaero, Tokyo, Japan) at 30 °C for 48 h. Following enumeration, higher dilution plates that contained 20 to 40 distinct colonies were selected, and each colony was assigned a number. Ten colonies from each of these plates, representing ≥ 25% of the colonies, were randomly selected and further purified by successive streaking on same agar medium used for enumeration.
2.3. Molecular Characterization and Identification of Bacterial Isolates
Three representative clones were randomly selected from each plate, and a total of 30 bacterial isolates were used for molecular characterization. Colony samples were submitted to GENEWIZ (South Plainfield, NJ, USA) and underwent a crude NaOH lysis to be directly used in polymerase chain reaction (PCR). PCR was performed according to standard operating procedures, and primer extension sequencing was performed by GENEWIZ using Applied Biosystems BigDye version 3.1 (Waltham, MA, USA). A proprietary pool of universal 16S rRNA primers and species-specific primers were used to amplify a wide variety of species in clonal samples. The reactions were later run on Applied Biosystems’s 3730xl DNA Analyzer. The primer set amplifies regions V1–V9 of the 16S rRNA gene (~1400 bp amplicon). Internal sequencing primers were utilized to generate a consensus sequence with the forward and reverse traces.
2.4. Construction of Phylogenetic Tree Construction
Alignment and phylogenetic reconstructions were performed using the function “build” of ETE3 v3.1.1 [27] as implemented on GenomeNet (https://www.genome.jp/tools/ete/, accessed on 28 February 2022). The tree was constructed using FastTree v2.1.8 with default parameters [28]. Values at nodes are SH-like local support. FastTree infers approximately maximum-likelihood phylogenetic trees from alignments of nucleotide or protein sequences. FastTree can handle alignments with up to a million sequences in a reasonable amount of time and memory. For large alignments, FastTree is 100–1000 times faster than PhyML 3.0 or RAxML 7. FastTree is more accurate than PhyML 3 with default settings and much more accurate than the distance-matrix methods that are traditionally used for large alignments. FastTree uses the Jukes–Cantor or generalized time-reversible (GTR) models of nucleotide evolution and the JTT [29], WAG [30], or LG [31] models of amino acid evolution. To account for the varying rates of evolution across sites, FastTree uses a single rate for each site (the “CAT” approximation). To quickly estimate the reliability of each split in the tree, FastTree computes local support values with the Shimodaira–Hasegawa test (these are the same as PhyML 3’s “SH-like local supports”).
2.5. RNA Structure Prediction
The RNA secondary structure was predicted through RNAfold web server. The RNAfold web server predicts secondary structures of single-stranded RNA sequences. Current limits are 7500 nt for partition function calculations and 10,000 nt for minimum free energy predictions. To predict a secondary structure, the RNAfold web server combines the following four separate prediction and analyzes algorithms: calculating a partition function, predicting a minimum free energy (MFE) structure, finding structures with maximum expected accuracy, and pseudoknot prediction. This server takes a sequence of RNA and creates a highly probable, probability-annotated group of secondary structures, starting with the lowest free energy structure and including others with varied probabilities of correctness. Other structures are included because the minimum free energy structure may not be the correct one. It is important to note that if SHAPE constraints are specified, they will be applied to the annotated structures, and this will lead to the generation of a second group of SHAPE constrained, which is distinct from the probability annotated structure group as described (http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi, accessed on 28 February 2022).
2.6. Similarity Analysis of 16S rRNA Sequences
The similarity matrix of rRNA was built based on the modified A-RISC Index Analysis method [32,33]. When comparing two 16S rRNA gene sequences, the compared RNA is divided into three groups: identical, similar, and dissimilar. Similar is defined as “similar” = “same” + “similar but not identical”. Only identical and similar amino acids are responsible for cross-reactivity. The following equation describes a subset of all amino acids that may interact with cross-reactive antibodies: I + S − I2 = I + S2. In addition, it provides the formula for calculation of the A-RISC index. The levels of relatedness obtained for the type strains ranging from 99 to 100% were high (Table 1). The levels of similarity for many type strains were >98.0% high, and their species status could not be confirmed based on this data alone. The 16S rRNAs of the subspecies are virtually identical (level of similarity, 99.9%). Values in the same range were also found when the sequences of the type strains were compared.

Table 1.
Colony species in salt-mediated fermented cassava.
