Abstract
Muscular atrophy is a chronic muscle disease characterized by a loss of muscle mass and muscle weakness due to excessive protein breakdown relative to protein synthesis. Apoptosis is a major factor in sarcopenia and the final stage of muscle atrophy that occurs via various mechanisms. In this study, we evaluated the protective effects of cell-free supernatants (CFSs) from different lactic acid bacteria (LAB) strains in dexamethasone (DEX)-treated C2C12 cells, followed by probiotic properties. We found that Limosilactobacillus fermentum (L. fermentum) MG4263 and MG5091 and Lactococcus lactis (Lc. lactis) MG4668 and MG5474 inhibited muscle atrophy F-box (atrogin-1) and muscle-specific RING-finger protein-1 (MuRF-1) in DEX-treated C2C12 cells. In addition, LAB strains inhibited the expression of apoptotic proteins, such as Bcl-2-associated X (Bax)/Bcl-2 and caspase-3 in DEX-treated C2C12 cells. L. fermentum MG5091, Lc. lactis MG4668, and MG5474 showed high survival rates in gastrointestinal (GIT) conditions and high adhesion rate to HT-29 cells. The LAB strains were also assessed for hemolysis and toxicity in HT-29 cells to confirm their stability. The LAB strains showed no hemolytic activity and toxicity to HT-29 cells. Therefore, L. fermentum MG5091, Lc. lactis MG4668, and MG5474 suggest their potential as probiotics to be used as functional foods for the inhibition of muscular atrophy.
1. Introduction
The elderly population is rapidly increasing worldwide due to aging [1]. Loss of skeletal muscle mass and muscle strength is the most prominent feature of aging in humans. Skeletal muscle is the largest tissue in the body and the most important protein reservoir [2]. Maintaining appropriate muscle protein levels is important to perform normal body functions [3]. Skeletal muscle mass is determined by the balance between muscle protein synthesis and breakdown [4]. Imbalance in protein metabolism leads to sarcopenia, which is a type of muscle atrophy characterized by a loss of skeletal muscle mass and function, physical disability, and reduced quality of life [5]. The World Health Organization designates sarcopenia as an official disease that is associated with excessive protein degradation due to aging and various chronic diseases, such as cancer, chronic obstructive pulmonary disease, and chronic heart disease [6,7]. Sarcopenia causes a loss in strength and muscle mass and increases the risk of physical disability and mortality in affected patients [8]. The incidence of sarcopenia is positively correlated with age, suggesting the need for medical and social services for the elderly; it further increases after the age of 50 years (from 5–13% at age 70 to 11–50% at age 80) [9]. Interestingly, sarcopenia exhibits a two-fold higher prevalence in females than in males, detrimentally affecting both the quality of life and lifespan of the affected individuals [10].
Lactic acid bacteria (LAB) are living microorganisms; in particular, some species, including Lactobacillus and Lactococcus, are used as probiotics and adhere to the intestinal epithelial cells to remove harmful microorganisms and exert beneficial effects on the host [11,12]. The intricate interplay between the gut microbiota and host physiological homeostasis is crucial for the regulation of various essential processes, including the modulation of nutrient absorption, control of inflammatory responses, management of oxidative stress, regulation of immune function, and maintenance of anabolic balance in the body [13]. Although many reports have demonstrated the impact of LAB on intestinal health, recent studies have highlighted their potential in alleviating diabetes, obesity, heart disease, and liver disease [14]. Recent studies have also revealed the potential benefits of LAB in promoting gut microbiome balance and skeletal muscle growth [15]. Intestinal dysbiosis and consequent changes in the microbiome can lead to significant changes in skeletal muscle metabolism [16]. Consuming probiotics is one of the solutions that can restore gut microbiome balance and prevent gut microbiome imbalance [17]. Lactiplantibacillus plantarum, Limosilactobacillus reuteri, Lactobacillus gasseri, and Lacticaseibacillus casei have been reported to restore muscle-related parameters [18,19,20]. Although various LAB strains inhibit skeletal muscle atrophy, studies on some species are limited. Therefore, in this study, we aimed to evaluate the efficacies of various LAB strains in inhibiting dexamethasone (DEX)-induced skeletal muscle atrophy.
2. Materials and Methods
2.1. Preparation of Cell-Free Supernatants (CFSs) from Lactobacillus and Lactococcus Strains
Limosilactobacillus fermentum (L. fermentum) MG4263, MG5091, and MG5159 and Lactococcus lactis (Lc. lactis) MG4668, MG5049, MG5052, and MG5474 were obtained from MEDIOGEN (Jecheon, Republic of Korea). All LAB strains were cultured in the de Man, Rogosa and Sharp (MRS) broth (BD Bioscience, Franklin Lakes, NJ, USA) at 37 °C for 24 h. Then, the LAB strains were adjusted to an OD600 of 1.0 (108–109 CFU/mL) and sub-cultured at 37 °C for 18 h. CFS was obtained via centrifugation at 4000× g for 15 min at 4 °C and filtered using a 0.22 μm polytetrafluoroethylene membrane filter (ADVANTEC, Tokyo, Japan). All LAB strains were confirmed via 16S rRNA gene sequencing (SolGent Co., Ltd., Daejeon, Republic of Korea) and registered on the National Center for Biotechnology Information database using the Basic Local Alignment Search Tool (Table 1).

