Antibacterial and Immunostimulatory Activity of Potential Probiotic Lactic Acid Bacteria Isolated from Ethiopian Fermented Dairy Products
Abstract
1. Introduction
2. Materials and Method
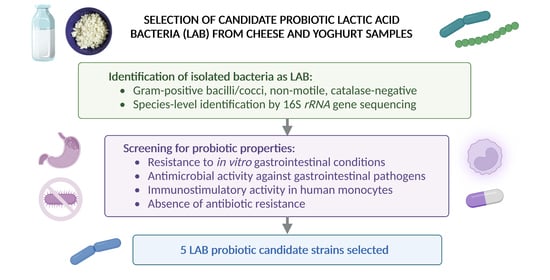
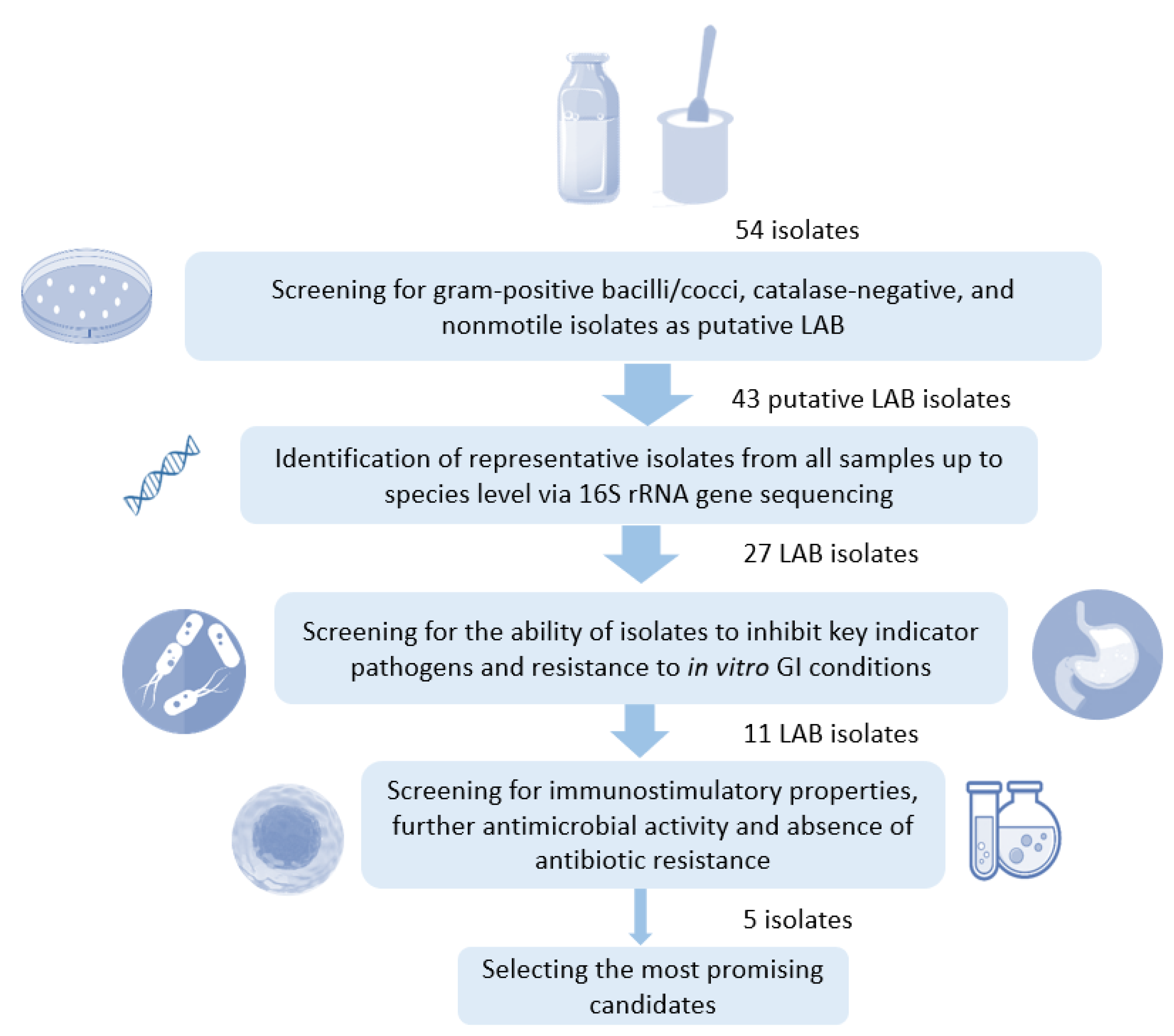
2.1. Isolation and Characterization of LAB Strains
2.2. Molecular Identification of LAB Isolates
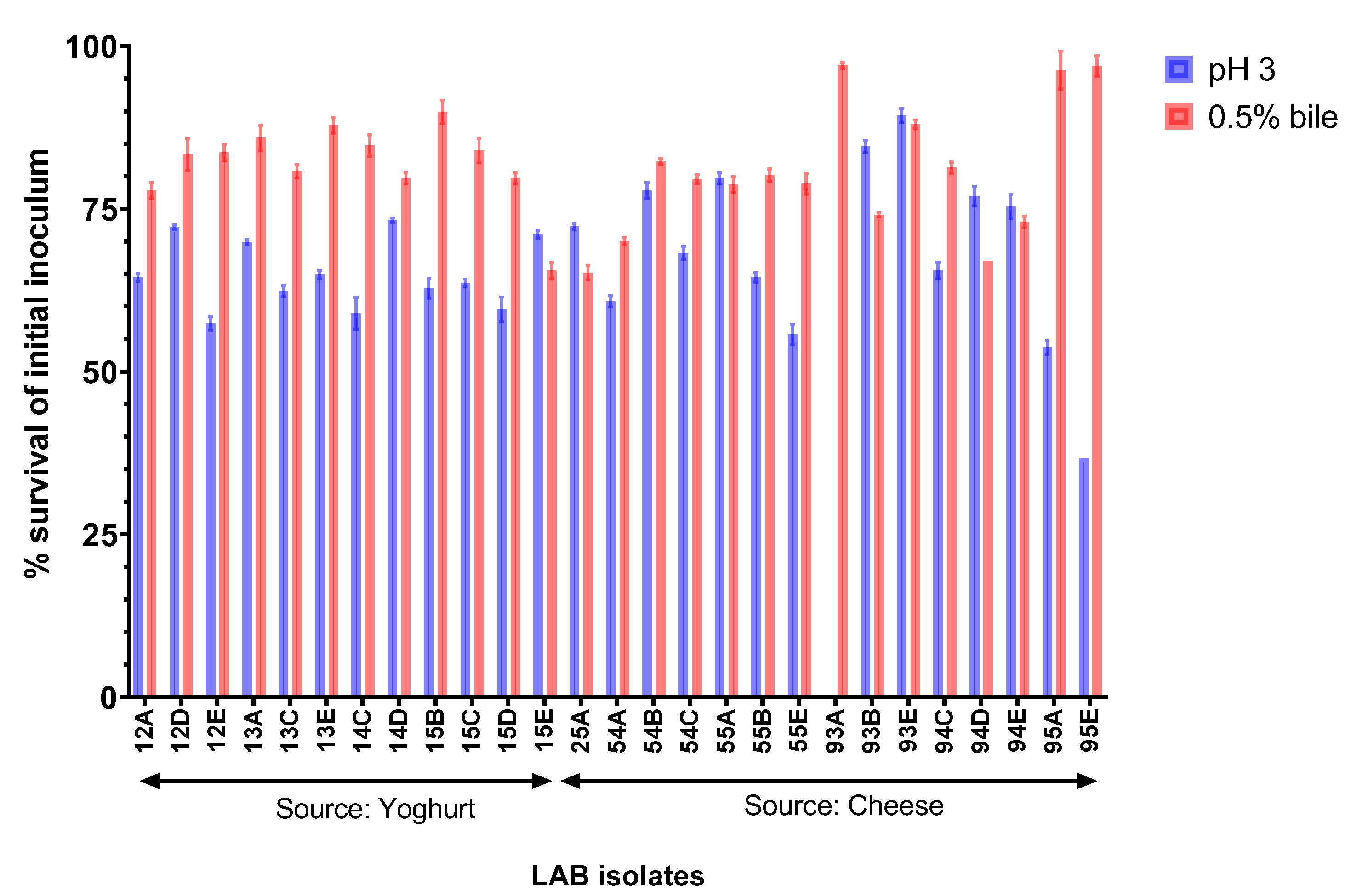
2.3. Resistance of LAB Isolates to Gastrointestinal Conditions In Vitro
2.4. Antagonistic Activity of LAB Isolates against Indicator Pathogens
2.4.1. Spot Overlay Assay
2.4.2. Radial Diffusion Assay
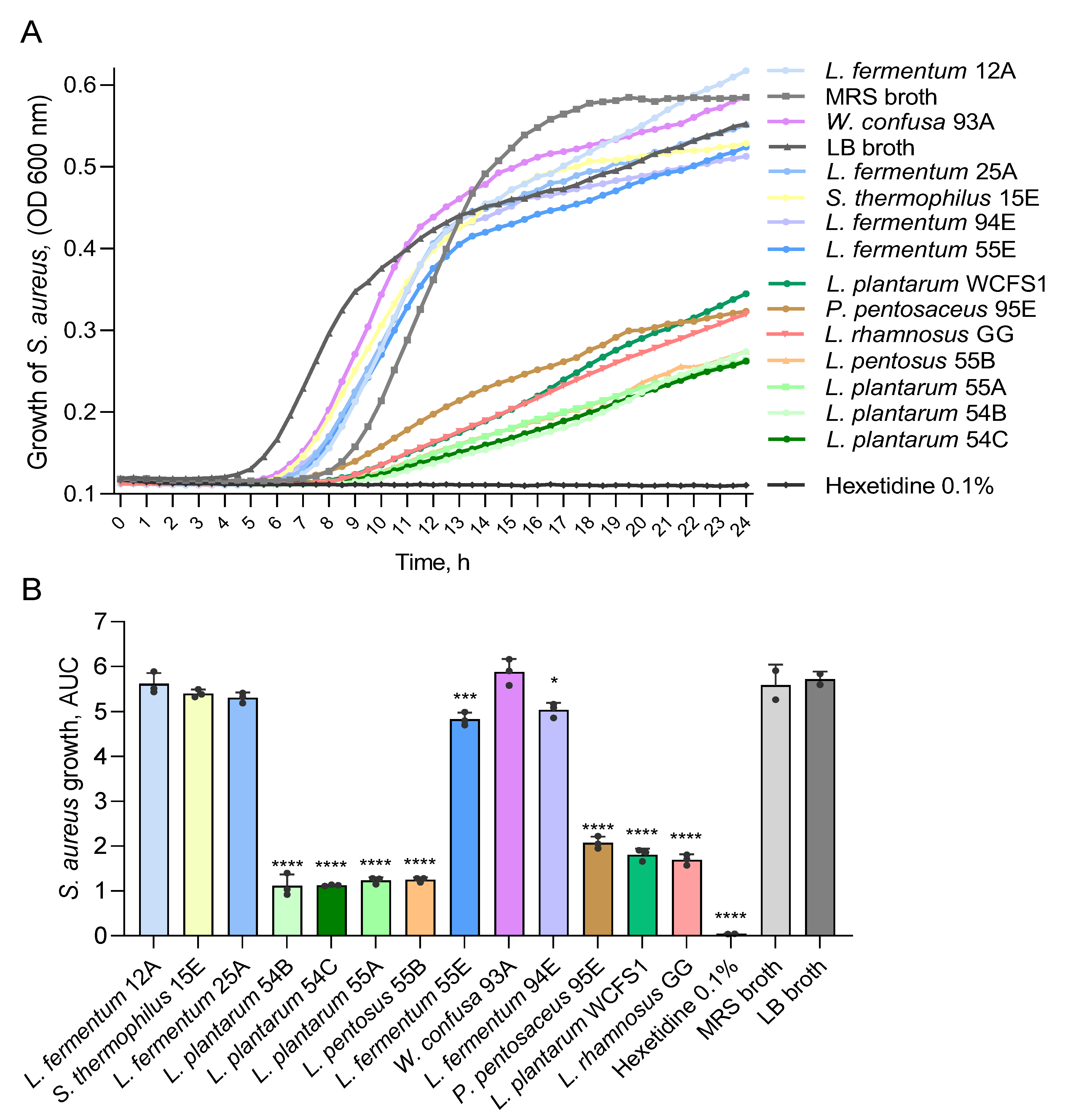
2.4.3. Antimicrobial Activity Screening of Cell-Free Culture Supernatants in Liquid Culture Assays
2.5. Assessment of Immunostimulatory Activity of LAB Isolates
2.6. Antibacterial Susceptibility Testing of LAB Isolates
2.7. Statistical Analysis
3. Results
3.1. Selected LAB Isolates from Ethiopian Fermented Dairy Products Predominantly Belong to the Genus Limosilactobacillus
3.2. Selected Isolates Show High In Vitro GI Resistance
3.3. LAB Isolates from Ethiopian Fermented Foods Inhibit Indicator Foodborne Pathogens
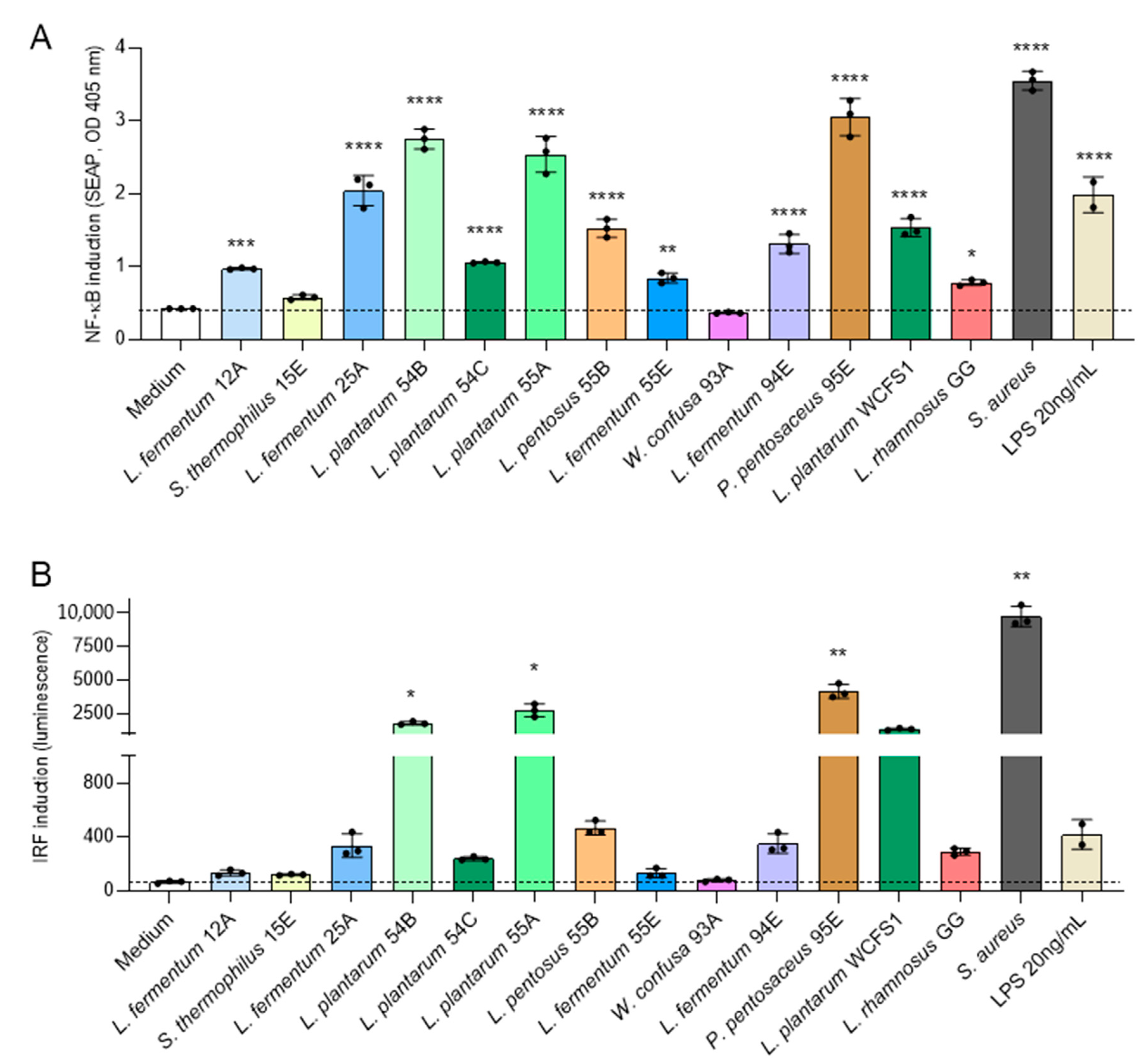
3.4. Selected Ethiopian Dairy LAB Isolates Activate NF-κB and IRF Pathways in Human Monocytes
3.5. Antibiotic Susceptibility Profile of Select LAB Isolates as Candidate Probiotic Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hutkins, R.W. Microbiology and Technology of Fermented Foods, 2nd ed.; IFT Press series; Wiley Blackwell: Hoboken, NJ, USA; Chichester, UK, 2019; ISBN 978-1-119-02756-0. [Google Scholar]
- Bell, V.; Ferrão, J.; Pimentel, L.; Pintado, M.; Fernandes, T. One Health, Fermented Foods, and Gut Microbiota. Foods 2018, 7, 195. [Google Scholar] [CrossRef] [PubMed]
- Marco, M.L.; Sanders, M.E.; Gänzle, M.; Arrieta, M.C.; Cotter, P.D.; De Vuyst, L.; Hill, C.; Holzapfel, W.; Lebeer, S.; Merenstein, D.; et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) Consensus Statement on Fermented Foods. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Koricha, A.D.; Han, D.; Bacha, K.; Bai, F. Diversity and Distribution of Yeasts in Indigenous Fermented Foods and Beverages of Ethiopia. J. Sci. Food Agric. 2020, 100, 3630–3638. [Google Scholar] [CrossRef] [PubMed]
- Ashenafi, M. Review Article: A Review on the Microbiology of Indigenous Fermented Foods and Beverages of Ethiopia. Ethiop. J. Biol. Sci. 2008, 5, 189–245. [Google Scholar] [CrossRef]
- Fentie, E.G.; Emire, S.A.; Demsash, H.D.; Dadi, D.W.; Shin, J.-H. Cereal- and Fruit-Based Ethiopian Traditional Fermented Alcoholic Beverages. Foods 2020, 9, 1781. [Google Scholar] [CrossRef]
- Lee, M.; Regu, M.; Seleshe, S. Uniqueness of Ethiopian Traditional Alcoholic Beverage of Plant Origin, Tella. J. Ethn. Foods 2015, 2, 110–114. [Google Scholar] [CrossRef]