3. Results
The pH of the control and fermented cassava samples (day 50) dropped from 6.94–6.20 to 4.41–3.81, respectively, indicating the acidification of the medium. Titratable acidity ranged from 0.027–0.036 to 0.006–0.25%, respectively.
3.1. Colony Sequencing and Similarity Detection Analysis
The BLAST results from the randomly selected and sequenced colony are presented in Table 1. Two bacterial genera, Lactiplantibacillus and Limosilactobacillus, were identified in the 0–15% salt-treated fermented cassava, with Lactiplantibacillus being dominant. In the control ferment (0%), 10%, and 15% samples, 100% of the colonies identified were Lactiplantibacillus. In the 5% salt-treated sample, 80% of the colonies identified were Lactiplantibacillus, and the remaining 20% were Limosilactobacillus.
Of the two bacterial genera identified in the 0–15% salt-treated fermented cassava, only Lactiplantibacillus was identified in the 20 and 25% salt treatments. As the salt concentration increased to 20 and 25%, an additional bacterial genus, Weissella, was detected, indicating high salt tolerance. To our knowledge, this is the first study that has identified Weissella in salt-mediated fermented cassava. The two bacterial genera found in the 20% salt-treated fermented cassava were Lactiplantibacillus (32%) and Weissella (78%). This was made up of four species of Lactiplantibacillus (L. fabifermentans, L. pentosus, L. plantarum, and L. paraplantarum) and eight species of Weissella (W. bombi, W. cibaria, W. confusa, W. hellenica, W. jogaejeotgali, W. paramesenteroides, W. thailandensis, and W. soli). A high population of Lactiplantibacillus (60%) was found in the 25% salt-treated fermented cassava, and the remaining 40% was identified as Weissella.
3.2. Colony Classification Based on Sequence Similarity
The sequencing results showed that the 30 isolates were classified into four groups (A–D) with identical features (Figure 1). Group A included 23 isolates (LC0001, LC0002, LC0003, LC0004, LC0005, LC0201, LC0202, LC0203, LC0204, LC0401, LC0402, LC0403, LC0404, LC0405, LC0601, LC0602, LC0603, LC0604, LC0605, LC0803, LC1002, LC1003, and LC1004). Group B, five isolates (LC0801, LC0802, LC0804, LC0805, and LC1001), and Groups C and D, one isolate each, LC1005 and LC0205, respectively. The results clearly showed that Group A is the dominant colony in the bacterial population in fermented cassava, with 23 of 30 isolates being of the same strain and accounting for 76.67% of all colonies. Groups B, C, and D accounted for 16.67, 3.33, and 3.33% of all the colonies, respectively.
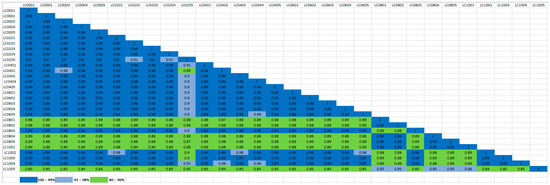
Figure 1.
Similarity analysis of different 16S rRNA copies. Different colors were used to represent different similarities, where green, light blue, and blue denote 80–90, 91–98, and 99–100% similarities, respectively.
3.3. Strain Identification
To identify the strain of these colonies, sequences of all 30 colonies were searched in the NCBI database. After sequence analysis, 17 strains were identified (Table 2), made up of two (2), one (1), five (5), and nine (9) kinds of strains from Groups A, B, C, and D isolates, respectively. The results clearly showed that the bacterial populations/strains in the four colony groups were from three different genera. All colonies in Group A were from Limosilactobacillus, which is a thermophilic and heterofermentative genus of LAB that was created from Lactiplantibacillus. Bacterial strains in Groups B and C were from a Gram-positive bacterium, Weissella, which was placed within the family Lactobacillaceae [34] and was formerly considered a species of the Leuconostoc paramesenteroides group, while bacterial strains in Group D were from Limosilactobacillus. The RNA type of all these strains was 16S ribosomal RNA.

Table 2.
List of strains identified in each group.