Table 1.
Accession numbers and origins of the LAB strains used in this study.
2.2. Cell Culture and Myotube Differentiation
C2C12 cells (American Type Culture Collection, Manassas, VA, USA) were cultured in the Roswell Park Memorial Institute-1640 medium (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin (P/S; Gibco) at 37 °C and 5% CO2. C2C12 cells were cultured in a cell culture plate (1 × 105 cells/mL) for 24 h. To induce C2C12 cell differentiation into myotubes, the medium was replaced with a differentiation medium containing 2% horse serum and 1% P/S after five days. The differentiation medium was replaced every two days.
2.3. Cell Viability
Briefly, C2C12 cells were grown in a 96-well plate with the growth medium for 24 h, after which the medium was changed to a differentiation medium. After five days, cells were treated with 10% CFS of LAB and DEX (100 μM; Sigma-Aldrich, St. Louis, MO, USA) for 24 h. Cytotoxicity of the LAB strains was determined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay [21]. MTT solution (0.25 mg/mL; Sigma-Aldrich) was added for 2 h. The formazan product was dissolved in dimethyl sulfoxide. Absorbance was measured at 550 nm using a microplate reader (BioTek, Winooski, VT, USA).
2.4. Western Blotting
C2C12 cells were seeded in a 12-well plate with the growth medium for 24 h, after which the medium was changed to differentiation medium. After five days, the cells were treated with CFS and exposed to DEX, as described in Section 2.3. Whole-cell lysates were prepared as previously described [22]. Cells were subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and proteins were transferred to polyvinylidene fluoride membranes (0.45 μm; GenDEPOT, Katy, TX, USA). The membrane was washed thrice with Tris-buffered saline with 0.1% Tween-20 (TBS-T; GenDEPOT) and blocked with smart-block 5 min fast blocking buffer (Biomax, Guri-si, Gyeonggi-do, Korea). After blocking, the membrane was incubated with the muscle atrophy F-box (MAFbx/atrogin-1; Santa Cruz Biotechnology, Dallas, TX, USA), muscle-specific RING-finger protein-1 (MuRF-1; Santa Cruz Biotechnology), Bcl-2-associated X (Bax; Santa Cruz Biotechnology), Bcl-2 (Santa Cruz Biotechnology), glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Santa Cruz Biotechnology), pro-caspase 3 (Cell Signaling Technology Beverly, MA, USA), and cleaved-caspase-3 (Cell Signaling Technology) primary antibodies diluted to 1:1000 overnight at 4 °C. After washing thrice with TBS-T, the membranes were incubated with secondary antibodies (1:5000 dilution; GenDEPOT) for 1 h at room temperature. Western blotting images were visualized using LuminoGraph III Lite (ATTO, Tokyo, Japan) with EzWestLumi plus mid-femto ECL solution (ATTO). Quantitative analysis was performed using ImageJ software (version 1.52a for Windows; NIH, Rockville, MD, USA).
2.5. Morphological Characterization of LAB Strains
Morphological characteristics of the LAB strains were determined using scanning electron microscopy (SEM; Hitachi S-4300SE, Tokyo, Japan), as previously described [23]. The cells were observed at 10,000× magnification.
2.6. Characterization of Carbohydrate Fermentation
To assess the availability of carbohydrates, the LAB strains were cultured on MRS agar (Difco, Sparks, MD, USA) for 18 h at 37 °C. Then, the carbohydrate fermentation profiles of the LAB strains were determined using the API 50 CHL Kit (BioMerieux, Craponne, France), according to the manufacturer’s instructions. The extent of substrate hydrolysis was evaluated based on the intensity of color.
2.7. Hemolyltic Activity
Hemolytic activities of the LAB strains were assessed using Columbia agar (Oxoid, Basingstoke, UK) supplemented with 5% sheep blood (MB cell, Seoul, Republic of Korea). LAB strains were streaked onto agar plates and incubated at 37 °C for 48 h. Assessment of hemolytic activity involved the examination of red blood cell lysis in the vicinity of bacterial colonies on agar plates. Hemolysis was categorized into three types based on distinct visual characteristics: α-hemolysis: characterized by the presence of a greenish zone around the colonies, indicating partial lysis of red blood cells; β-hemolysis: characterized by a transparent zone, indicating the complete destruction of red blood cells; and γ-hemolysis: characterized by no noticeable zone, suggesting the complete absence of hemolysis [24].
2.8. Lactate Dehydrogenase (LDH) Release
Human colon adenocarcinoma (HT-29) cells (Korean Cell Line Bank, Seoul, Republic of Korea) were cultured in Dulbecco’s modified Eagle’s medium (Gibco) containing 10% FBS and 1% P/S at 37 °C in 5% CO2. Cytotoxicity of the LAB strains was determined using the Quanti-LDH PLUS Cytotoxicity Assay Kit (Biomax, Seoul, Republic of Korea). HT-29 cells were cultured in a 96-well plate at a density of 2.5 × 104 cells/well. Subsequently, the LAB strains were treated with 106, 107, or 108 CFU/mL for 24 h. The culture medium (50 μL) was transferred to a fresh 96-well plate, and 50 μL of LDH substrate mix was added. After 30 min of incubation at 37 °C, the absorbance was measured at 450 nm using a microplate reader. LDH release was calculated using the following equation:
LDH (%) = (Sample − Low control)/(High control − Background control) × 100
Low control: cell culture supernatant of cells only; high control: cell culture supernatant of cells after lysis; background control: medium only.