- Minten, B.; Habte, Y.; Tamru, S.; Tesfaye, A. The Transforming Dairy Sector in Ethiopia. PLoS ONE 2020, 15, e0237456. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A Taxonomic Note on the Genus Lactobacillus: Description of 23 Novel Genera, Emended Description of the Genus Lactobacillus Beijerinck 1901, and Union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Dejene, F.; Regasa Dadi, B.; Tadesse, D. In Vitro Antagonistic Effect of Lactic Acid Bacteria Isolated from Fermented Beverage and Finfish on Pathogenic and Foodborne Pathogenic Microorganism in Ethiopia. Int. J. Microbiol. 2021, 2021, 5370556. [Google Scholar] [CrossRef]
- EFSA Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Hilbert, F.; et al. Update of the List of QPS-recommended Biological Agents Intentionally Added to Food or Feed as Notified to EFSA 13: Suitability of Taxonomic Units Notified to EFSA until September 2020. EFSA J. 2021, 19, e06377. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Binda, S.; Hill, C.; Johansen, E.; Obis, D.; Pot, B.; Sanders, M.E.; Tremblay, A.; Ouwehand, A.C. Criteria to Qualify Microorganisms as “Probiotic” in Foods and Dietary Supplements. Front. Microbiol. 2020, 11, 1662. [Google Scholar] [CrossRef]
- Bermudez-Brito, M.; Plaza-Díaz, J.; Muñoz-Quezada, S.; Gómez-Llorente, C.; Gil, A. Probiotic Mechanisms of Action. Ann. Nutr. Metab. 2012, 61, 160–174. [Google Scholar] [CrossRef]
- Ashraf, R.; Shah, N.P. Immune System Stimulation by Probiotic Microorganisms. Crit. Rev. Food Sci. Nutr. 2014, 54, 938–956. [Google Scholar] [CrossRef]
- Wilkins, T.; Sequoia, J. Probiotics for Gastrointestinal Conditions: A Summary of the Evidence. Am. Fam. Physician 2017, 96, 170–178. [Google Scholar]
- WHO. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015; World Health Organization: Geneva, Switzerland, 2015; ISBN 978-92-4-156516-5.
- Havelaar, A.H.; Kirk, M.D.; Torgerson, P.R.; Gibb, H.J.; Hald, T.; Lake, R.J.; Praet, N.; Bellinger, D.C.; de Silva, N.R.; Gargouri, N.; et al. World Health Organization Global Estimates and Regional Comparisons of the Burden of Foodborne Disease in 2010. PLoS Med. 2015, 12, e1001923. [Google Scholar] [CrossRef]
- Misganaw, A.; Melaku, Y.A.; Tessema, G.A.; Deribew, A.; Deribe, K.; Abera, S.F.; Dessalegn, M.; Lakew, Y.; Bekele, T.; Haregu, T.N.; et al. National Disability-Adjusted Life Years (DALYs) for 257 Diseases and Injuries in Ethiopia, 1990–2015: Findings from the Global Burden of Disease Study 2015. Popul. Health Metr. 2017, 15, 28. [Google Scholar] [CrossRef]
- Eshetie, S.; Tarekegn, F.; Moges, F.; Amsalu, A.; Birhan, W.; Huruy, K. Methicillin Resistant Staphylococcus Aureus in Ethiopia: A Meta-Analysis. BMC Infect. Dis. 2016, 16, 689. [Google Scholar] [CrossRef]
- Hussen, S.; Mulatu, G.; Yohannes Kassa, Z. Prevalence of Shigella Species and Its Drug Resistance Pattern in Ethiopia: A Systematic Review and Meta-Analysis. Ann. Clin. Microbiol. Antimicrob. 2019, 18, 22. [Google Scholar] [CrossRef]
- WHO. Global Action Plan on Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2015; ISBN 978-92-4-150976-3. [Google Scholar]
