Abstract
Thermophilic endoglucanases have become of significant interest for effectively catalyzing the hydrolysis of cellulose. Myceliophthora thermophila is an ideal source of thermophilic enzymes. Interestingly, different hosts differently express the same enzymes. In this study, we successfully overexpressed endoglucanase (MtEG5-1) from M. thermophila in the methylotrophic yeast, Pichia pastoris GS115, via electroporation. We found that purified MtEG5-1 exhibited optimum activity levels at pH 5 and 70 °C, with 88% thermal stability after being incubated at 70 °C for 2 h. However, we observed that purified MtEG5-1 had a molecular weight of 55 kDa. The Km and Vmax values of purified MtEG5-1 were approximately 6.11 mg/mL and 91.74 μmol/min/mg at 70 °C (pH 5.0), respectively. Additionally, the optimum NaCl concentration of purified MtEG5-1 was found to be 6 g/L. Furthermore, we observed that the activity of purified MtEG5-1 was significantly enhanced by Mn2+ and was inhibited by K+. These results indicated that MtEG5-1 expressed by P. pastoris GS115 is more heat-tolerant than that expressed by A. niger and P. pastoris X33. These properties of MtEG5-1 make it highly suitable for future academic research and industrial applications.
1. Introduction
Currently, 88% of global primary energy use is still derived from fossil fuels [1]. However, due to their depleting reserves and serious environmental impact, it is imperative to find sustainable and renewable alternatives to fossil energy sources [2]. Cellulose is considered to be one of the best alternatives to fossil energy, as it is a renewable energy source that is abundantly available in many forms, such as corn straw and bagasse [3]. Nevertheless, the widespread use of cellulose has been limited by the high energy consumption and pollution associated with chemical and physical pretreatment methods [4]. Fortunately, the emergence of synthetic biology provides novel methods to address this problem. Cellulose can be efficiently and gently degraded by cellulase, providing more opportunities to apply it in a wide range of industrial processes.
Cellulose is a macromolecular polysaccharide composed of repeating glucose units with β-1,4-glycosidic bonds [5]. The complete conversion of cellulose to glucose requires the combined action of three enzymes: cellobiohydrolases (CBHs, EC3.2.1.9), endoglucanases (EGs, EC 3.2.1.4), and β-glucosidases (BGLs, EC 3.2.1.21) [6,7]. The principle of the cellulase synergistic decomposition of cellulose is shown in Figure 1. EGs, as the most important rate-limiting enzyme in cellulose degradation, focus on the hydrolysis of β-1, 4-glucosidic glucan linkages [8]. This result in the conversion of long cellulase chains into short chains, thereby increasing the amount of reducing ends exposed in the cellulose chain [9]. Consequently, EGs are not only a prerequisite for the quick initial liquefaction of biomass, but they are also a key biocatalyst for cellulose biodegradation [10]. Increasing the system temperature for cellulose degradation may reduce the risk of contamination with mesophilic microorganisms and increase the reaction rate [11]. In addition, thermostable enzymes can be transferred and stored more easily than mesophilic enzymes can [12]. Therefore, it is necessary to screen for thermostable EGs and apply them to cellulose degradation.
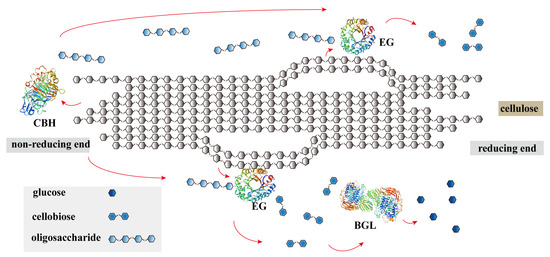
Figure 1.
The synergistic effect of cellulase for cellulose decomposition [6,7]. CBH: cellobiohydrolase, EG: endoglucanase, BGL: beta-glucosidase. The red arrows indicate the sequence of cellulase synergy required for the complete degradation of cellulose into monosaccharides.
Myceliophthora thermophila is a thermophilic fungus with a good cellulose-degrading ability. Its cellulase products are generally recognized as safe [13]. Additionally, EGs possess a high catalytic efficiency and thermal stability, making them ideal candidates for industrial cellulose degradation. However, when cellulase is overexpressed in M. thermophila, several endogenous enzymes, such as xylanase [14], are overexpressed, which complicates the isolation of cellulose and makes it difficult to study the enzymatic properties and structure of the enzyme.
Pichia pastoris is a methylotrophic strain that is widely used as a potent heterologous protein expression system, owing to its high cell density and low levels of endogenous protein expression [11]. Furthermore, the expressed protein always exhibits higher stability for glycosylation and other posttranslational modifications [15]. Consequently, P. pastoris is becoming increasingly popular for academic or industrial applications. Although MtEG5-1 has been successfully overexpressed in Aspergillus niger and P. pastoris X33, the results indicate that MtEG5-1 has poor thermal stability and a higher molecular weight [16,17]. In contrast, an expression system constructed using P. pastoris GS115 as the host strain can produce a thermally stable MtEG5-1.
In this study, we used the enzyme obtained from M. thermophila and analyzed some enzymatic properties of MtEG5-1 overexpressed in P. pastoris GS115. Therefore, this work provides a thermo-acid-stable enzyme for the utilization of cellulose resources.
2. Materials and Methods
2.1. Strains and Culture Conditions
M. thermophila ATCC 42464 and the P. pastoris GS115 strain were obtained at the Tianjin Institute of Industrial Biotechnology. M. thermophila was routinely grown in/on GY broth/agar (30 g/L glucose and 8 g/L yeast extract) at 45 °C. P. pastoris GS115 was grown in/on YPD broth/agar (10 g/L yeast extract, 20 g/L peptone, 20 g/L glucose, and 15 g/L agarose) at 30 °C. Escherichia coli DH5α (Biomed, Beijing, China) was used for gene cloning and vector construction, and it was grown in/on LB broth/agar (10 g/L NaCl, 10 g/L peptone, and 5 g/L yeast extract) at 37 °C.