2.9. Survival in the Gastrointestinal Tract (GIT)
Survival of LAB strains under conditions resembling the GIT environment was evaluated as previously described [25]. For simulated gastric fluid (SGF), LAB strains were treated with phosphate-buffered saline (PBS; pH 2.5) and 0.3% pepsin (Sigma-Aldrich) for 2 h at 37 °C. For simulated intestinal fluid (SIF), LAB strains were treated with PBS (pH 7.4) and 1% pancreatin-bile salt (Sigma-Aldrich) for 2.5 h at 37 °C. Following incubation, the viability of LAB strains was determined via live colony counts on the MRS agar plates.
2.10. Adhesion Assay on Intestinal Epithelial Cells
To evaluate the adhesion of the LAB strains to HT-29 cells, an adhesion assay was performed, as previously reported, with some modifications [26]. Briefly, HT-29 cells were seeded at 1.5 × 105 cells/well in a 12-well plate until a cellular monolayer was formed. Then, the cells were treated with LAB (1 × 108 CFU/mL) for 2 h, washed thrice, and lysed with 1 mL of PBS. The adhesion rate (%) was calculated by comparing the number of adherent cells to the initial number of viable cells using the following equation: log (adherent counts) CFU/mL/log (initial counts) CFU/mL × 100.
2.11. Statistical Analysis
All results are expressed as the mean ± standard error of three independent experiments. Statistical analysis was conducted using the Student’s t-test with Prism 5.02 software (GraphPad Software, San Diego, CA, USA). Statistical significance was set at p < 0.05.
3. Results
3.1. Cytotoxicity of the LAB Strains in DEX-Treated C2C12 Cells
DEX treatment significantly reduced the cell viability in a concentration-dependent manner (Figure 1A). After determining that 200 μM DEX drastically decreased the cell viability, subsequent experiments were performed using 100 μM DEX. As shown in Figure 1B, 10% CFS of all LAB strains exhibited no cytotoxicity (≥0.91-fold of the control). As shown in Figure 1C, L. fermentum MG4263, MG5091, and MG5159 and Lc. lactis MG4668, MG5049, MG5052, and MG5474 restored the cell viability (0.66–0.82-fold of the control) of DEX-treated C2C12 cells. Based on their cytotoxicity in DEX-treated C2C12 cells, four LAB strains (L. fermentum MG4263 and MG5091 and Lc. lactis MG4668 and MG5474) were selected to confirm muscle atrophy protein expression.
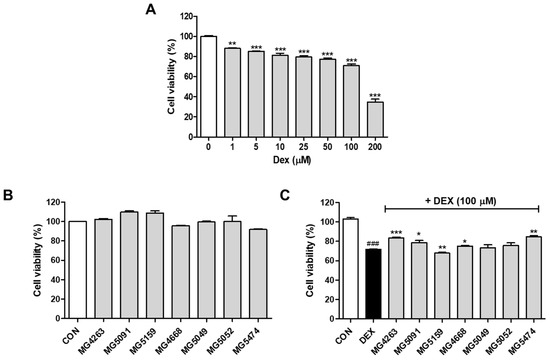
Figure 1.
Cytotoxicity of the cell-free supernatants (CFSs) of different LAB strains with or without dexamethasone (DEX) treatment. C2C12 cells were treated with CFS of LAB strains (0–200 μM; (A)). Cell viability with 10% CFS without DEX (B) and with DEX (C). Data were analyzed using the Student’s t-test. ### p < 0.001 vs. CON and * p < 0.05, ** p < 0.01, and *** p < 0.01 vs. DEX treatment.
3.2. Inhibitory Effects of the LAB Strains on Muscle Atrophy Protein Expression Levels in DEX-Treated C2C12 Cells
Western blotting was performed to confirm the effects of CFSs from the selected LAB strains on muscle atrophy protein expression. Protein expression levels of MuRF-1 and atrogin-1 were significantly increased in DEX-treated C2C12 cells compared to the control cells (Figure 2). Expression levels of MuRF-1 were significantly decreased by all LAB strains (0.39–0.57-fold of DEX treatment; Figure 2A). In addition, the expression levels of atrogin-1 were significantly decreased by all LAB strains (0.47–0.80-fold of DEX treatment, p < 0.01), except L. fermentum MG4263 (Figure 2B).
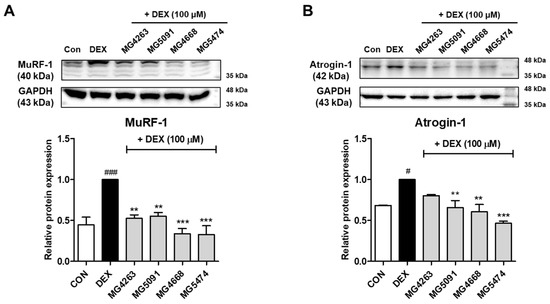
Figure 2.
LAB strains inhibit muscle-specific RING-finger protein-1 (MuRF-1) (A) and muscle atrophy F-box (MAFbx/atrogin-1) (B) protein expression in DEX-treated C2C12 cells. Protein expression is normalized to that of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Data were analyzed using the Student’s t-test. # p < 0.05 and ### p < 0.001 vs. CON and ** p < 0.01 and *** p < 0.01 vs. DEX treatment.