- Jubeh, B.; Breijyeh, Z.; Karaman, R. Resistance of Gram-Positive Bacteria to Current Antibacterial Agents and Overcoming Approaches. Molecules 2020, 25, 2888. [Google Scholar] [CrossRef]
- Lebeer, S.; Vanderleyden, J.; De Keersmaecker, S.C.J. Genes and Molecules of Lactobacilli Supporting Probiotic Action. Microbiol. Mol. Biol. Rev. 2008, 72, 728–764. [Google Scholar] [CrossRef] [PubMed]
- Shanahan, F. 99th Dahlem Conference on Infection, Inflammation and Chronic Inflammatory Disorders: Host-Microbe Interactions in the Gut: Target for Drug Therapy, Opportunity for Drug Discovery: Mining the Microbiota. Clin. Exp. Immunol. 2010, 160, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yao, M.; Lv, L.; Ling, Z.; Li, L. The Human Microbiota in Health and Disease. Engineering 2017, 3, 71–82. [Google Scholar] [CrossRef]
- Lebeer, S.; Vanderleyden, J.; De Keersmaecker, S.C.J. Host Interactions of Probiotic Bacterial Surface Molecules: Comparison with Commensals and Pathogens. Nat. Rev. Microbiol. 2010, 8, 171–184. [Google Scholar] [CrossRef]
- Wells, J.M.; Loonen, L.M.P.; Karczewski, J.M. The Role of Innate Signaling in the Homeostasis of Tolerance and Immunity in the Intestine. Int. J. Med. Microbiol. 2010, 300, 41–48. [Google Scholar] [CrossRef]
- Jensen, H.; Drømtorp, S.M.; Axelsson, L.; Grimmer, S. Immunomodulation of Monocytes by Probiotic and Selected Lactic Acid Bacteria. Probiotics Antimicrob. Proteins 2015, 7, 14–23. [Google Scholar] [CrossRef]
- Spacova, I.; De Boeck, I.; Bron, P.A.; Delputte, P.; Lebeer, S. Topical Microbial Therapeutics against Respiratory Viral Infections. Trends Mol. Med. 2021, 27, 538–553. [Google Scholar] [CrossRef]
- Kawashima, T.; Kosaka, A.; Yan, H.; Guo, Z.; Uchiyama, R.; Fukui, R.; Kaneko, D.; Kumagai, Y.; You, D.-J.; Carreras, J.; et al. Double-Stranded RNA of Intestinal Commensal but Not Pathogenic Bacteria Triggers Production of Protective Interferon-β. Immunity 2013, 38, 1187–1197. [Google Scholar] [CrossRef]
- Spacova, I.; De Boeck, I.; Cauwenberghs, E.; Delanghe, L.; Bron, P.A.; Henkens, T.; Simons, A.; Gamgami, I.; Persoons, L.; Claes, I.; et al. Development of a Live Biotherapeutic Throat Spray with Lactobacilli Targeting Respiratory Viral Infections. Microb. Biotechnol. 2023, 16, 99–115. [Google Scholar] [CrossRef]
- De MAN, J.C.; Rogosa, M.; Sharpe, M.E. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 1960, 23, 130–135. [Google Scholar] [CrossRef]
- da Silva, N. Microbiological Examination Methods of Food and Water: A Laboratory Manual, 2nd ed.; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2013; ISBN 978-1-138-05711-1. [Google Scholar]
- Lagier, J.-C.; Bilen, M.; Cadoret, F.; Drancourt, M.; Fournier, P.-E.; La Scola, B.; Raoult, D. Naming Microorganisms: The Contribution of the IHU Méditerranée Infection, Marseille, France. New Microbes New Infect. 2018, 26, S89–S95. [Google Scholar] [CrossRef]