3.4. rRNA Structure and Phylogenetic Analysis
To confirm the bacterial strains identified, phylogenetic analysis was carried out using L. fabifermentans (NR_042676.1) as a reference strain. The results showed that all 17 strains clearly fell into three clusters (Figure 2). In cluster I, nine strains, NR_119069.1, NR_024994.1, NR_028810.1, NR_118032.1, NR_134066.1, NR_104927.1, NR_118978.1, NR_041566.1, and NR_029084.1 belonging to Limosilactobacillus were determined and were in Group D (Table 1 and Figure 2). Two strains; NR_042676.1 and NR_104573.1. associated with Limosilactobacillus bacteria were observed in cluster II and again were classified as Limosilactobacillus bacteria in Group A colonies (Table 1 and Figure 2). In cluster III, NR_025642.1, NR_036924.1, NR_040822.1, NR_104568.1, NR_136437.1, and NR_145896.1, were determined and found to be Weissella from Groups B and C (Table 1 and Figure 2). RNA secondary structure plays an important role in cell fate determination. Especially for non-coding RNAs, the secondary structure is very important for the RNA to function properly. RNA secondary structure plays an important regulatory function in the life activities of bacteria, fungi, viruses, and mammals. One of the applications of bioinformatics is to search the genomes to predict RNA secondary structures as functional forms of RNA rather than coding.
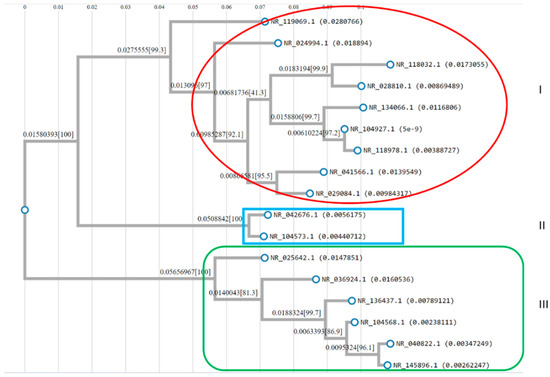
Figure 2.
Phylogenetic tree showing relationships between the 17 bacterial strains identified in fermented cassava. The red, blue, and green shapes correspond to cluster I, II, and III, respectively. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches.
To better understand the functional mechanism of those 17 16S rRNA identified, the secondary structure was further analyzed by RNAfold. The results showed that NR_104927.1, NR_041566.1, and NR_029084.1 found in cluster I had almost the same structures, including NR_134066.1 and NR_118978.1. They were all characterized by long stem-loop structures interrupted by small inner loops, with NR_118978 showing more bulges, while NR_118978.1 showed an un-typical clover structure 1 (Figure 3). RNA secondary structure plays an important role in cell fate determination, especially for non-coding RNAs, while the secondary structure is very important for the proper function of RNA. For instance, in cluster I, the stem and loop structure in NR_119069.1 and NR_024994.1 were similar, confirming the rRNA main structure.
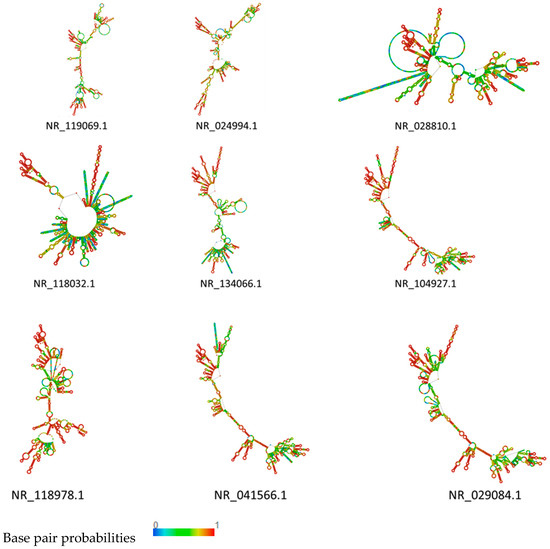
Figure 3.
Plain sequence centroid secondary structure of rRNA in cluster I.
In cluster I, NR_119069.1 and NR_024994.1 showed a similar structure. The stem and loop were the main structure in these two rRNA. The long stem-loop structures of NR_028810.1 and NR_118032.1 were interrupted by small inner loops characterized by several big bulges (Figure 3). In cluster II, the structure of NR_042676.1 and NR_104573.1 were very similar, indicating a typical structure of rRNA with long stem-loop structures interrupted by small inner loops and several bulges along the main stem. The only difference was the upper part, where more loops were predicted in NR_104573.1. In NR_042676, the stem showed a typical structure at the upper part (Figure 4). The rRNA in clusters I and II were compared to the structure of rRNA in cluster III and cluster III rRNA, which were characterized by more big bulges and long stem-loop structures interrupted by small inner loops and showed more diversity (Figure 5). A pseudoknot structure was predicted in NR_036924.1 (Figure 5), and, on further analysis and strain identification, all six strains in cluster III belong to Weissella. The rRNA structure analysis results confirmed the classification of the strain identification section.