2.2. Chemicals
The SE Seamless Cloning and Assembly Kit was purchased from Beijing Zoman Biotechnology Co., Ltd. (Beijing, China). All restriction endonucleases were purchased from Guangzhou Chuangrong Biotechnology Co., Ltd. (Guangzhou, China). The pre-stained protein marker and the DNA maker were purchased from Beijing Yishan Huitong Technology Co., Ltd. (Beijing, China). PCR primers were prepared by QingKe Biotechnology Company (Beijing, China). The DNA agarose recycling kit and the plasmid extraction kits were purchased from Omega (Norcross, GA, USA). The total RNA Extraction Kit was purchased from Shenzhen Yibaishun Technology Co., Ltd. (Shenzhen, China). SuperScript II reverse transcriptase was purchased from Thermo Fisher Scientific (Wilmington, MA, USA). All chemicals used were of analytical grade.
2.3. Bioinformatics Analysis of MtEG5-1
The evolutionary location of MtEG5-1 was analyzed using MEGA 7.0 software [18]. The signal peptide of MtEG5-1 was predicted using the Signal P online website [19]. The tertiary structure of MtEG5-1 was predicted using the SWISS-MODEL online website and PyMOL software [20].
2.4. Expression Vector Construction
M. thermophila was inoculated into GY liquid medium at 45 °C and 180 rpm for 24 h with shock culture. After filtration, mycelium was collected and stored in liquid nitrogen. The total RNA of M. thermophila was extracted using the total RNA Extraction Kit, and cDNA was obtained using reverse transcriptase. The entire coding region of mtEG5 in M. thermophila was amplified via PCR from cDNA as a template using the primer EcoRI-mtEG5-F/R. The PCR program was set up with an initial denaturation step of 95 °C for 5 min, followed by 38 cycles of 95 °C for 10 s, 55 °C for 15 s, and 72 for 60 s, and a final extension at 72 °C for 10 min. The PCR product was purified using the glue recovery kit according to the manufacturer’s protocol. The mtEG5 PCR product with the EcoRI fragment was ligated to the EcoRI-digested and dephosphorylated vector, pPIC9K, at a 5:1 M ratio of insert: vector. The recombinant plasmid was transformed into E. coli DH5α. The correct vector was verified by sequencing after verification by performing colony PCR with the primers YmtEG5-F/R. All the primers are listed in Table 1.

Table 1.
Primers were used in this study.
2.5. Transformation and Selection of Positive Transformants
The PmeI-linearized pPIC9K-mtEG5 plasmid and pPIC9K plasmid were transformed into the P. pastoris GS115 strain via electroporation (BTX, ECM630 at 1.5 kV, 200 Ω, 25 µF, and 5–10 ms with a 0.2 cm cuvette) [21]. After the transformation, 100 µL cuvette contents was spread on minimal dextrose agar plates (0.2 mg/mL biotin, 20 g/L glucose, 15 g/L agarose, and 13.4 g/L yeast nitrogen base, with ammonium sulfate and without amino acids) and incubated at 30 °C for 36 h. As a negative control, the plasmid of linearized pPIC9K was transferred onto the plates. Positive transformants were screened with YPD medium containing 1.75 mg/mL G418 and confirmed via PCR. One transformer, P. pastoris GS115, expressing the highest recombinant MtEG5-1 activity level was selected for the analysis of recombinant enzymatic properties.
2.6. Fermentation of MtEG5-1 in P. pastoris GS115
A positive colony of P. pastoris GS115 expressing MtEG5-1 was cultured in 100 mL of buffered glycerol-complex medium (10 g/L yeast extract, 20 g/L peptone, 13.4 g/L yeast nitrogen base with ammonium sulfate and without amino acids, 0.2 mg/mL biotin, 100 mM (pH 6.0) potassium phosphate, and 1.0% glycerol) grown at 30 °C and 180 pm for 48 h. The yeast cells were resuspended in 100 mL of buffered methanol-complex medium via the natural sinking method (10 g/L yeast extract, 20 g/L peptone, 13.4 g/L yeast nitrogen base with ammonium sulfate and without amino acids, 0.2 mg/mL biotin, 100 mM (pH 6.0) potassium phosphate, and 1.0% methanol). The suspension was grown in a 1 L baffled flask at 30 °C and 200 rpm for 48 h and supplemented with 1% methanol every 24 h for induction. To determine the time course of MtEG5-1 expression in P. pastoris GS115, the fermented supernatant was collected daily (0–7 d), and the total MtEG5-1 concentration (BCA Protein Assay Reagent) and the activity of MtEG5-1 were analyzed [22]. Three replicates of each experiment were performed to obtain accurate data.
2.7. Enzymatic Assay of MtEG5-1
The activity of MtEG5-1 was determined via the DNS method [23], which is based on the generation of reducing sugar ends from carboxymethyl cellulose (CMC) as a substrate and its reaction with dinitro salicylic acid. The activity of MtEG5-1 was determined in an assay using 1% CMC in 50 mM citrate buffer (pH 4.8) at 50 °C for 30 min, and then the reaction was stopped with the addition of 3 mL of DNS reagent. The resulting mixture was placed in boiling water for 5 min for color development, and the amount of reducing ends was determined by measuring absorbance at 540 nm. As a negative control, the enzyme was replaced by the pPIC9K vector in P. pastoris. An endoglucanase unit (IU) is defined as the amount of enzyme that generates the equivalent to 1 μmol of glucose per minute in the abovementioned reaction conditions. Cell-free supernatant (CFS) was extracted from a 1% methanol-induced P. pastoris GS115 converter for 1–7 days, enzyme activity was measured daily, and 2 mL of CFS was collected and stored at −20 °C. Finally, CFS was run through SDS-PAGE from 1 to 7 days.
2.8. Activity Was Determined via Congo-Red Staining
P. pastoris GS115 transformant cultures were centrifuged at 12,000 rpm for 10 min, and the clarified CFS obtained after passing it through a 0.2 μm filter was used as the source of extracellular enzymes for the agar well diffusion assay. A total of 1% agarose gel supplemented with 1 g/L CMC and Congo-red was cast on a plate, and wells were made [24]. After adding 100 μL of CFS to each well, the CMC plate was incubated at 50 °C for 1 h. Gels were stained for 1 h, followed by de-staining with 1 mol/L NaCl for another 10 min (they were washed until there was no color).