- Panya, M.; Lulitanond, V.; Rattanachaikunsopon, P.; Srivoramas, T.; Chaiwong, T. Isolation, Identification, and Evaluation of Novel Probiotic Strains Isolated from Feces of Breast-Fed Infants. J. Med. Assoc. Thail. Chotmaihet Thangphaet 2016, 99, S28–S34. [Google Scholar]
- Argyri, A.A.; Zoumpopoulou, G.; Karatzas, K.-A.G.; Tsakalidou, E.; Nychas, G.-J.E.; Panagou, E.Z.; Tassou, C.C. Selection of Potential Probiotic Lactic Acid Bacteria from Fermented Olives by in Vitro Tests. Food Microbiol. 2013, 33, 282–291. [Google Scholar] [CrossRef]
- Garcia, E.F.; Luciano, W.A.; Xavier, D.E.; da Costa, W.C.A.; de Sousa Oliveira, K.; Franco, O.L.; de Morais Júnior, M.A.; Lucena, B.T.L.; Picão, R.C.; Magnani, M.; et al. Identification of Lactic Acid Bacteria in Fruit Pulp Processing Byproducts and Potential Probiotic Properties of Selected Lactobacillus Strains. Front. Microbiol. 2016, 7, 1371. [Google Scholar] [CrossRef]
- Van Beeck, W.; Verschueren, C.; Wuyts, S.; van den Broek, M.F.L.; Uyttendaele, M.; Lebeer, S. Robustness of Fermented Carrot Juice against Listeria Monocytogenes, Salmonella Typhimurium and Escherichia Coli O157:H7. Int. J. Food Microbiol. 2020, 335, 108854. [Google Scholar] [CrossRef]
- van den Broek, M.F.L.; De Boeck, I.; Claes, I.J.J.; Nizet, V.; Lebeer, S. Multifactorial Inhibition of Lactobacilli against the Respiratory Tract Pathogen Moraxella Catarrhalis. Benef. Microbes 2018, 9, 429–439. [Google Scholar] [CrossRef]
- EFSA. Guidance on the Assessment of Bacterial Susceptibility to Antimicrobials of Human and Veterinary Importance EFSA Panel on Additives and Products or Substances Used in Animal Feed (FEEDAP). EFSA J. 2012, 10, 2740. [Google Scholar] [CrossRef]
- CLSI (Ed.) Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard, 9th ed.; Clinical and Laboratory Standards Institute; CLSI: Wayne, PA, USA, 2012; ISBN 978-1-56238-784-6. [Google Scholar]
- Kankainen, M.; Paulin, L.; Tynkkynen, S.; von Ossowski, I.; Reunanen, J.; Partanen, P.; Satokari, R.; Vesterlund, S.; Hendrickx, A.P.A.; Lebeer, S.; et al. Comparative Genomic Analysis of Lactobacillus rhamnosus GG Reveals Pili Containing a Human- Mucus Binding Protein. Proc. Natl. Acad. Sci. USA 2009, 106, 17193–17198. [Google Scholar] [CrossRef]
- Kleerebezem, M.; Boekhorst, J.; van Kranenburg, R.; Molenaar, D.; Kuipers, O.P.; Leer, R.; Tarchini, R.; Peters, S.A.; Sandbrink, H.M.; Fiers, M.W.E.J.; et al. Complete Genome Sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 2003, 100, 1990–1995. [Google Scholar] [CrossRef]
- Ram, Y.; Dellus-Gur, E.; Bibi, M.; Karkare, K.; Obolski, U.; Feldman, M.W.; Cooper, T.F.; Berman, J.; Hadany, L. Predicting Microbial Growth in a Mixed Culture from Growth Curve Data. Proc. Natl. Acad. Sci. USA 2019, 116, 14698–14707. [Google Scholar] [CrossRef]
- FAO. WHO Guidelines for the Evaluation of Probiotics in Food; Report of a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food; FAO: Rome, Italy, 2002. [Google Scholar]
- Monteagudo-Mera, A.; Rodríguez-Aparicio, L.; Rúa, J.; Martínez-Blanco, H.; Navasa, N.; García-Armesto, M.R.; Ferrero, M.Á. In Vitro Evaluation of Physiological Probiotic Properties of Different Lactic Acid Bacteria Strains of Dairy and Human Origin. J. Funct. Foods 2012, 4, 531–541. [Google Scholar] [CrossRef]