3.3. Inhibitory Effects of the LAB Strains on Apoptotic Protein Expression Levels in DEX-Treated C2C12 Cells
To assess the inhibitory effects of CFS on apoptotic protein expression, the expression levels of Bax/Bcl-2 and caspase-3 were determined in DEX-treated C2C12 cells (Figure 3). DEX treatment significantly upregulated the expression levels of Bax/Bcl-2 and cleaved-caspase-3. Expression levels of Bax/Bcl-2 were reduced by LAB strains (0.43–0.83-fold of DEX treatment), except L. fermentum MG4263 (Figure 3A). In addition, the expression levels of cleaved-caspase-3 were significantly decreased by all LAB strains (0.38–0.67-fold of DEX treatment; p < 0.05; Figure 3B).
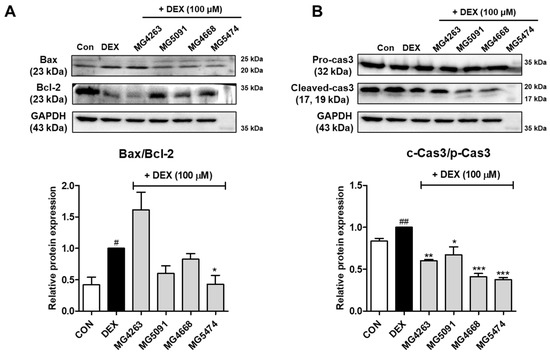
Figure 3.
LAB strains inhibit Bax/Bcl-2 (A) and caspase-3 (B) expression in DEX-treated C2C12 cells. Protein expression is normalized to that of GAPDH. Data were analyzed using the Student’s t-test. # p < 0.05 and ## p < 0.01 vs. CON and * p < 0.05, ** p < 0.01, and *** p < 0.001 vs. DEX treatment.
3.4. Identification of the LAB Strains
The selected LAB strains were further assessed using SEM, which confirmed that all strains were in the form of bacilli (L. fermentum MG5091) and cocci (Lc. lactis MG4668 and MG5474, Figure 4). Evaluation of the carbohydrate fermentation profiles of the LAB strains using the API web software revealed that they all used L-arabinose, D-ribose, D-galactose, D-glucose, D-fructose, esculin, D-maltose, D-melibiose, D-sucrose, D-trehalose, and D-raffinose (Table 2). Each strain had a different ability to use residual carbohydrates, and the strains were identified based on 16S rRNA gene sequencing results.
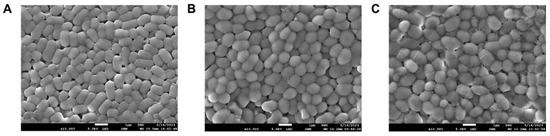
Figure 4.
Scanning electron microscopy (SEM) micrographs of the Limosilactobacillus fermentum MG5091, (A) Lactococcus lactis MG4668 (B), and Lc. lactis MG5474 (C) at 10,000× magnification.

Table 2.
Carbohydrate fermentation profiles of the LAB strains.
3.5. Cytotoxicity of the LAB Strains in HT-29 Cells
To determine the safety of L. fermentum MG5091 and Lc. lactis MG4668 and MG5474, which inhibited muscle atrophy protein expression in C2C12 cells, their cytotoxicity was assessed in HT-29 cells. L. fermentum MG5091 and Lc. lactis MG4668 and MG5474 exhibited no cytotoxicity (0.95–1.15-fold of control) in HT-29 cells up to 1 × 108 CFU/mL (Figure 5).
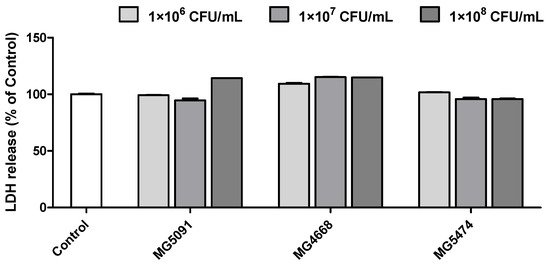
Figure 5.
Cytotoxicity of the LAB strains in HT-29 cells. HT-29 cells were treated with LAB strains (106–108 CFU/mL). Cytotoxicity was subsequently confirmed via LDH release.
3.6. Viability in Simulated Gastrointestinal Fluid and Adhesion of LAB Strains to HT-29 Cells
To confirm the maintenance of viability during gastrointestinal transit, viability of the selected strains was assessed by culturing them in SGF and SIF (Table 3). The viability of the strains in SGF ranged from 36.6 to 98.2%. L. fermentum MG5091 and Lc. lactis MG4668 and MG5474 exhibited viabilities ranging from 37.5 to 96.8% in SIF. L. fermentum MG5091 showed higher survival rates than Lc. lactis MG4668 and MG5474. Next, we confirmed the ability of these cells to adhere to intestinal epithelial cells. L. fermentum MG5091 and Lc. lactis MG4668 and MG5474 adhered to HT-29 cells at 74.0–89.7% compared to the initial count (Table 3).

Table 3.