2.9. Scanning Electron Microscopy (SEM) Analysis
As a natural lignocellulose material, corn straw has a dense and stable structure, which inhibits the decomposition and utilization of polysaccharide, thereby reducing the treatment efficiency and greatly increasing the production cost of industrial ethanol and other products [25]. To remove lignin and solubilize hemicellulose to break down the natural barrier of lignocellulose and increase the contact area with enzymes and microorganisms, corn straw pretreatment is necessary, which can greatly improve the availability of corn straw [26]. Dilute acid pretreatment can break the structure of lignocellulose and expose cellulose, thus providing hydrolysis sites for cellulase [27]. Zhang et al. pretreated rice straw with HCl, and the cellulose content of the pretreated straw increased from 38.10 to 52.54~56.09% [28]. In the study, corn straw was cut into long strips of 0.5 × 1.0 cm, pretreated with 2 mol/L HCl at 28 °C for 24 h, washed with distilled water until neutral, and dried at 60 °C. Pretreated corn straw was reacted with 1.5 mL of crude enzyme solution and sterile water in a water bath at 70 °C for 7 days. At the same time, untreated corn straw was also reacted with 1.5 mL sterile water in a water bath at 70 °C for 7 days as a blank control. The samples were sputter coated with platinum nanoparticles in an ion-sputter coater before viewing via SEM at 10 mm and 3 kV acceleration voltage and 1000× magnification [29].
2.10. Purification of Recombinant MtEG5-1 from P. pastoris GS115
In the process of protein purification, liquids used were filtered through a 0.22 μm filter membrane and ultrasonicated for more than 15 min. MtEG5-1 was purified via Ni2+ affinity chromatography [30]. The system was balanced with 3–5 times more of the volume of Tris-HCl (pH 7.0). The supernatant after centrifugation was taken and slowly allowed to flow through the Ni column at a flow rate of 1 mL/min. The protein was eluted with the same buffer at a flow rate of 5 mL/min, the system pressure was no more than 1 MPa, and 2 mL of eluent was collected in each tube using AKTA Purifier 10. The weakly bound hetero proteins were washed with 10 mM imidazole, and the proteins were eluted with 60 mM imidazole. The target protein was eluted with 400 mM imidazole. The purified recombinant protein MtEG5-1 was run using an SDS-PAGE gel.
2.11. Enzyme Characterization
The optimum pH for MtEG5-1 was determined via the standard assay at different pH values (3–11) using either 50 mM sodium citrate buffer at pH 2–5, 50 mM phosphate buffer at pH 6–8, 100 mM Tris-HCl buffer at pH 9, or 50 mM NaHCO3-NaOH buffer at pH 9–11. On the other hand, the stability at different pH conditions was determined after the incubation of MtEG5-1 in the abovementioned buffers at 4 °C for 24 h and the measurement of the remaining activity after the standard assay. The optimal temperature of MtEG5-1 was estimated by using the standard assay procedure at varying temperatures (30–90 °C) for 2 h. The highest activity level obtained under standard conditions was defined as 100% activity. To determine temperature stability, 0.38 mg of purified MtEG5-1 was incubated at various temperatures (50, 60, and 70 °C) for different time intervals, and the residual activity level was measured via the standard assay procedure. The thermostability of MtEG5-1 was tested at an optimal pH value. The optimal NaCl concentration of MtEG5-1 was estimated via the standard assay procedure at NaCl final concentrations of 1–10 g/L at 50 °C for 2 h. The influence of metal ions on purified MtEG5-1 activity was determined by incorporating metal ions such as 0.33 mol/L Na2SO4, MgSO4, MnSO4, CuSO4, K2SO4, FeSO4, and NiSO4 solutions during the standard assay procedure; however, the reaction duration was 2 h.
2.12. Enzymatic Kinetics of MtEG5-1
CMC (0.2–2.0%) was used as a substrate to measure enzyme kinetics under standard conditions (50 mM sodium citrate buffer, pH 5.0, 70 °C). Michaelis–Menten constants (Km) and the maximum velocity (Vm) of purified MtEG5-1 were determined by measuring the rate of CMC hydrolysis using a Lineweaver—Burk plot, as shown in Equation (1), where V is the reaction rate and [S] is the substrate concentration [31].
3. Results and Discussion
3.1. Bioinformatics Analysis of MtEG5-1 and Construction of pPIC9K-mtEG5
The mtEG5 gene (NCBI: XP_003659014.1) was obtained from M. thermophila ATCC 42464, which is also known as Sporotrichum thermophile ATCC 42464, Thielavia heterothallica ATCC 42464, and Thermothelomyces thermophilus ATCC 42464 (https://www.uniprot.org/taxonomy/573729 (accessed on 8 May 2023)). This species can grow at 45–55 °C [32]. The enzyme expressed by the mtEG5 gene in P. pastoris GS115 was named MtEG5-1 in this study. MtEG5-1 (UniProtKB: G2Q5D8) has 389 amino acids and an 18-aa N-terminal signal peptide, as depicted in Figure 2a. Carbohydrate-active enzymes are classified based on their protein sequence and structure [33]. MtEG5-1 belongs to glycoside hydrolase family 5 (GH5), one of the largest families among all GHs. The two catalytic residues in GH5 enzymes are both glutamic acids: one is a nucleophile and the other is a catalytic proton donor [34]. The carbohydrate-active enzyme database (http://cazy.org (accessed on 8 May 2023)) lists 53 sub-families, with approximately 80% of all known GH5 sequences falling into one of these subfamilies [35]. MtEG5-1 belongs to the GH5-1 subfamily. The 3D structure of MtEG5-1 was predicted to be a classical (α/β)8 TIM-barrel, which was connected to eight parallel β-strands and eight parallel α-helices through αβ or βα loops [33] (Figure 2b). These loops are typically found on the protein surface and play a key role in the interactions between the catalytic core and substrate [36,37]. During protein folding, the (α/β)8 barrel protein provides stability for hydrogen bonds and non-covalent interactions between amino acid residues to resist the local tendency for unfolding [38]. A mutation in these loops can alter their flexibility and improve the biochemical properties of cellulase [39]. The neighbor-joining method was used to further analyze the evolutionary position of MtEG5-1, and we found that it shares 99% identity with homologous proteins from Precursor Humicola insolens (Figure 2c). Finally, the expression vector pPIC9K-mtEG5 was constructed following the described procedure (Figure 2d).