- Szutowska, J.; Gwiazdowska, D. Probiotic Potential of Lactic Acid Bacteria Obtained from Fermented Curly Kale Juice. Arch. Microbiol. 2021, 203, 975–988. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Castilho, N.P.A.; Todorov, S.D.; Nero, L.A. Beneficial Properties of Lactic Acid Bacteria Naturally Present in Dairy Production. BMC Microbiol. 2018, 18, 219. [Google Scholar] [CrossRef] [PubMed]
- Taye, Y.; Degu, T.; Fesseha, H.; Mathewos, M. Isolation and Identification of Lactic Acid Bacteria from Cow Milk and Milk Products. Sci. World J. 2021, 4697445. [Google Scholar] [CrossRef]
- Park, S.; Steinegger, M.; Cho, H.-S.; Chun, J. Metagenomic Association Analysis of Gut Symbiont Limosilactobacillus Reuteri without Host-Specific Genome Isolation. Front. Microbiol. 2020, 11, 585622. [Google Scholar] [CrossRef]
- Del Piano, M.; Carmagnola, S.; Ballarè, M.; Sartori, M.; Orsello, M.; Balzarini, M.; Pagliarulo, M.; Tari, R.; Anderloni, A.; Strozzi, G.P.; et al. Is Microencapsulation the Future of Probiotic Preparations? The Increased Efficacy of Gastro-Protected Probiotics. Gut Microbes 2011, 2, 120–123. [Google Scholar] [CrossRef]
- Reuben, R.C.; Roy, P.C.; Sarkar, S.L.; Rubayet Ul Alam, A.S.M.; Jahid, I.K. Characterization and Evaluation of Lactic Acid Bacteria from Indigenous Raw Milk for Potential Probiotic Properties. J. Dairy Sci. 2020, 103, 1223–1237. [Google Scholar] [CrossRef]
- Al-Madboly, L.A.; Abdullah, A.K. Potent Antagonistic Activity of Egyptian Lactobacillus Plantarum against Multiresistant and Virulent Food-Associated Pathogens. Front. Microbiol. 2015, 6, 347. [Google Scholar] [CrossRef]
- Spacova, I.; O’Neill, C.; Lebeer, S. Lacticaseibacillus Rhamnosus GG Inhibits Infection of Human Keratinocytes by Staphylococcus Aureus through Mechanisms Involving Cell Surface Molecules and PH Reduction. Benef. Microbes 2020, 11, 703–715. [Google Scholar] [CrossRef]
- Spacova, I.; Van Beeck, W.; Seys, S.; Devos, F.; Vanoirbeek, J.; Vanderleyden, J.; Ceuppens, J.; Petrova, M.; Lebeer, S. Lactobacillus Rhamnosus Probiotic Prevents Airway Function Deterioration and Promotes Gut Microbiome Resilience in a Murine Asthma Model. Gut Microbes 2020, 11, 1729–1744. [Google Scholar] [CrossRef]
- Lorea Baroja, M.; Kirjavainen, P.V.; Hekmat, S.; Reid, G. Anti-Inflammatory Effects of Probiotic Yogurt in Inflammatory Bowel Disease Patients. Clin. Exp. Immunol. 2007, 149, 470–479. [Google Scholar] [CrossRef]
- De Boeck, I.; Cauwenberghs, E.; Spacova, I.; Gehrmann, T.; Eilers, T.; Delanghe, L.; Wittouck, S.; Bron, P.A.; Henkens, T.; Gamgami, I.; et al. Randomized, double-blind, placebo-controlled trial of a throat spray with selected lactobacilli in COVID-19 outpatients. Microbiol. Spectr. 2022, 10, e0168222. [Google Scholar] [CrossRef]