Survival rates of LAB strains in simulated gastrointestinal fluid and their adhesion to HT-29 cells.
4. Discussion
With the global population transitioning to a super-aged society, the interest in geriatric diseases, including sarcopenia, hypertension, and degenerative neurological disorders, is rapidly increasing. Muscular atrophy is a chronic progressive muscle disease that reduces the quality of life of affected patients and is characterized by a loss of muscle mass, muscle weakness, and disability due to excessive protein breakdown relative to protein synthesis [13]. Therefore, maintenance of muscle health, the principal reservoir of proteins within the body, promotes healthy aging, ensures sufficient energy supply, and alleviates the risk of metabolic disorders [2,27]. Understanding and managing this condition are crucial for improving the quality of life of individuals with muscular atrophy worldwide. Currently, only non-pharmacological therapies (exercise, physical therapy, and diet) are used concurrently for treatment, as there are no US Food and Drug Administration-approved drugs for treating sarcopenia [28]. Due to the lack of effective medications, it is essential to identify safe materials that can maintain skeletal muscle mass by controlling muscle degradation without any side effects. Probiotic supplementation is effective to change the proportion and distribution of the gut microbiome. L. fermentum and Lc. lactis, generally recognized as safe (GRAS)-status microorganisms, are known to be safe when ingested up to concentrations of 109–1011 CFU/day [29]. Recent studies have reported that gut probiotics can influence skeletal muscle metabolism through the formation of ATP through metabolic activity and may play a key role in maintaining energy production during exercise [20]. Also, ingestion of LAB improves muscle mass and exercise capacity in healthy people and reduces muscle loss in cancer patients [30]. Previous studies have demonstrated the beneficial effects of the oral administration of Lactobacillus strains on muscle-related parameters in animal models. For instance, oral administration of L. plantarum TWK10 increases the muscle mass and improves the exercise capacity of the Institute of Cancer Research mice [20]. Similarly, L. reuteri 100-23 and L. gasseri 311,476 significantly reduce the expression levels of muscle atrophy markers, such as MuRF-1 and atrogin-1 [19]. L. casei LC122 enhances muscle strength in C57BL/6 mice [18]. These studies highlight the therapeutic potential of Lactobacillus species in alleviating muscle atrophy by promoting muscle mass gain.
Muscle atrophy primarily occurs due to multiple factors, including muscle cell damage induced by elevated levels of oxidative substances, reduced production and regeneration of muscle proteins due to decreased expression of stress proteins, and promotion of muscle protein degradation via the activation of the ubiquitin–proteasome pathway [31]. The ubiquitin–proteasome pathway is primarily involved in skeletal muscle protein degradation [32]. MuRF-1 and atrogin-1 are skeletal muscle-specific ubiquitin ligases, whose activation induces the loss of muscle mass, that are used as markers of muscle atrophy [33]. DEX, a potent anti-inflammatory and immunosuppressive agent, is used for its biological effects. However, high doses and prolonged use of DEX impair protein synthesis and induce muscle mass loss [34]. DEX has been extensively used in both in vitro and in vivo models to elucidate the fundamental mechanisms underlying muscle atrophy [35]. In this study, DEX increased, whereas L. fermentum MG5091, Lc. lactis MG4668, and MG5474 significantly decreased the expression levels of MuRF-1 and atrogin-1.
Skeletal muscle apoptosis is a major indicator of sarcopenia that inhibits cell proliferation and induces apoptosis via various mechanisms [36]. Apoptosis is the final stage of muscle loss and atrophy [9]. During early apoptosis, Bax activation leads to an increase in the Bax/Bcl-2 ratio, resulting in the initiation of the mitochondrial apoptotic signaling pathway and activation of the final effector caspases [37]. Caspases, also known as cysteine-aspartic proteases, are essential enzymes involved in the regulation of apoptosis and skeletal muscle wasting under diverse conditions [9]. Caspases in the nucleus and mitochondrial membranes of cells play important roles in determining the morphological characteristics of apoptosis [38]. Activated caspase-3 induces DNA cleavage by stimulating apoptosis in cells [39]. In this study, L. fermentum MG5091, Lc. lactis MG4668, and MG5474 inhibited the expression of the pro-apoptotic protein, Bax, and the activation of the apoptotic protein, caspase-3, in DEX-treated C2C12 cells. In particular, the levels of these apoptotic factors were significantly decreased by Lc. lactis MG5474 compared to that by L. fermentum MG5091 and Lc. lactis MG4668. These results suggest that LAB strains suppress sarcopenia by inhibiting muscle atrophy and apoptotic protein expression in C2C12 cells.
Probiotics have beneficial properties, such as tolerance to gastrointestinal conditions, adherence to intestinal epithelial cells, and safety [12]. Hemolysis is a major virulence factor for pathogenic bacteria and a vital safety property of probiotics [40]. We proved the safety of Lc. lactis MG5474 in our previous study [12]. Here, L. fermentum MG5091 and Lc. lactis MG4668 did not exhibit any hemolytic activity (Figure S1) and were not cytotoxic to HT-29 cells. Thus, L. fermentum MG5091, Lc. lactis MG4668, and MG5474 were confirmed to be safe for practical use. When consumed, probiotics must survive in the human stomach and gastrointestinal tract and adhere to the colon to provide health benefits to the host [41]. L. fermentum MG5091 and Lc. lactis MG5474 cells exhibited excellent adhesion to HT-29 cells in both SGF and SIF. In contrast, the survival rate of Lc. lactis MG4668 was lower than that of L. fermentum MG5091 and Lc. lactis MG5474. In our previous study, cryoprotectants, such as sodium alginate and pumpkin powder, improved the survival rate of strains in SGF and SIF [42]. Therefore, probiotic protectants may aid in increasing the survival rate of Lc. lactis MG4668 in GIT; however, this requires validation in future studies.