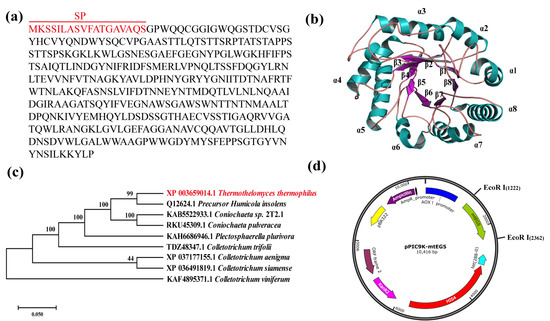
Figure 2.
Bioinformatics analysis of MtEG5-1. (a) The number and location of MtEG5-1’s signal peptide according to the Signal P 5.0 online website. (b) Homology-based (3D) model showing the barrel (α/β)8 fold in mtEG5. The labels α1-α8 are the eight parallel α-helices located on the outside of the TIM-barrel, and the labels β1-β8 are the eight parallel β-strands located on the inside of the TIM barrel. (c) Neighbor-joining phylogenetic tree constructed using MtEG5-1’s amino acid sequence along with other cellulase sequences retrieved from the NCBI database using MEGA 7.0 software. The digit at each branch point represents the percentage of bootstrap support calculated from 1000 replicates. The deduced amino acid sequence of MtEG5-1 exhibited maximum homology with the endoglucanase of P. Humicola insolens. (d) Construction map of the recombinant expression vector pPIC9K-mtEG5 in P. pastoris GS115 obtained using SnapGene 6.0.2 software.
3.2. Selection of P. pastoris GS115 Positive Transformants and Heterologous Expression
The sequencing of the mtEG5 gene fragment revealed that it consists of 1174 base pairs. In order to identify positive transformants, eight random samples were selected, all of which showed the same band expansion as the target band in PCR amplification. Based on this result, transformants 1, 3, 6, and 8, which produced wide and bright bands, were selected for further experiments, as depicted in Figure 3a. Four transformants were then tested every 24 h, and with MtEG5-1 activity induced by 1% methanol, and positive transformants with high copy numbers were selected for subsequent experiments. No corresponding bands were amplified in the CK control group. The hydrolysis of cellulose was observed in cellulose plates inoculated with the CFS of MtEG5-1 culture, indicating the production of MtEG5-1 by P. pastoris GS115 transformants. Cellulose hydrolysis was detected on the first day, indicating that MtEG5-1 is capable of decomposing cellulose, as shown in Figure 3b. Daily detection of the CFS of cultured P. pastoris GS115 via 12% SDS-PAGE revealed a protein with a molecular weight of approximately 55 kDa, compared to that of wild-type yeast. The protein bands were clear from day 1 to day 7, indicating that MtEG5-1 had good stability (Figure 3c). The concentration of MtEG5-1 was 0.95 g/L on day 1, and the highest concentration was observed on day 5 at 1.15 g/L. The activity level of MtEG5-1 was 133.76 IU/mL on day 1, with the highest activity level being observed on day 5 at 162.39 IU/mL. Furthermore, the enzyme activity level was maintained at above 100 IU/mL from days 1–7, providing further evidence of the stability of MtEG5-1, as shown in Figure 3d.
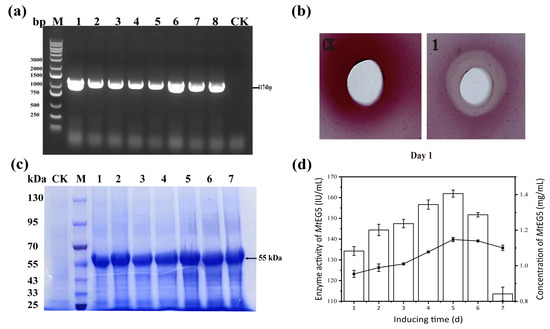
Figure 3.
Positive transformants were screened for molecular identification and activity detection. (a) P. pastoris GS115 transformants of mtEG5 verified by PCR. Lane M, marker 10,000 kb; Lanes 1–8, different mtEG5 colonies; Lane 9, pPIC9K plasmid. (b) The CFS of P. pastoris transformed with pPIC9K-mtEG5 was stained with Congo-red on the CMC plate for the first day. The CK well was supplemented with 100 μL of CFS from pPIC9K yeast transformation, while well 1 was supplemented with 100 μL of CFS from pPIC9K-mtEG5 yeast transformation. (c) The expression of MtEG5-1 induced for different times was detected via 12% SDS-PAGE. Lane M, molecular weight marker; Lane CK, fermentation broth of P. pastoris transformed with pPIC9K induced by methanol; Lanes 1–7, fermentation broth of P. pastoris transformed with pPIC9K-mtEG5 induced by methanol for the first day to the seventh day. Twenty microliters of pretreated sample were loaded in every lane. (d) The expression of MtEG5-1 on different induction days was measured via the BCA method, and the activity of MtEG5-1 on different induction days was measured via the DNS method.
3.3. Scanning Electron Microscopy of Hydrolyzed Corn Straw
We further studied whether the crude enzyme solution of MtEG5-1 has an obvious ability to degrade corn straw. SEM can help to identify morphological changes in pretreated lignocellulosic materials [40]. SEM was used to compare the morphological changes in pretreated corn straw samples before and after catalysis by MtEG5-1. Prior to pretreatment, the fiber surface of corn straw was smooth, with a dense and rigid structure and only a few cracks (Figure 4a). However, after the HCl pretreatment, the fiber of corn straw broke and disconnected at many points, which exposed the cellulose fibrils partially to the surface, thereby increasing the size of the contact site of cellulose with cellulase (Figure 4b). In contrast to the previous two images, the surface of the corn straw decomposed as a result of the use of MtEG5-1 and displayed more fibrous nematocytic filaments, as demonstrated in Figure 4c. The findings indicate that MtEG5-1 decomposed the pretreated corn straw. Nevertheless, future research is required to determine the specific decomposition capacity.