- Brown, K.D.; Claudio, E.; Siebenlist, U. The Roles of the Classical and Alternative Nuclear Factor-KappaB Pathways: Potential Implications for Autoimmunity and Rheumatoid Arthritis. Arthritis Res. Ther. 2008, 10, 212. [Google Scholar] [CrossRef]
- Grinberg-Bleyer, Y.; Caron, R.; Seeley, J.J.; De Silva, N.S.; Schindler, C.W.; Hayden, M.S.; Klein, U.; Ghosh, S. The Alternative NF-ΚB Pathway in Regulatory T Cell Homeostasis and Suppressive Function. J. Immunol. 2018, 200, 2362–2371. [Google Scholar] [CrossRef]
- Miraghazadeh, B.; Cook, M.C. Nuclear Factor-KappaB in Autoimmunity: Man and Mouse. Front. Immunol. 2018, 9, 613. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-ΚB Signaling in Inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Weiss, G.; Maaetoft-Udsen, K.; Stifter, S.A.; Hertzog, P.; Goriely, S.; Thomsen, A.R.; Paludan, S.R.; Frøkiær, H. MyD88 Drives the IFN-β Response to Lactobacillus acidophilus in Dendritic Cells through a Mechanism Involving IRF1, IRF3, and IRF7. J. Immunol. 2012, 189, 2860–2868. [Google Scholar] [CrossRef]
- Coleman, J.L.; Hatch-McChesney, A.; Small, S.D.; Allen, J.T.; Sullo, E.; Agans, R.T.; Fagnant, H.S.; Bukhari, A.S.; Karl, J.P. Orally Ingested Probiotics, Prebiotics, and Synbiotics as Countermeasures for Respiratory Tract Infections in Nonelderly Adults: A Systematic Review and Meta-Analysis. Adv. Nutr. 2022, 13, 2277–2295. [Google Scholar] [CrossRef]
- Nawaz, M.; Wang, J.; Zhou, A.; Ma, C.; Wu, X.; Moore, J.E.; Cherie Millar, B.; Xu, J. Characterization and Transfer of Antibiotic Resistance in Lactic Acid Bacteria from Fermented Food Products. Curr. Microbiol. 2011, 62, 1081–1089. [Google Scholar] [CrossRef]
| Source | Strain | Identified by 16S rRNA as: | Pairwise Similarity (%) | Selected (Yes) |
|---|---|---|---|---|
| Commercially fermented yogurt | 12A | Limosilactobacillus fermentum | 99.92 | Yes |
| 12D | Limosilactobacillus fermentum | 100 | ||
| 12E | Limosilactobacillus fermentum | 100 | ||
| 13A | Limosilactobacillus fermentum | 99.91 | ||
| 13C | Limosilactobacillus fermentum | 100 | ||
| 13E | Limosilactobacillus fermentum | 100 | ||
| 14C | Limosilactobacillus fermentum | 100 | ||
| 14D | Limosilactobacillus fermentum | 100 | ||
| 15B | Limosilactobacillus fermentum | 100 | ||
| 15C | Limosilactobacillus fermentum | 100 | ||
| 15D | Limosilactobacillus fermentum | 100 | ||
| 15E | Streptococcus thermophilus | 99.92 | Yes | |
| Commercially fermented cheese | 25A | Limosilactobacillus fermentum | 99.92 | Yes |
| Spontaneously fermented cheese | 54A | Enterococcus lactis | 99.77 | |
| 54B | Lactiplantibacillusplantarum | 100 | Yes | |
| 54C | Lactiplantibacillusplantarum | 100 | Yes | |
| 55A | Lactiplantibacillus plantarum | 100 | Yes | |
| 55B | Lactiplantibacillus pentosus | 100 | Yes | |
| 55E | Limosilactobacillus fermentum | 100 | Yes | |
| 93A | Weissella confusa | 100 | Yes | |
| 93B | Limosilactobacillus fermentum | 99.92 | ||
| 93E | Limosilactobacillus fermentum | 99.85 | ||
| 94C | Limosilactobacillus fermentum | 99.85 | ||
| 94D | Limosilactobacillus fermentum | 99.84 | ||