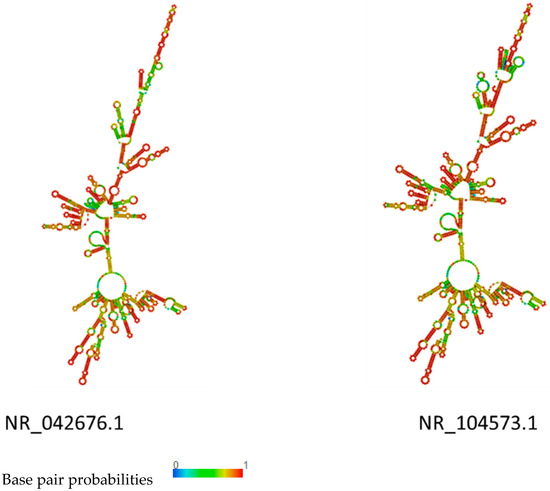
Figure 4.
Plain sequence centroid secondary structure of rRNA in cluster II.
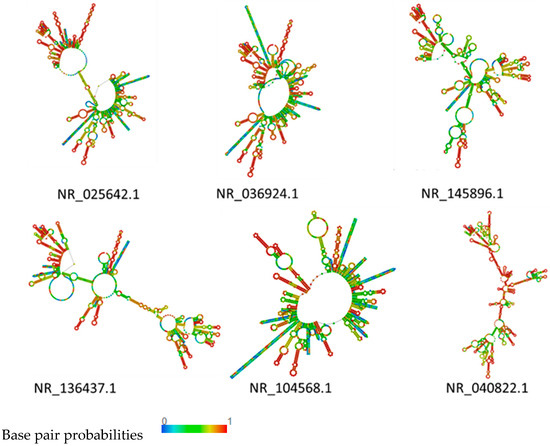
Figure 5.
Plain sequence centroid secondary structure of rRNA in cluster III.
4. Discussion
The study revealed insights of the microbial dynamics in salt-mediated fermented cassava. In recent years, the demand for high-quality and healthier food options has significantly increased among consumers. This growing awareness is driving the exploration of new uses of fermented cassava as both complete food and ingredients for functional food development. As part of food processing, physical and chemical treatments are employed to eliminate or reduce the presence of microorganisms in food products [4,18,35].
The significantly higher population of Lactiplantibacillus in both salt-treated and untreated samples compared to other microorganisms present in fermented cassava suggest that these LAB strains play a crucial role in cassava fermentation and are resilient in salt concentrations within the range tested. Salt content is an environmental factor that can significantly impact microbial growth and survival [18]. Different bacteria species and strains have varying abilities to tolerate or thrive in high-salt conditions. Some bacteria, such as the Limosilactobacillus genus, that stopped growing as salt concentration increased above 5% are likely more sensitive to elevated salt levels. High salt concentrations can affect the osmotic balance within cells, disrupt cellular processes, and impair the growth and metabolism of these salt-sensitive bacteria. On the other hand, the species that were favored or continued to grow at higher salt content (L. argentoratensis, L. paraplantarum, L. fabifermentans, L. paraplantarum, L. pentosus, and L. plantarum) are likely more halotolerant or capable of adapting to high salt conditions. They may possess mechanisms to regulate osmotic balance, transport and metabolize specific salts, or produce protective compounds against high salt stress [36,37]. Orike et al. (2018) observed a high tolerance in L. plantarum A2 and L. bulgaricus C2 to bile salts (0.4% w/v) and survival at low pH (pH 2) [38]. These characteristics are indicative of the potential of these strains to withstand the harsh conditions of the gastrointestinal tract and colonize the gut and have potential use in food fermentation, providing potential health benefits and promising probiotics. The presence of Lactiplantibacillus species in fermented cassava can contribute to the softening of cassava tubers and inhibit the growth of pathogens that may cause foul odor during fufu production [35].