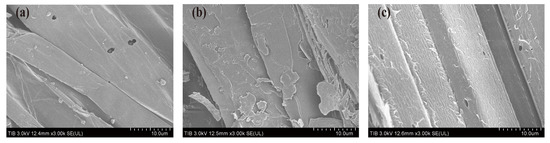
Figure 4.
Analysis of the results of scanning electron microscopy. (a) Corn straw without any treatment. (b) Corn straw pretreated with 2 mol/L HCl for 24 h. (c) The CFS of 1.5 mL crude enzyme solution was mixed with the pretreated corn straw at 70 °C for seven days.
3.4. Characterization of Purified MtEG5-1
In this study, the purified recombinant, MtEG5-1, demonstrates a clear band in the SDS-PAGE results, with an approximate size of 55 kDa (Figure 5a), which differs from the theoretical value of 43 kDa. Typically, cellulases are glycosylated enzymes [41]. Enzymatic modifications can increase the actual molecular weight beyond the theoretical value, as demonstrated by Karnaouri et al., who obtained a purified recombinant enzyme with a molecular weight of 83 kDa by using a purified protein expressed by yeast; deglycosylation reduced the molecular weight to 66 kDa [42].
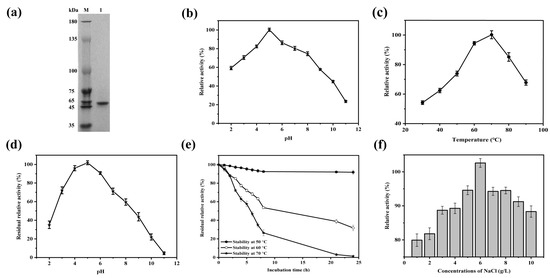
Figure 5.
Effects of different factors on the activity of purified MtEG5-1. (a) Purified MtEG5-1 expression was detected via 12% SDS-PAGE. Lane M, molecular weight marker; Lane 1, a purified MtEG5-1 protein. (b) Effects of assay pH on MtEG5-1 activity. The standard assay was used, except the reaction pH is shown in the graph. (c) Effects of assay temperature on MtEG5-1 activity. The standard assay was used, except the reaction temperature is shown in the graph. (d) Effects of pH on MtEG5-1 stability. The enzyme was incubated for 24 h at 4 °C, and the pH values are indicated, followed by an activity assay under standard conditions. (e) Effects of temperature on MtEG5-1 stability. The enzyme was incubated for the times plotted at the temperatures indicated and at pH 5 for 24 h, followed by an activity assay under standard conditions (black circle is 50 °C, white circle is 60 °C, and black pentagram is 70 °C). (f) Effects of NaCl concentration on MtEG5-1 activity. The enzyme was incubated for 2 h at 50 °C and various NaCl concentrations indicated, followed by an activity assay under standard conditions.
Figure 5b, and c demonstrates that the recombinant enzyme reaches its maximum activity at an optimum pH of 5 and a temperature of 70 °C. After incubation in different pH buffers for 24 h, MtEG5-1 was most stable at pH 5 and least stable at pH 11, with approximately 30% at pH 2. Therefore, MtEG5-1 is an acidic and highly stable protein at pH 4–6 (Figure 5d). Furthermore, MtEG5-1 also exhibited good thermal stability, retaining a relative activity level of above 88% at all temperatures until it was incubated for 2 h. Subsequently, the relative activity levels decreased to approximately 97, 85, and 72% at temperatures of 50, 60 and 70 °C, respectively, after 3 h of incubation (Figure 5e). The glycosylation of the protein may contribute to its thermal stability. Moreover, the optimization of glycosylation sites may serve as an effective and feasible strategy to enhance the enzymatic activity and thermostability [43]. In addition, the relative activity of MtEG5-1 incubated with NaCl (1–10 g/L) at 50 °C for 2 h was above 80%, and the optimal salt concentration of MtEG5-1 was 6 g/L (Figure 5f).
Metal ions have been found to cause structural changes in enzymes [44], resulting in alterations to their activity. This study investigates the effect of Na+, Mg2+, Mn2+, Cu2+, K+, Fe2+, and Ni2+ on the activity level of MtEG5-1, which was incubated at 50 °C for 2 h. The results showed that Mn2+ increased the activity level of MtEG5-1 by 142.6%, while K+ decreased its activity level by 76.1% (Table 2). These results indicate that MtEG5-1 has remarkable tolerance to harsh conditions, such as high salt concentrations, high temperatures, and low pH, making it an ideal candidate for cellulose degradation.

Table 2.
Effect of various metal ions on MtEG5-1 activity. The standard assay was used, except the reaction duration was 2 h.
Interestingly, the experimental results differ from those reported in another published paper. While MtEG5-1 was successfully expressed in P. pastoris X33 [17] and A. niger [16], the properties of MtEG5-1 expressed by P. pastoris GS115, as displayed in Table 3, were dissimilar. The optimal temperature and pH of MtEG5-1 expressed by the three strains were 70 °C and 5, respectively. However, the relative activity levels of the enzyme expressed by A. niger, P. pastoris X33, and P. pastoris GS115 were determined at different temperatures and pH 5 levels. The results showed that the relative enzyme activity levels were still above 80% in the temperature ranges of 60–70 °C, 60–75 °C, and 60–80 °C. The relative activity levels of the enzyme expressed by P. pastoris X33 and P. pastoris GS115 were measured at 4 °C and different pH values for 24 h, while the relative activity level of the enzyme expressed by A. niger was measured at 37 °C and different pH values for 2 h. The results showed that the relative enzyme activity levels were still above 80% in the pH ranges of 4–6, 4–6, and 5.5–6.5. Notably, although there was no significant difference in the optimal temperature and pH of the expressed MtEG5-1 among the three hosts, MtEG5-1 exhibited a greater tolerance to high temperatures than the enzymes expressed by P. pastoris X33 and A. niger did. For instance, the thermal stability of MtEG5-1 expressed by P. pastoris GS115 remained at the 88% residual activity level after 2 h at 70 °C, compared to the 25% residual activity level of the enzyme expressed in P. pastoris X33 after 2 h at 65 °C and the complete inactivation of the enzyme expressed in A. niger after 2 h at 60 °C. Altogether, the enzyme expressed by P. pastoris GS115 was resistant to a wider range of temperatures than the enzyme expressed by A. niger and P. pastoris X33 was. Moreover, the molecular weight of MtEG5-1 expressed by P. pastoris X33 was 75 kDa, while the molecular weight of MtEG5-1 expressed by P. pastoris GS115 was smaller (55 kDa). Generally, larger molecular weight proteins are less stable as they are more susceptible to denaturation as a result of chemical and physical factors [45,46]. Although A. niger, P. pastoris X33, and P. pastoris GS115 expressed the enzyme encoded by the mtEG5 gene, the enzymatic properties and molecular weight of the expressed enzymes were different. Future analysis of the crystal structure of MtEG5-1 could help to explain its characteristics. Therefore, the diverse expression of MtEG5-1 in different hosts requires further investigation. The microbial cell factory constructed in our study can be applied more widely in the industry. As the properties of MtEG5-1 were further verified and improved, this study provides an effective strategy for the utilization of cellulose resources.