| 94E | Limosilactobacillus fermentum | 99.84 | Yes | |
| 95A | Limosilactobacillus fermentum | 99.85 | ||
| 95E | Pediococcus pentosaceus | 100 | Yes |
| Zone of Inhibition (mm) 1, Data Are Mean Values ± SD, (n = 3) | Zone of Inhibition (mm)2, Data Are Mean Values ± SD, (n = 3) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L. monocytogenes ATCC 19115 | S. aureus ATCC 25923 | E. coli ATCC 25922 | methicillin-resistant S. aureus | E. coli O157:H7 BRMSID188 | S. enterica subsp. enterica var. Typhimurium NTCT 13347 | S. flexneri LMG 10472 | L. monocytogenes MB2022 | S. aureus MI/1310/1938 | ||||||
| LAB strain (Source) | Spot overlay | Spot overlay | Spot overlay | Spot overlay | Radial diffusion | Spot overlay | Radial diffusion | Spot overlay | Radial diffusion | Spot overlay | Radial diffusion | Spot overlay | Radial diffusion | Spot overlay |
| Limosilactobacillus fermentum 12A (1) | +++ | + | +++ | +++ | ++ | − | ++ | − | + | − | + | ++ | − | + |
| Streptococcus thermophilus 15E (1) | ++ | + | ++ | − | ++ | − | ++ | − | – | − | – | − | − | − |
| L. fermentum 25A (2) | ++ | ++ | +++ | ++ | ++ | − | ++ | − | + | − | ++ | ++ | − | ++ |
| Lactiplantibacillus plantarum 54B (5) | ++ | + | ++ | ++ | ++ | ++ | ++ | ++ | +++ | ++ | +++ | +++ | − | +++ |
| L. plantarum 54C (5) | ++ | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | +++ | ++ | +++ | − | ++ |
| L. plantarum 55A (5) | ++ | ++ | ++ | ++ | +++ | ++ | ++ | ++ | +++ | ++ | +++ | +++ | − | ++ |
| Lactiplantibacillus pentosus 55B (5) | +++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | +++ | − | +++ |
| L. fermentum 55E (5) | ++ | ++ | ++ | +++ | ++ | − | ++ | − | − | − | ++ | ++ | − | + |
| Weissella confusa 93A (9) | − | + | ++ | ++ | ++ | ++ | ++ | ++ | − | ++ | − | +++ | − | ++ |
| L. fermentum 94E (9) | ++ | +++ | ++ | ++ | +++ | − | ++ | − | ++ | − | ++ | ++ | − | − |
| Pediococcus pentosaceus 95E (9) | +++ | ++ | +++ | ++ | ++ | ++ | ++ | ++ | ++ | +++ | ++ | +++ | − | ++ |
| Chlorhexidine 0.2% | ++ | + | + | + | ||||||||||
| Lacticaseibacillus rhamnosus GG | +++ | +++ | ++ | ++ | ++ | +++ | +++ | +++ | − | +++ | ||||
| L. plantarum WCFS1 | +++ | ++ | ++ | ++ | ++ | +++ | +++ | ++ | − | ++ | ||||
| Hexetidine 0.1% | ++ | ++ | + | − | ++ | + | +++ | +++ | +++ | +++ | ||||
| Isolate | Amp | Gent | Kana | Strep | Eryth | Clind | TTC | CAF |
|---|---|---|---|---|---|---|---|---|
| L. fermentum 12A | S | S | R | S | S | S | S | S |
| S. thermophilus 15E | S | S | S | S | S | S | S | S |
| L. fermentum 25A | S | S | R | S | S | S | S | S |
| L. plantarum 54B | S | S | R | n.r | S | S | S | S |
| L. plantarum 54C | S | S | R | n.r | S | S | S | S |
| L. plantarum 55A | S | R | R | n.r | S | S | S | S |
| L. pentosus 55B | S | S | S | n.r | S | S | S | S |
| L. fermentum 55E | S | S | R | S | S | S | S | S |
| W. confusa 93A | S | S | R | S | S | S | S | S |
| L. fermentum 94E | S | S | R | S | S | S | S | S |
| P. pentosaceus 95E | S | S | R | S | S | S | S | S |