The shift in microbial dominance when salt content reached 20%, with Weissella accounting for 68% of the total population and surpassing Lactiplantibacillus (32%), suggests that the former is a more halophilic and halotolerant bacterial strain at that salt content. The near doubling of the population of Lactiplantibacillus at 25% salt may be ascribed to the uniqueness of this bacteria, and further investigation is warranted. The change in microbial composition observed in response to change in salt content towards species that are better equipped to survive and thrive in the presence of elevated salt levels could be attributed to the effect of salt on the functional processes of the bacterial species and the selective pressure it exerts on the bacterial community. It is important to note that the response to salt concentration can vary among bacterial species and strains, and the specific mechanisms and adaptations involved can be complex and strain specific. Further studies into the specific mechanisms and genetic traits of the bacteria in question can provide more insights into their responses to salt stress.
Conversely, the absence of salt may have influenced the survival and growth of certain LAB strains, resulting in different dominant strains compared to our findings [39,40]. The acidification of the medium due to the drop in pH is a significant influence on the fermentation process. A drop in pH to 3.32 on the fourth day of cassava fermentation was previously reported [24] and this may explain the identification of 76.67% of strains belonging to the genus Limosilactobacillus. In another study, Limosilactobacillus reuteri was isolated and identified using 16S rRNA gene sequencing [41]. The L. reuteri PSC102 identified exhibited tolerance toward low pH and bile salt, resistant to gastrointestinal conditions and did not exhibit hemolytic or gelatinase activities or produce undesirable enzymes. Additionally, the bacteria also exhibited antioxidant and certain antibacterial activities. The ameliorative effect and probiotic properties of other species in this genus suggests their valuable contribution as potential probiotics for functional food and as alternative to synthetic antioxidants.
The clustering of the strains into three distinct groups during phylogenetic analysis identified specific strains within these dominant genera, and it is indicative of the significant effect of salt treatment on the microbial community in cassava fermentation. While it remains to be investigated, this observation provides the idea that the microbial ecology during cassava fermentation can be affected by salt concentration. This also suggests that genes related to nutrient and energy metabolism, as well as signal transduction were expressed [42]. The phylogenetic analysis also reveals the diversity detected during cassava fermentation, which reflects the real bacterial abundance when sequences are amplified. Other studies have found high to low or no redundant sequences [42,43]. Our results can be interpreted as relative bacterial diversity and may indicate limited bacterial diversity caused by selective influences of salt concentration.
This study also revealed six strains belonging to the genus Weissella in cluster III. Weissella bacteria are fermentative but can also be opportunistic pathogens. Members of the genus Weissella have been isolated from different sources, including fresh vegetables, fermented silage, meat, or meat products [44,45]. Their isolate possesses a high capacity for industrial use due to their ability to produce high amounts of dextran compared to other strains reported [45]. Their health-promoting effects, including probiotic and prebiotic, antioxidant, and antibacterial properties, as well as a wide range of industrial applications have also been reported [44,45,46]. The majority of LAB belonging to the genus Weissella have been introduced to wheat sourdough baking for in situ production of exopolysaccharides, but from this study, we determined that Weissella can also be produced from cassava using a high salt concentration. Based on molecular characterization, the strains identified in this study showed relationships with other bacteria strains. Further insights into the potentials of Weissella strains identified in this study to determine their biochemical characteristics and biotechnological application in functional food development and modification of techno-functional properties of foods and beverages are warranted.
The 16S rRNA structures indicate that the salt concentrations used in this study did not cause significant changes to the rRNA structure. This suggests that the functional mechanism of salt within the fermentation process was not impaired. The stability of the rRNA structure further supports the notion that salt concentration primarily influences the microbial composition rather than affecting the fundamental functional processes within the fermentation.
These findings contribute to our understanding of the microbial dynamics during cassava fermentation and highlight the importance of salt concentration in shaping the microbiota. Further studies in this area, including the optimization of salt concentration, can facilitate the production of high-quality and healthy fermented cassava-based foods and ingredients.