In this study, L. fermentum MG5091, Lc. lactis MG4668, and MG5474 were evaluated for their safety, including adhesion to intestinal epithelial cells and hemolytic activity. Our results indicate that these strains are safe for use as probiotics. Moreover, we confirmed the effects of L. fermentum MG5091, Lc. lactis MG4668, and MG5474 strains on muscle atrophy and apoptotic protein expression in C2C12 cells. However, the specific mechanism of sarcopenia remains unknown and requires further investigation in future studies. Moreover, animal studies and clinical trials are necessary to verify the inhibitory effects of these probiotic strains on muscle atrophy.
5. Conclusions
In summary, LAB strains suppressed muscle atrophy and apoptotic proteins in DEX-treated C2C12 cells. L. fermentum MG5091 and MG4668 and Lc. lactis MG5474 showed high viability in GIT conditions and high adhesion to HT-29 cells. These strains are also safe to be used as probiotics, with no hemolytic activity. Our results suggest L. fermentum MG5091 and MG4668 and Lc. lactis MG5474 as potential probiotics for the prevention of sarcopenia.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation9070659/s1, Figure S1: Hemolysis activity of LAB strains.
Author Contributions
Conceptualization, C.-H.K.; methodology, J.-Y.P. and Y.K.; investigation, J.-Y.P.; resources, C.-H.K.; data curation, J.-Y.P. and J.Y.L. writing—original draft preparation, J.-Y.P.; writing—review and editing, J.-Y.P., J.Y.L. and C.-H.K.; project administration, C.-H.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was conducted with support from the project for Collabo R&D between Industry and Academy funded by Korea Ministry of SMEs and Startups in 2023 (Project No.RS-2023-00225041).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Abiri, B.; Vafa, M. The role of nutrition in attenuating age-related skeletal muscle atrophy. In Reviews on New Drug Targets in Age-Related Disorders, 1st ed.; Guest, P.C., Ed.; Springer: Berlin/Heidelberg, Germany, 2020; Volume 1260, pp. 297–318. [Google Scholar]
- Shiota, C.; Abe, T.; Kawai, N.; Ohno, A.; Teshima-Kondo, S.; Mori, H.; Terao, J.; Tanaka, E.; Nikawa, T. Flavones inhibit LPS-induced atrogin-1/MAFbx expression in mouse C2C12 skeletal myotubes. J. Nutr. Sci. Vitaminol. 2015, 61, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-W.; Chang, Y.B.; Park, C.W.; Han, S.H.; Suh, H.J.; Ahn, Y. Protein hydrolysate from Spirulina platensis prevents dexamethasone-induced muscle atrophy via Akt/Foxo3 signaling in C2C12 myotubes. Mar. Drugs 2022, 20, 365. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.P. Molecular regulation of skeletal muscle mass. Clin. Exp. Pharmacol. Physiol. 2010, 37, 378–384. [Google Scholar] [CrossRef]
- Lee, J.H.; Jang, J.Y.; Kwon, Y.H.; Lee, S.H.; Park, C.; Choi, Y.H.; Kim, N.D. Effects of Rosemary Extract on C2C12 Myoblast Differentiation and 5-Aminoimidazole-4-carboxamide Ribonucleoside (AICAR)-Induced Muscle Cell Atrophy. Appl. Sci. 2023, 13, 986. [Google Scholar] [CrossRef]
- Frontera, W.R. What is it about old muscles? J. Physiol. 2017, 595, 4581. [Google Scholar] [CrossRef] [PubMed]
- Miki, Y.; Morioka, T.; Shioi, A.; Fujimoto, K.; Sakura, T.; Uedono, H.; Kakutani, Y.; Ochi, A.; Mori, K.; Shoji, T. Oncostatin M induces C2C12 myotube atrophy by modulating muscle differentiation and degradation. Biochem. Biophys. Res. Commun. 2019, 516, 951–956. [Google Scholar] [CrossRef]
- Hah, Y.-S.; Lee, W.K.; Lee, S.; Kim, E.J.; Lee, J.H.; Lee, S.-J.; Ji, Y.H.; Kim, S.G.; Lee, H.-H.; Hong, S.Y. β-Sitosterol attenuates dexamethasone-induced muscle atrophy via regulating FoxO1-dependent signaling in C2C12 cell and mice model. Nutrients 2022, 14, 2894. [Google Scholar] [CrossRef]
- Yin, L.; Li, N.; Jia, W.; Wang, N.; Liang, M.; Yang, X.; Du, G. Skeletal muscle atrophy: From mechanisms to treatments. Pharmacol. Res. 2021, 172, 105807. [Google Scholar] [CrossRef]