| L. rhamnosus GG | S | R | R | S | S | S | S | S |
| L. plantarum WCFS1 | S | S | S | S | S | S | S | S |
| Property Tested | Good Candidate LAB Strains | Poor candidate LAB Strains | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L. plantarum 54B | L. plantarum 54C | L. plantarum 55A | L. pentosus 55B | P. pentosaceus 95E | W. confusa 93A | L. fermentum 12A | S. thermophilus 15E | L. fermentum 25A | L. fermentum 55E | L. fermentum 94E | ||
| Antipathogenic activity against | L. monocytogenes ATCC 19115 | √ | √ | √ | √ | √ | − | √ | √ | √ | √ | √ |
| S. aureus ATCC 25923 | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | |
| E. coli ATCC 25922 | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | |
| Methicillin-resistant S. aureus (MRSA) | √ | √ | √ | √ | √ | √ | √ | − | √ | √ | √ | |
| L. monocytogenes MB2022 | √ | √ | √ | √ | √ | − | √ | − | √ | √ | √ | |
| S. enterica subsp. enterica var. Typhimurium NTCT 13347 | √ | √ | √ | √ | √ | √ | − | − | − | − | − | |
| E. coli O157:H7 BRMSID188 | √ | √ | √ | √ | √ | √ | − | − | − | − | − | |
| S. aureus MI/1310/1938 | √ | √ | √ | √ | √ | − | − | − | − | − | − | |
| S. flexneri LMG 10472 | √ | √ | √ | √ | √ | − | − | − | − | − | − | |
| In vitro GI conditions resistance | pH= 3 | √ | √ | √ | √ | √ | − | √ | √ | √ | √ | √ |
| Bile salt 0.5% | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | |
| NF-κB activation | √ | √ | √ | √ | √ | − | √ | − | √ | √ | √ | |
| IRF induction | √ | ns | ns | ns | √ | − | − | − | ns | − | ns | |
| AST | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gizachew, S.; Van Beeck, W.; Spacova, I.; Dekeukeleire, M.; Alemu, A.; Woldemedhin, W.M.; Mariam, S.H.; Lebeer, S.; Engidawork, E. Antibacterial and Immunostimulatory Activity of Potential Probiotic Lactic Acid Bacteria Isolated from Ethiopian Fermented Dairy Products. Fermentation 2023, 9, 258. https://doi.org/10.3390/fermentation9030258
Gizachew S, Van Beeck W, Spacova I, Dekeukeleire M, Alemu A, Woldemedhin WM, Mariam SH, Lebeer S, Engidawork E. Antibacterial and Immunostimulatory Activity of Potential Probiotic Lactic Acid Bacteria Isolated from Ethiopian Fermented Dairy Products. Fermentation. 2023; 9(3):258. https://doi.org/10.3390/fermentation9030258
Chicago/Turabian StyleGizachew, Seyoum, Wannes Van Beeck, Irina Spacova, Max Dekeukeleire, Ashenafi Alemu, Wude Mihret Woldemedhin, Solomon H. Mariam, Sarah Lebeer, and Ephrem Engidawork. 2023. "Antibacterial and Immunostimulatory Activity of Potential Probiotic Lactic Acid Bacteria Isolated from Ethiopian Fermented Dairy Products" Fermentation 9, no. 3: 258. https://doi.org/10.3390/fermentation9030258
APA StyleGizachew, S., Van Beeck, W., Spacova, I., Dekeukeleire, M., Alemu, A., Woldemedhin, W. M., Mariam, S. H., Lebeer, S., & Engidawork, E. (2023). Antibacterial and Immunostimulatory Activity of Potential Probiotic Lactic Acid Bacteria Isolated from Ethiopian Fermented Dairy Products. Fermentation, 9(3), 258. https://doi.org/10.3390/fermentation9030258