Table 3.
Differences between MtEG5-1 in this experiment and other experiments.
3.5. Enzyme Kinetic Parameters of Purified MtEG5-1
For different carbon source substrates, GH5 family cellulases have different substrate binding manners [47]. They are known to hydrolyze cellulose and non-cellulose substrates, usually acting on β-1,4 linkages [48]. In most cases, CMC is one of the best substrates for fungal cellulase [49]. For example, Ma et al. showed that Cel-5A was highly effective at hydrolyzing CMC [50]. By studying the cellulose decomposition activity of endoglucanase caused by bacterial flora, Salehi et al. suggested that endoglucanase could decompose all three types of substrates, including CMC, cellulose, and Avicel. The results showed that CMC was the best substrate for the purified enzyme, which was considered to have 100% activity. There was no significant difference between the specificity of the enzyme in the presence of CMC and cellulose. However, the activity levels recorded were 90% in the presence of cellulose and 35% in the presence of Avicel [51]. Based on the Lineweaver—Burk double inverse method, the values of Km and Vmax for purified MtEG5-1 were approximately 6.11 mg/mL and 91.74 μmol/min/mg at 70 °C (pH 5), respectively (Figure 6). The kcat value was found to be 84.94/s.
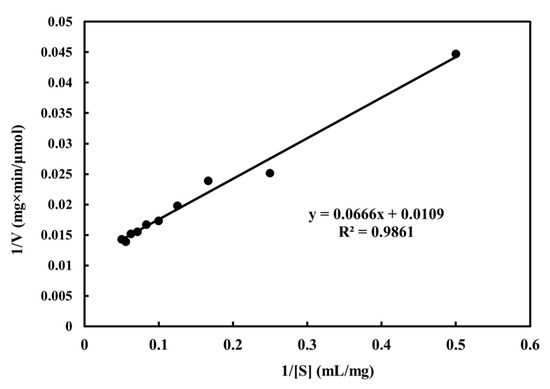
Figure 6.
Lineweaver—Burk plot for MtEG5-1 with CMC as substrate.
4. Conclusions
In this study, MtEG5-1 was successfully overexpressed in P. pastoris GS115, and the enzymatic properties were characterized. Recombinant protein, MtEG5-1, was stable in extreme environments, such as an acidic pH (4–6), a high temperature (60–80 °C), and the presence of salt (6 g/L), in our study. MtEG5-1 expressed by P. pastoris GS115 is more tolerant to high temperatures than the enzyme expressed by P. pastoris X33 and A. niger. These unique characteristics suggest that MtEG5-1 has a lot of potential for practical applications under extreme environmental conditions, including varying pH levels, osmolarities, and temperatures.
Author Contributions
Conceptualization, D.L.; methodology, W.Z. and S.T.; formal analysis, W.Z. and W.C.; investigation, W.Z.; validation, W.Z.; writing—original draft, W.Z., S.T.; writing—review and editing, S.T., F.R.A., J.C. and D.L.; conceptualization, S.T.; project administration, S.T., W.C., J.C. and D.L.; supervision, J.C. and D.L.; funding acquisition, D.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Strategic Pilot Science and Technology Project of the Chinese Academy of Sciences (XDA28030301). Many thanks to Xu Jian for his valuable suggestions for this article.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are included in the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Correa, D.F.; Beyer, H.L.; Fargione, J.E.; Hill, J.D.; Possingham, H.P.; Thomas-Hall, S.R.; Schenk, P.M. Towards the implementation of sustainable biofuel production systems. Renew. Sustain. Energy Rev. 2019, 107, 250–263. [Google Scholar] [CrossRef]
- Zhou, Z.; Liu, D.; Zhao, X. Conversion of lignocellulose to biofuels and chemicals via sugar platform: An updated review on chemistry and mechanisms of acid hydrolysis of lignocellulose. Renew. Sustain. Energy Rev. 2021, 146, 111169. [Google Scholar] [CrossRef]
- Xia, Z.; Li, J.; Zhang, J.; Zhang, X.; Zheng, X.; Zhang, J. Processing and valorization of cellulose, lignin and lignocellulose using ionic liquids. J. Bioresour. Bioprod. 2020, 5, 79–95. [Google Scholar] [CrossRef]
- Tan, J.; Li, Y.; Tan, X.; Wu, H.; Li, H.; Yang, S. Advances in Pretreatment of Straw Biomass for Sugar Production. Front. Chem. 2021, 9, 696030. [Google Scholar] [CrossRef]
- Kadowaki, M.A.S.; Higasi, P.; de Godoy, M.O.; Prade, R.A.; Polikarpov, I. Biochemical and structural insights into a thermostable cellobiohydrolase from Myceliophthora thermophila. FEBS J. 2018, 285, 559–579. [Google Scholar] [CrossRef] [PubMed]
- Himmel, M.E.; Ruth, M.F.; Wyman, C.E. Cellulase for commodity products from cellulosic biomass. Curr. Opin. Biotechnol. 1999, 10, 358–364. [Google Scholar] [CrossRef]
- Yang, W.; Fan, H.; Zhou, M.; Zhou, Z.; Yan, L.; Ju, X.; Li, L. Synergistic effect of ionic liquid and surfactant for enzymatic hydrolysis of lignocellulose by Paenibacillus sp. LLZ1 cellulase. Biomass Bioenergy 2020, 142, 105760. [Google Scholar] [CrossRef]
- Hamalainen, V.; Barajas Lopez, J.D.; Berlina, Y.; Alvarez Rafael, R.; Birikh, K. New thermostable endoglucanase from Spirochaeta thermophila and its mutants with altered substrate preferences. Appl. Microbiollogy Biotechnol. 2021, 105, 1133–1145. [Google Scholar] [CrossRef]
- Singh, B. Myceliophthora thermophila syn. Sporotrichum thermophile: A thermophilic mould of biotechnological potential. Crit. Rev. Biotechnol. 2016, 36, 59–69. [Google Scholar] [CrossRef]