- Bann, D.; Chen, H.; Bonell, C.; Glynn, N.W.; Fielding, R.A.; Manini, T.; King, A.C.; Pahor, M.; Mihalko, S.L.; Gill, T.M. Socioeconomic differences in the benefits of structured physical activity compared with health education on the prevention of major mobility disability in older adults: The LIFE study. J. Epidemiol. Community Health 2016, 70, 930–933. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Lee, J.Y.; Park, J.Y.; Jeong, Y.; Kang, C.H. Anti-Inflammatory Response in TNFα/IFNγ-Induced HaCaT Keratinocytes and Probiotic Properties of Lacticaseibacillus rhamnosus MG4644, Lacticaseibacillus paracasei MG4693, and Lactococcus lactis MG5474. J. Microbiol. Biotechnol. 2023, 33, 1–11. [Google Scholar]
- Wang, Y.; Liu, Q.; Quan, H.; Kang, S.-G.; Huang, K.; Tong, T. Nutraceuticals in the Prevention and Treatment of the Muscle Atrophy. Nutrients 2021, 13, 1914. [Google Scholar] [CrossRef] [PubMed]
- Azad, M.A.K.; Sarker, M.; Li, T.; Yin, J. Probiotic species in the modulation of gut microbiota: An overview. BioMed Res. Int. 2018, 2018. [Google Scholar] [CrossRef]
- Zhao, J.; Huang, Y.; Yu, X. A narrative review of gut-muscle axis and sarcopenia: The potential role of gut microbiota. Int. J. Gen. Med. 2021, 14, 1263–1273. [Google Scholar] [CrossRef] [PubMed]
- Mancin, L.; Wu, G.D.; Paoli, A. Gut microbiota–bile acid–skeletal muscle axis. Trends Microbiol. 2022, 31, 254–269. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-K.; Guevarra, R.B.; Kim, Y.-T.; Kwon, J.; Kim, H.; Cho, J.H.; Kim, H.B.; Lee, J.-H. Role of probiotics in human gut microbiome-associated diseases. JMB 2019, 29, 1335–1340. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Yang, X.; Zheng, L.; Wang, Z.; Wu, L.; Jiang, J.; Yang, T.; Ma, L.; Fu, Z. Lactobacillus and Bifidobacterium improves physiological function and cognitive ability in aged mice by the regulation of gut microbiota. Mol. Nutr. Food Res. 2019, 63, 1900603. [Google Scholar] [CrossRef]
- Bindels, L.B.; Beck, R.; Schakman, O.; Martin, J.C.; De Backer, F.; Sohet, F.M.; Dewulf, E.M.; Pachikian, B.D.; Neyrinck, A.M.; Thissen, J.-P. Restoring specific lactobacilli levels decreases inflammation and muscle atrophy markers in an acute leukemia mouse model. PLoS ONE 2012, 7, e37971. [Google Scholar] [CrossRef]
- Chen, Y.-M.; Wei, L.; Chiu, Y.-S.; Hsu, Y.-J.; Tsai, T.-Y.; Wang, M.-F.; Huang, C.-C. Lactobacillus plantarum TWK10 supplementation improves exercise performance and increases muscle mass in mice. Nutrients 2016, 8, 205. [Google Scholar] [CrossRef]
- Lee, K.; Kim, H.J.; Kim, S.A.; Park, S.D.; Shim, J.J.; Lee, J.L. Exopolysaccharide from lactobacillus plantarum HY7714 protects against skin aging through skin–gut axis communication. Molecules 2021, 26, 1651. [Google Scholar] [CrossRef]
- Park, J.-Y.; Lee, J.Y.; Kim, Y.; Kang, C.-H. Latilactobacillus sakei Wikim0066 Protects Skin through MMP Regulation on UVB-Irradiated In Vitro and In Vivo Model. Nutrients 2023, 15, 726. [Google Scholar] [CrossRef]
- Kang, C.-H.; Han, S.H.; Kim, J.-S.; Kim, Y.; Jeong, Y.; Park, H.M.; Paek, N.-S. Inhibition of nitric oxide production, oxidative stress prevention, and probiotic activity of lactic acid bacteria isolated from the human vagina and fermented food. Microorganisms 2019, 7, 109. [Google Scholar] [CrossRef] [PubMed]
- Buxton, R. Blood agar plates and hemolysis protocols. Am. Soc. Microbiol. 2005, 15, 1–9. [Google Scholar]
- Lee, J.; Kim, S.; Kang, C.-H. Immunostimulatory Activity of Lactic Acid Bacteria Cell-Free Supernatants through the Activation of NF-κB and MAPK Signaling Pathways in RAW 264.7 Cells. Microorganisms 2022, 10, 2247. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.; Kim, J.-S.; Kwon, H.-J.; Kang, C.-H. The Effect of a Glutathione (GSH)-Containing Cryo-Protectant on the Viability of Probiotic Cells Using a Freeze-Drying Process. Fermentation 2023, 9, 187. [Google Scholar] [CrossRef]
- Bonaldo, P.; Sandri, M. Cellular and molecular mechanisms of muscle atrophy. Dis. Model. Mech. 2013, 6, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Sani, A.; Hasegawa, K.; Yamaguchi, Y.; Panichayupakaranant, P.; Pengjam, Y. Inhibitory effects of curcuminoids on dexamethasone-induced muscle atrophy in differentiation of C2C12 cells. Phytomed. Plus 2021, 1, 100012. [Google Scholar] [CrossRef]