5. Conclusions
The spectra and characteristics of bacteria involved in submerged spontaneous cassava fermentation at different salt concentrations and the potential interactions between other bacteria were investigated using 16S rRNA analysis and bioinformatics. Both unsalted and salted (0–25%) treatments led to increased numbers of Lactiplantibacillus in the microbial communities exhibiting varying levels of tolerance. A new Weissella genus was detected in the 20 and 25% salt-mediated fermented cassava. Classification of the colonies based on sequence similarity revealed that all 17 isolates distinctly belonged to three clusters, of which 76.67% were Limosilactobacillus. 16S rRNA sequencing accurately distinguished Weissella species from Lactiplantibacillus and Limosilactobacillus strains. Weissella was resistant to high salt content during the fermentation period. The results showed that the phenotypic diversity of the bacteria flora during cassava fermentation depended on the amount of added salt. It also suggests that salt concentration plays a significant role in shaping the composition of the microbial community during cassava fermentation and highlights the importance of considering salt as a factor in optimizing fermentation processes. This study provides a good reference for the identification and characterization of microbiota in salt-mediated cassava fermentation and other similar materials. Fermented cassava (20 and 25% salt) could be considered a potential source for the genus Weissella.
Author Contributions
W.Z.: data curation, formal analysis; A.N.A.A.: conceptualization, funding acquisition, supervision, writing—original draft, and writing—review and editing; A.A.: writing—review and editing; D.O.U.: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported in part by the U.S. Department of Agriculture, National Institute of Food and Agriculture (Grant #2021-67022-34148).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Acknowledgments
The authors would like to thank James Owusu-Kwarteng and Christabel Aheto for their help with the experiment.
Conflicts of Interest
The authors declare that the study was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Falade, K.O.; Akingbala, J.O. Utilization of cassava for food. Food Rev. Int. 2010, 27, 51–83. [Google Scholar] [CrossRef]
- FAOSTAT. Food and Agriculture Organization of the United Nations (FAO); FAOSTAT: Rome, Italy, 2020. [Google Scholar]
- Awoyale, W.; Alamu, E.O.; Chijioke, U.; Tran, T.; Takam Tchuente, H.N.; Ndjouenkeu, R.; Kegah, N.; Maziya-Dixon, B. A review of cassava semolina (gari and eba) end-user preferences and implications for varietal trait evaluation. Int. J. Food Sci. Technol. 2021, 56, 1206–1222. [Google Scholar] [CrossRef] [PubMed]
- Komen, J.; Tripathi, L.; Mkoko, B.; Ofosu, D.O.; Oloka, H.; Wangari, D. Biosafety regulatory reviews and leeway to operate: Case studies from sub-Sahara Africa. Front. Plant Sci. 2020, 11, 130. [Google Scholar] [PubMed]
- Dimidi, E.; Cox, S.R.; Rossi, M.; Whelan, K. Fermented foods: Definitions and characteristics, impact on the gut microbiota and effects on gastrointestinal health and disease. Nutrients 2019, 11, 1806. [Google Scholar] [CrossRef]
- Jimenez, M.E.; O’Donovan, C.M.; de Ullivarri, M.F.; Cotter, P.D. Microorganisms present in artisanal fermented food from South America. Front. Microbiol. 2022, 13, 941866. [Google Scholar] [CrossRef]
- Owusu-Kwarteng, J.; Agyei, D.; Akabanda, F.; Atuna, R.A.; Amagloh, F.K. Plant-Based Alkaline Fermented Foods as Sustainable Sources of Nutrients and Health-Promoting Bioactive Compounds. Front. Sustain. Food Syst. 2022, 6, 885328. [Google Scholar] [CrossRef]