- Chen, X.T.; Li, W.G.; Ji, P.; Zhao, Y.; Hua, C.Y.; Han, C. Engineering the conserved and noncatalytic residues of a thermostable beta-1,4-endoglucanase to improve specific activity and thermostability. Sci. Rep. 2018, 8, 2954. [Google Scholar] [CrossRef]
- Javanmard, A.S.; Matin, M.M.; Bahrami, A.R. Polycistronic cellulase gene expression in Pichia pastoris. Biomass Convers. Biorefinery 2021, 7, 1–15. [Google Scholar] [CrossRef]
- Kumar, S.; Nussinov, R. How do thermophilic proteins deal with heat? Cell. Mol. Life Sci. CMLS 2001, 58, 1216–1233. [Google Scholar] [CrossRef] [PubMed]
- Da Rosa-Garzon, N.G.; Laure, H.J.; Rosa, J.C.; Cabral, H. Valorization of agricultural residues using Myceliophthora thermophila as a platform for production of lignocellulolytic enzymes for cellulose saccharification. Biomass Bioenergy 2022, 161, 106452. [Google Scholar] [CrossRef]
- Wang, J.; Wu, Y.; Gong, Y.; Yu, S.; Liu, G. Enhancing xylanase production in the thermophilic fungus Myceliophthora thermophila by homologous overexpression of Mtxyr1. J. Ind. Microbiol. Biotechnol. 2015, 42, 1233–1241. [Google Scholar] [CrossRef]
- Saxena, S.; Shrivastava, S.; Arora, R.; Hussain, S.; Jena, S.C.; Kumar, M.; Vasu, R.K.; Srivastava, S.; Sharma, P.; Kumar, N.; et al. Development of Real-Time PCR Assays for Detecting Matrix Metalloproteinases-2 & 9 Over-expression in Canine Mammary Tumours. Adv. Anim. Vet. Sci. 2016, 4, 342–345. [Google Scholar] [CrossRef]
- Tambor, J.H.; Ren, H.; Ushinsky, S.; Zheng, Y.; Riemens, A.; St-Francois, C.; Tsang, A.; Powlowski, J.; Storms, R. Recombinant expression, activity screening and functional characterization identifies three novel endo-1,4-beta-glucanases that efficiently hydrolyse cellulosic substrates. Appl. Microbiol. Biotechnol. 2012, 93, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Karnaouri, A.; Muraleedharan, M.N.; Dimarogona, M.; Topakas, E.; Rova, U.; Sandgren, M.; Christakopoulos, P. Recombinant expression of thermostable processive MtEG5 endoglucanase and its synergism with MtLPMO from Myceliophthora thermophila during the hydrolysis of lignocellulosic substrates. Biotechnol. Biofuels 2017, 10, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Malik, Y.S.; Sircar, S.; Bhat, S.; Sharun, K.; Dhama, K.; Dadar, M.; Tiwari, R.; Chaicumpa, W. Emerging novel coronavirus (2019-nCoV)—Current scenario, evolutionary perspective based on genome analysis and recent developments. Vet. Q. 2020, 40, 68–76. [Google Scholar] [CrossRef]
- Reynolds, S.M.; Käll, L.; Riffle, M.E.; Bilmes, J.A.; Noble, W.S. Transmembrane Topology and Signal Peptide Prediction Using Dynamic Bayesian Networks. PLOS Comput. Biol. 2008, 4, e1000213. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, 252–258. [Google Scholar] [CrossRef]
- Karbalaei, M.; Rezaee, S.A.; Farsiani, H. Pichia pastoris: A highly successful expression system for optimal synthesis of heterologous proteins. J. Cell. Physiol. 2020, 235, 5867–5881. [Google Scholar] [CrossRef] [PubMed]
- Hill, H.D.; Straka, J.G. Protein determination using bicinchoninic acid in the presence of sulfhydryl reagents. Anal. Biochem. 1988, 170, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugars. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Gavande, P.V.; Basak, A.; Sen, S.; Lepcha, K.; Murmu, N.; Rai, V.; Mazumdar, D.; Saha, S.P.; Das, V.; Ghosh, S. Functional characterization of thermotolerant microbial consortium for lignocellulolytic enzymes with central role of Firmicutes in rice straw depolymerization. Sci. Rep. 2021, 11, 3032. [Google Scholar] [CrossRef]
- Sun, N.; Rahman, M.; Qin, Y.; Maxim, M.L.; Rodríguez, H.; Rogers, R.D. Complete dissolution and partial delignification of wood in the ionic liquid 1-ethyl-3-methylimidazolium acetate. Green Chem. 2009, 11, 646–655. [Google Scholar] [CrossRef]
- Sorn, V.; Chang, K.-L.; Phitsuwan, P.; Ratanakhanokchai, K.; Dong, C.-D. Effect of microwave-assisted ionic liquid/acidic ionic liquid pretreatment on the morphology, structure, and enhanced delignification of rice straw. Bioresour. Technol. 2019, 293, 121929. [Google Scholar] [CrossRef]
- Sahoo, D.; Ummalyma, S.B.; Okram, A.K.; Pandey, A.; Sankar, M.; Sukumaran, R.K. Effect of dilute acid pretreatment of wild rice grass (Zizania latifolia) from Loktak Lake for enzymatic hydrolysis. Bioresour. Technol. 2018, 253, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, J.; Wang, Y.; Sun, J.; Huang, P.; Chang, K. Effect of ultrasound on ionic liquid-hydrochloric acid pretreatment with rice straw. Biomass Convers. Biorefinery 2020, 11, 1749–1757. [Google Scholar] [CrossRef]
- Ghorbani, S.; Eyni, H.; Bazaz, S.R.; Nazari, H.; Asl, L.S.; Zaferani, H.; Kiani, V.; Mehrizi, A.A.; Soleimani, M. Hydrogels Based on Cellulose and its Derivatives: Applications, Synthesis, and Characteristics. Polym. Sci. Ser. A 2018, 60, 707–722. [Google Scholar] [CrossRef]
- Dalal, S.; Raghava, S.; Gupta, M. Single-step purification of recombinant green fluorescent protein on expanded beds of immobilized metal affinity chromatography media. Biochem. Eng. J. 2008, 42, 301–307. [Google Scholar] [CrossRef]