- de Lacerda, J.R.M.; da Silva, T.F.; Vollú, R.E.; Marques, J.M.; Seldin, L. Generally recognized as safe (GRAS) Lactococcus lactis strains associated with Lippia sidoides Cham. are able to solubilize/mineralize phosphate. Springerplus 2016, 5, 1–7. [Google Scholar] [CrossRef]
- Lee, M.-C.; Tu, Y.-T.; Lee, C.-C.; Tsai, S.-C.; Hsu, H.-Y.; Tsai, T.-Y.; Liu, T.-H.; Young, S.-L.; Lin, J.-S.; Huang, C.-C. Lactobacillus plantarum TWK10 improves muscle mass and functional performance in frail older adults: A randomized, double-blind clinical trial. Microorganisms 2021, 9, 1466. [Google Scholar] [CrossRef]
- Chang, Y.B.; Ahn, Y.; Suh, H.J.; Jo, K. Yeast hydrolysate ameliorates dexamethasone-induced muscle atrophy by suppressing MuRF-1 expression in C2C12 cells and C57BL/6 mice. J. Funct. Foods 2022, 90, 104985. [Google Scholar] [CrossRef]
- Choi, R.-Y.; Kim, B.S.; Ban, E.-J.; Seo, M.; Lee, J.H.; Kim, I.-W. Mealworm Ethanol Extract Enhances Myogenic Differentiation and Alleviates Dexamethasone-Induced Muscle Atrophy in C2C12 Cells. Life 2022, 13, 58. [Google Scholar] [CrossRef]
- Li, F.; Luo, T.; Rong, H.; Lu, L.; Zhang, L.; Zheng, C.; Yi, D.; Peng, Y.; Lei, E.; Xiong, X. Maternal rodent exposure to di-(2-ethylhexyl) phthalate decreases muscle mass in the offspring by increasing myostatin. J. Cachexia Sarcopenia Muscle 2022, 13, 2740–2751. [Google Scholar] [CrossRef]
- Ryu, B.; Oh, S.; Yang, H.-W.; Sosorburam, B.; Chung, D.-M.; Seo, M.; Park, S.-J.; Byun, K.; Jeon, Y.-J. Diphlorethohydroxycarmalol derived from Ishige okamurae improves behavioral and physiological responses of muscle atrophy induced by dexamethasone in an in-vivo model. Pharmaceutics 2022, 14, 719. [Google Scholar] [CrossRef]
- Kim, J.W.; Ku, S.-K.; Han, M.H.; Kim, K.Y.; Kim, S.G.; Kim, G.-Y.; Hwang, H.J.; Kim, B.W.; Kim, C.M.; Choi, Y.H. The administration of Fructus Schisandrae attenuates dexamethasone-induced muscle atrophy in mice. Int. J. Mol. Med. 2015, 36, 29–42. [Google Scholar] [CrossRef]
- Chen, C.; Yang, J.-S.; Lu, C.-C.; Chiu, Y.-J.; Chen, H.-C.; Chung, M.-I.; Wu, Y.-T.; Chen, F.-A. Effect of quercetin on dexamethasone-induced C2C12 skeletal muscle cell injury. Molecules 2020, 25, 3267. [Google Scholar] [CrossRef] [PubMed]
- Carotenuto, F.; Coletti, D.; Di Nardo, P.; Teodori, L. α-linolenic acid reduces TNF-induced apoptosis in C2C12 myoblasts by regulating expression of apoptotic proteins. Eur. J. Transl. Myol. 2016, 26, 6033. [Google Scholar] [CrossRef]
- Jin, Y.; Duan, L.X.; Xu, X.L.; Ge, W.J.; Li, R.F.; Qiu, X.J.; Song, Y.; Cao, S.S.; Wang, J.G. Mechanism of apoptosis induction in human hepatocellular carcinoma cells following treatment with a gecko peptides mixture. Biomed. Rep. 2016, 5, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Castañeda, F.; Kinne, R.K. Apoptosis induced in HepG2 cells by short exposure to millimolar concentrations of ethanol involves the Fas-receptor pathway. J. Cancer Res. Clin. Oncol. 2001, 127, 418–424. [Google Scholar] [CrossRef]
- Kim, H.; Kim, J.-S.; Kim, Y.; Jeong, Y.; Kim, J.-E.; Paek, N.-S.; Kang, C.-H. Antioxidant and probiotic properties of Lactobacilli and Bifidobacteria of human origins. Biotechnol. Bioprocess Eng. 2020, 25, 421–430. [Google Scholar] [CrossRef]
- Silva, C.; Domingos-Lopes, M.; Magalhães, V.; Freitas, D.; Coelho, M.; Rosa, H.; Dapkevicius, M. Latin-style fresh cheese enhances lactic acid bacteria survival but not Listeria monocytogenes resistance under in vitro simulated gastrointestinal conditions. J. Dairy Sci. 2015, 98, 4377–4383. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.; Kim, Y.; Kim, J.-S.; Jeong, Y.; Park, H.M.; Kim, J.W.; Kim, J.-E.; Kim, H.; Paek, N.-S.; Kang, C.-H. Evaluating the cryoprotective encapsulation of the lactic acid bacteria in simulated gastrointestinal conditions. Biotechnol. Bioprocess Eng. 2020, 25, 287–292. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).