- Aryee, A.N.A.; Owusu-Kwarteng, J.; Senwo, Z.; Alvarez, M.N. Characterizing fermented habanero pepper (Capsicum chinense L). Food Chem. Adv. 2022, 1, 100137. [Google Scholar] [CrossRef]
- Hawashi, M.; Widjaja, T.; Gunawan, S. Solid-State Fermentation of Cassava Products for Degradation of Anti-Nutritional Value and Enrichment of Nutritional Value. In New Advances on Fermentation Processes; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef]
- Seesaard, T.; Wongchoosuk, C. Recent Progress in Electronic Noses for Fermented Foods and Beverages Applications. Fermentation 2022, 8, 302. [Google Scholar] [CrossRef]
- Theron, C. The Advantages and Disadvantages of Spontaneous Fermentation—Wineland Media; Wineland Media: Paarl, South Africa, 2021. [Google Scholar]
- Sivamaruthi, B.S.; Kesika, P.; Chaiyasut, C. Toxins in fermented foods: Prevalence and preventions—A mini review. Toxins 2018, 11, 4. [Google Scholar] [CrossRef]
- El Sheikha, A.F.; Ray, R.C. Bioprocessing of horticultural wastes by solid-state fermentation into value-added/innovative bioproducts: A review. Food Rev. Int. 2022, 39, 1–57. [Google Scholar]
- Boonnop, K.; Wanapat, M.; Nontaso, N.; Wanapat, S. Enriching nutritive value of cassava root by yeast fermentation. Sci. Agric. 2009, 66, 629–633. [Google Scholar] [CrossRef]
- Kaczmarska, K.; Taylor, M.; Piyasiri, U.; Frank, D. Flavor and Metabolite Profiles of Meat, Meat Substitutes, and Traditional Plant-Based High-Protein Food Products Available in Australia. Foods 2021, 10, 801. [Google Scholar] [CrossRef] [PubMed]
- Frediansyar, A.; Kurniadi, M. Comparative influence of salinity and temperature on cassava flour product by Lactobacillus plantarum and Lactobacillus acidophilus during single culture fermentation. Nusant. Biosci. 2016, 8, 207–214. [Google Scholar] [CrossRef]
- Khanna, S. DigitalCommons@UMaine Effects of Salt Concentration on the Physicochemical Properties and Microbial Safety of Spontaneously Fermented Cabbage; The University of Maine: Orono, ME, USA, 2018. [Google Scholar]
- Barcenilla, C.; Álvarez-Ordóñez, A.; López, M.; Alvseike, O.; Prieto, M. Microbiological safety and shelf-life of low-salt meat products—A Review. Foods 2022, 11, 2331. [Google Scholar] [CrossRef] [PubMed]
- Meryandini, A.; Melani, V.; Sunarti, T.C. Addition of cellulolytic bacteria to improved the quality of fermented cassava flour. Afr. J. Food Sci. Technol. 2011, 2, 30–35. [Google Scholar]
- Anal, A.K. Quality ingredients and safety concerns for traditional fermented foods and beverages from Asia: A review. Fermentation 2019, 5, 8. [Google Scholar] [CrossRef]
- Tamang, J.P.; Cotter, P.D.; Endo, A.; Han, N.S.; Kort, R.; Liu, S.Q.; Mayo, B.; Westerik, N.; Hutkins, R. Fermented foods in a global age: East meets West. Compr. Rev. Food Sci. Food Saf. 2020, 19, 184–217. [Google Scholar] [CrossRef]
- Li, M.; Zhang, L.; Zhang, Q.; Zi, X.; Lv, R.; Tang, J.; Zhou, H. Impacts of citric acid and malic acid on fermentation quality and bacterial community of cassava foliage silage. Front. Microbiol. 2020, 11, 595622. [Google Scholar] [CrossRef]
- Balogun, O.B.; Adeleke, B.S.; Owoseni, I. Characterization of bacteria isolates from fermented cassava steeping water. Int. J. Appl. Biol. 2021, 5, 190–199. [Google Scholar]
- Flibert, G.; Abel, T.; Aly, S. African cassava Traditional Fermented Food: The Microorganism’s Contribution to their Nutritional and Safety Values—A Review. Int. J. Curr. Microbiol. Appl. Sci. 2016, 5, 664–687. [Google Scholar] [CrossRef]
- Rapoo, S.M.; Budeli, P.; Thaoge, M.L. Recovery of Potential Starter Cultures and Probiotics from Fermented Sorghum (Ting) Slurries. Microorganisms 2023, 11, 715. [Google Scholar] [PubMed]
- Oyeyinka, S.A.; Adeloye, A.A.; Olaomo, O.O.; Kayitesi, E. Effect of fermentation time on physicochemical properties of starch extracted from cassava root. Food Biosci. 2020, 33, 100485. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef]