- Lindmo, T.; Boven, E.; Cuttitta, F.; Fedorko, J.; Bunn, P.A., Jr. Determination of the immunoreactive function of radiolabeled monoclonal antibodies by linear extrapolation to binding at infinite antigen excess. J. Immunol. Methods 1984, 72, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Turner, P.; Mamo, G.; Karlsson, E.N. Potential and utilization of thermophiles and thermostable enzymes in biorefining. Microb. Cell Factories 2007, 6, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Wierenga, R. The TIM-barrel fold: A versatile framework for efficient enzymes. FEBS Lett. 2001, 492, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Aspeborg, H.; Coutinho, P.M.; Wang, Y.; Brumer, H.; Henrissat, B. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 2012, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Kumar, K.; Nath, P.; Goyal, A. Role of glycine 256 residue in improving the catalytic efficiency of mutant endoglucanase of family 5 glycoside hydrolase from Bacillus amyloliquefaciens SS35. Biotechnol. Bioeng. 2020, 117, 2668–2682. [Google Scholar] [CrossRef]
- Tu, T.; Pan, X.; Meng, K.; Luo, H.; Ma, R.; Wang, Y.; Yao, B. Substitution of a non-active-site residue located on the T3 loop increased the catalytic efficiency of endo-polygalacturonases. Process Biochem. 2016, 51, 1230–1238. [Google Scholar] [CrossRef]
- Zhai, X.; Amyes, T.L.; Richard, J.P. Role of loop-clamping side chains in catalysis by triosephosphate isomerase. J. Am. Chem. Soc. 2015, 137, 15185–15197. [Google Scholar] [CrossRef]
- Gromiha, M.M.; Pujadas, G.; Magyar, C.; Selvaraj, S.; Simon, I. Locating the stabilizing residues in (alpha/beta)8 barrel proteins based on hydrophobicity, long-range interactions, and sequence conservation. Proteins 2004, 55, 316–329. [Google Scholar] [CrossRef]
- Badieyan, S.; Bevan, D.R.; Zhang, C. Study and design of stability in GH5 cellulases. Biotechnol. Bioeng. 2012, 109, 31–44. [Google Scholar] [CrossRef]
- Silva, T.P.; de Albuquerque, F.S.; Nascimento Ferreira, A.; Santos, D.; Santos, T.V.D.; Meneghetti, S.M.P.; Franco, M.; Luz, J.; Pereira, H.J.V. Dilute acid pretreatment for enhancing the enzymatic saccharification of agroresidues using a Botrytis ricini endoglucanase. Biotechnol. Appl. Biochem. 2022, 70, 184–192. [Google Scholar] [CrossRef]
- Beckham, G.T.; Dai, Z.; Matthews, J.F.; Momany, M.; Payne, C.M.; Adney, W.S.; Baker, S.E.; Himmel, M.E. Harnessing glycosylation to improve cellulase activity. Curr. Opin. Biotechnol. 2012, 23, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Karnaouri, A.; Topakas, E.; Antonopoulou, I.; Christakopoulos, P. Genomic insights into the fungal lignocellulolytic system of Myceliophthora thermophila. Front. Microbiol. 2014, 5, 281. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Wang, Q.Q.; Sun, Y.X.; Yang, R.R.; Liu, M.Y.; Wang, S.Q.; Liu, Y.F.; Zhou, L.F.; Li, D.C. Improvement of the catalytic activity and thermostability of a hyperthermostable endoglucanase by optimizing N-glycosylation sites. Biotechnol. Biofuels 2020, 13, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Schiffmann, R.; Heine, A.; Klebe, G.; Klein, C.D. Metal ions as cofactors for the binding of inhibitors to methionine aminopeptidase: A critical view of the relevance of in vitro metalloenzyme assays. Angew. Chem. Int. Ed. 2005, 44, 3620–3623. [Google Scholar] [CrossRef]
- Shoichet, B.K.; Baase, W.A.; Kuroki, R.; Matthews, B.W. A relationship between protein stability and protein function. BIochemistry 1995, 92, 452–456. [Google Scholar] [CrossRef]
- Deller, M.C.; Kong, L.; Rupp, B. Protein stability: A crystallographer’s perspective. Acta Crystallogr. Sect. F 2016, 72, 72–95. [Google Scholar] [CrossRef]
- Bianchetti, C.M.; Brumm, P.; Smith, R.W.; Dyer, K.; Hura, G.L.; Rutkoski, T.J.; Phillips Jr, G.N. Structure, dynamics, and specificity of endoglucanase D from Clostridium cellulovorans. J. Mol. Biol. 2013, 425, 4267–4285. [Google Scholar] [CrossRef]
- Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991, 280, 309–316. [Google Scholar] [CrossRef]
- Niyonzima, F.N. Detergent-compatible fungal cellulases. Folia Microbiol. 2021, 66, 25–40. [Google Scholar] [CrossRef]
- Ma, L.; Aizhan, R.; Wang, X.; Yi, Y.; Shan, Y.; Liu, B.; Zhou, Y.; Lu, X. Cloning and characterization of low-temperature adapted GH5-CBM3 endo-cellulase from Bacillus subtilis 1AJ3 and their application in the saccharification of switchgrass and coffee grounds. AMB Express 2020, 10, 42. [Google Scholar] [CrossRef]
- Salehi, M.E.; Asoodeh, A. Extraction, Purification, and Biochemical Characterization of an Alkalothermophilic Endoglucanase from Bacterial Flora in Gastrointestinal Tract of Osphranteria coerulescens Larvae. Waste Biomass Valorization 2022, 14, 1251–1265. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).