- Koch, C.; Kroppenstedt, R.M.; Stackebrandt, E. Intrageneric relationships of the actinomycete genus Micromonospora. Int. J. Syst. Evol. Microbiol. 1996, 46, 383–387. [Google Scholar] [CrossRef]
- Chruszcz, M.; Kapingidza, A.B.; Dolamore, C.; Kowal, K. A robust method for the estimation and visualization of IgE cross-reactivity likelihood between allergens belonging to the same protein family. PLoS ONE 2018, 13, e0208276. [Google Scholar]
- Björkroth, J.; Holzapfel, W. Genera Leuconostoc, Oenococcus and Weissella. Prokaryotes 2006, 4, 267–319. [Google Scholar]
- Adesulu-Dahunsi, A.T.; Dahunsi, S.O.; Ajayeoba, T.A. Co-occurrence of Lactobacillus Species During Fermentation of African Indigenous Foods: Impact on Food Safety and Shelf-Life Extension. Front. Microbiol. 2022, 13, 684730. [Google Scholar] [PubMed]
- Papadimitriou, K.; Alegría, Á.; Bron, P.A.; De Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C. Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 837–890. [Google Scholar] [CrossRef] [PubMed]
- Yap, P.-C.; MatRahim, N.-A.; AbuBakar, S.; Lee, H.Y. Antilisterial potential of lactic acid bacteria in eliminating Listeria monocytogenes in host and ready-to-eat food application. Microbiol. Res. 2021, 12, 234–257. [Google Scholar]
- Orike, E.L.; Adeyemo, S.M.; Omafuvbe, B.O. Probiotic potentials of lactic acid bacteria isolated from fermenting cassava. Int. J. Probiot. Prebiot. 2018, 13, 69–76. [Google Scholar]
- Pang, H.; Xin, X.; He, J.; Cui, B.; Guo, D.; Liu, S.; Yan, Z.; Liu, C.; Wang, X.; Nan, J. Effect of NaCl Concentration on Microbiological Properties in NaCl Assistant Anaerobic Fermentation: Hydrolase Activity and Microbial Community Distribution. Front. Microbiol. 2020, 11, 589222. [Google Scholar] [CrossRef]
- Leska, A.; Nowak, A.; Rosicka-Kaczmarek, J.; Ryngajłło, M.; Czarnecka-Chrebelska, K.H. Characterization and Protective Properties of Lactic Acid Bacteria Intended to Be Used in Probiotic Preparation for Honeybees (Apis mellifera L.)—An In Vitro Study. Animals 2023, 13, 1059. [Google Scholar] [CrossRef]
- Ali, M.S.; Lee, E.-B.; Lim, S.-K.; Suk, K.; Park, S.-C. Isolation and Identification of Limosilactobacillus reuteri PSC102 and Evaluation of Its Potential Probiotic, Antioxidant, and Antibacterial Properties. Antioxidants 2023, 12, 238. [Google Scholar] [CrossRef]
- Li, M.; Zhou, H.; Pan, X.; Xu, T.; Zhang, Z.; Zi, X.; Jiang, Y. Cassava foliage affects the microbial diversity of Chinese indigenous geese caecum using 16S rRNA sequencing. Sci. Rep. 2017, 7, 45697. [Google Scholar] [CrossRef]
- Zhang, Y.; Lyu, J.; Ge, L.; Huang, L.; Peng, Z.; Liang, Y.; Zhang, X.; Fan, S. Probiotic Lacticaseibacillus rhamnosus GR-1 and Limosilactobacillus reuteri RC-14 as an Adjunctive Treatment for Bacterial Vaginosis Do Not Increase the Cure Rate in a Chinese Cohort: A Prospective, Parallel-Group, Randomized, Controlled Study. Front. Cell. Infect. Microbiol. 2021, 11, 669901. [Google Scholar] [CrossRef]
- Sharma, N.; Gupta, D.; Park, Y.-S. Genome analysis revealed a repertoire of oligosaccharide utilizing CAZymes in Weissella confusa CCK931 and Weissella cibaria YRK005. Food Sci. Biotechnol. 2023, 32, 553–564. [Google Scholar] [CrossRef]
- Nachtigall, C.; Hassler, V.; Wefers, D.; Rohm, H.; Jaros, D. Dextrans of Weissella cibaria DSM14295: Microbial production, structure and functionality. Int. J. Biol. Macromol. 2023, 246, 125631. [Google Scholar] [CrossRef] [PubMed]
- Kiran, F.; Demirhan, H.K.; Haliscelik, O.; Zatari, D. Metabolic profiles of Weissella spp. postbiotics with anti-microbial and anti-oxidant effects. J. Infect. Dev. Ctries. 2023, 17, 507–517. [Google Scholar] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).