Development of Digested Sludge-Assimilating and Biohydrogen-Yielding Microflorae
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
2.2. Preparation of DS
2.3. Heat Treatment of the Microflora and Vial-Scale Dark Fermentation
2.4. Bottle-Scale Dark Fermentation
2.5. Biogas Analysis
2.6. Community Fingerprinting of Microflora
2.7. DNA Sequencing of the PCR-DGGE Amplicons
2.8. Enzyme Assay
2.9. Statistical Analysis
3. Results
3.1. Heat Treatment of the DABYS Microflora
3.2. Enzyme Activity of the Heat-Treated DABYS Microflora
3.3. Bacterial Compositions of the Heat-Treated DABYS Microflorae
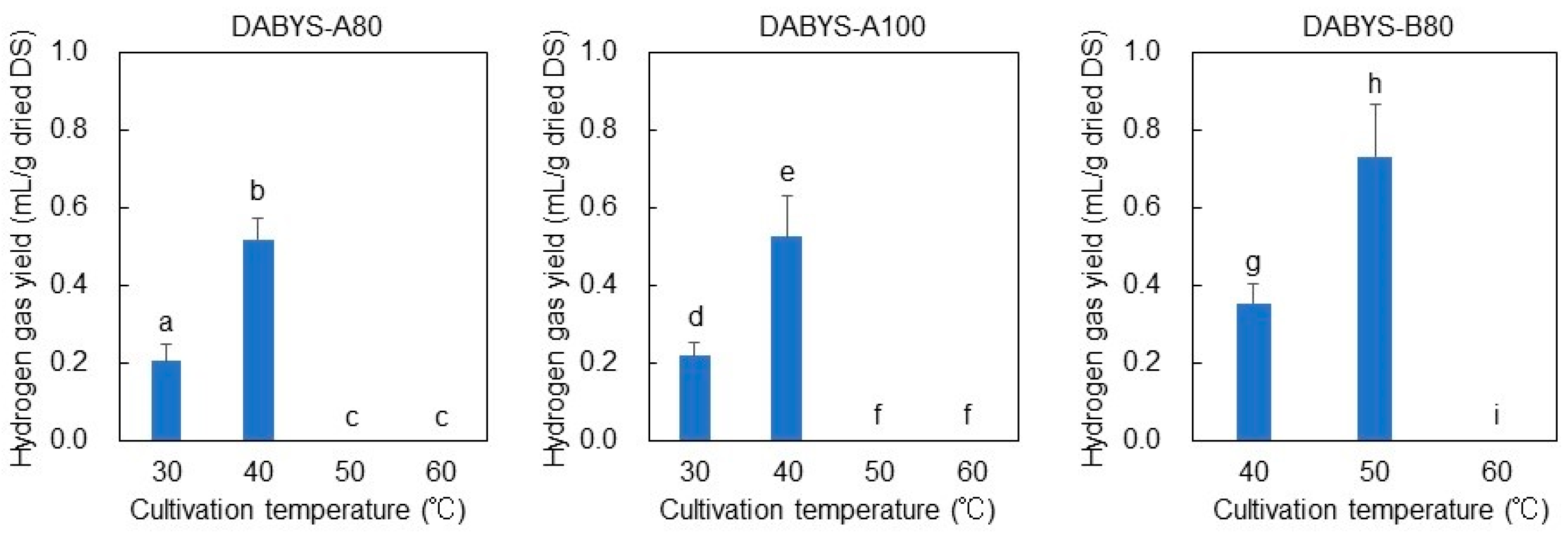
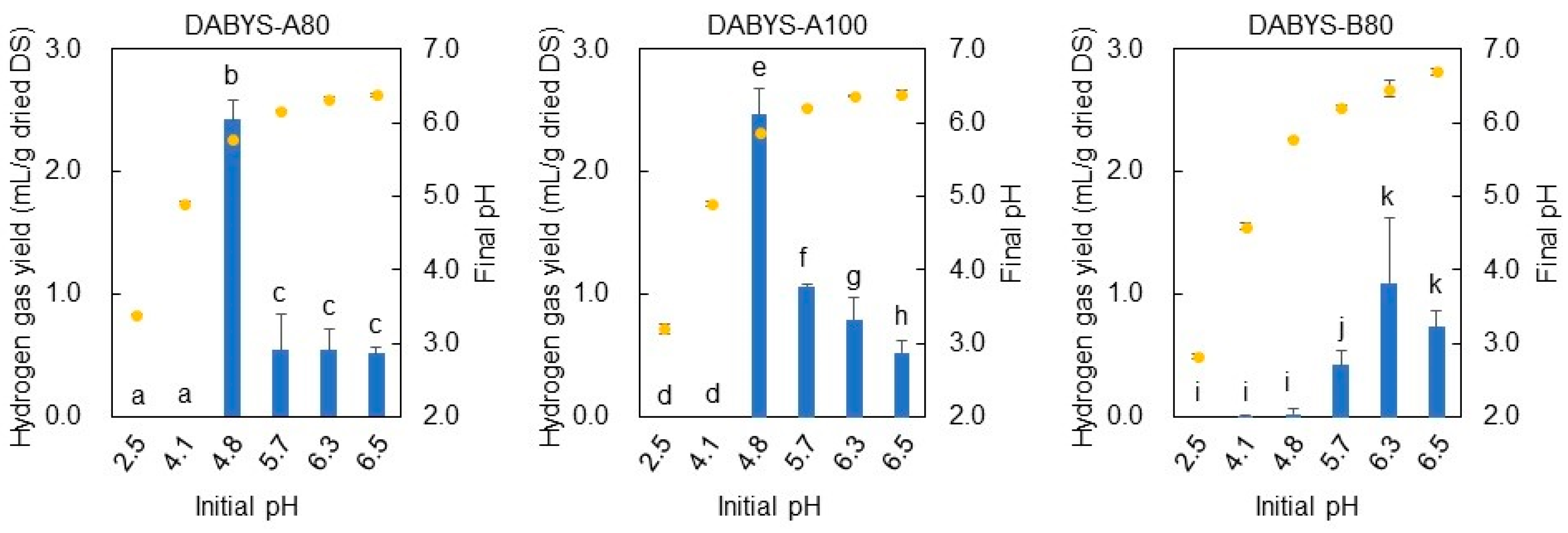
3.4. Influences of Temperature and Initial pH on the Hydrogen Gas-Producing Activity of the Microflorae in the Dark Fermentation
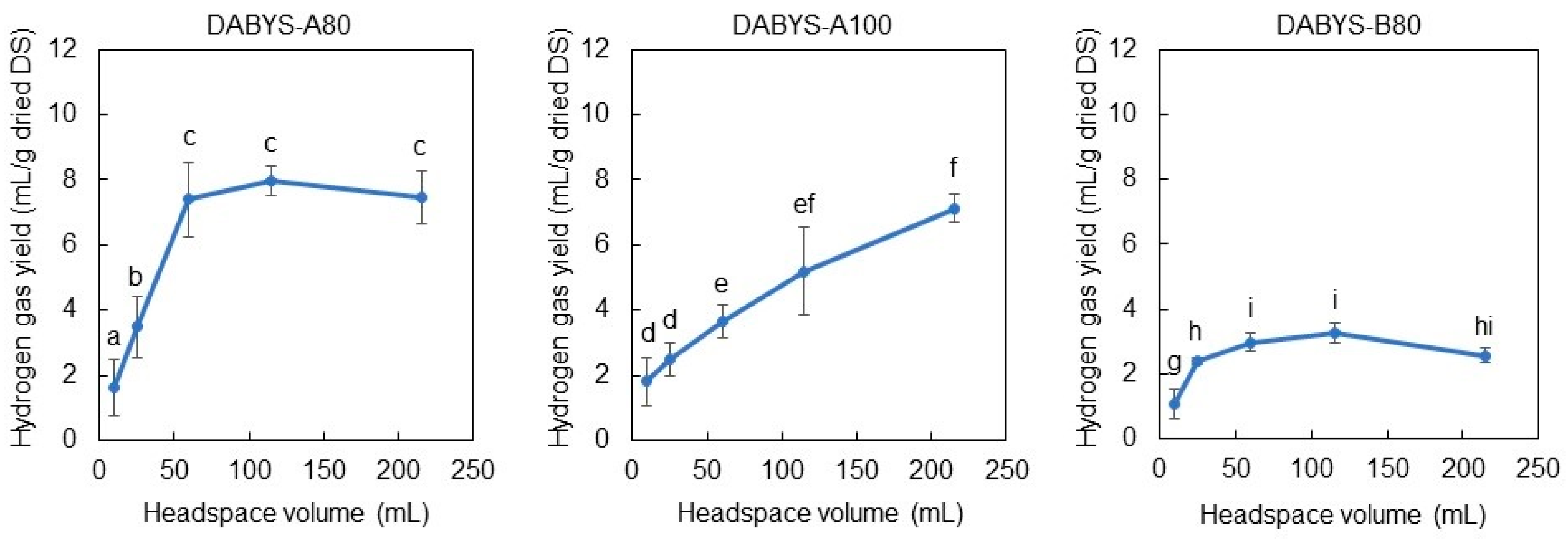
3.5. Influence of Headspace Volume on the Hydrogen-Producing Activity of the DABYS Microflorae in the Dark Fermentation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, Y.; Lu, Y.; Zheng, L.; Wang, Z.; Dai, X. Perspective on enhancing the anaerobic digestion of waste activated sludge. J. Hazard. Mater. 2020, 389, 121847. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Xin, X.; Qiu, W.; Li, D.; Liu, Z.; Ma, J. Waste sludge disintegration, methanogenesis and final disposal via various pretreatments: Comparison of performance and effectiveness. Environ. Sci. Ecotechnol. 2021, 8, 100132. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, T.; Duan, H.; Song, Y.; Lu, X.; Hu, S.; Yuan, Z.; Batstone, D.; Zheng, M. Post-treatment options for anaerobically digested sludge: Current status and future prospect. Water Res. 2021, 205, 117665. [Google Scholar] [CrossRef]
- Elalami, D.; Carrere, H.; Monlau, F.; Abdelouahdi, K.; Oukarroum, A.; Barakat, A. Pretreatment and co-digestion of wastewater sludge for biogas production: Recent research advances and trends. Renew. Sustain. Energy Rev. 2019, 114, 109287. [Google Scholar] [CrossRef]
- Nguyen, V.K.; Chaudhary, D.K.; Dahal, R.H.; Trinh, N.H.; Kim, J.; Chang, S.W.; Hong, Y.; La, D.D.; Nguyen, C.; Ngo, H.H.; et al. Review on pretreatment techniques to improve anaerobic digestion of sewage sludge. Fuel 2021, 285, 119105. [Google Scholar] [CrossRef]
- Kon, A.; Omata, S.; Hayakawa, Y.; Aburai, N.; Fujii, K. Microflora communities which can convert digested sludge to biogas. Environ. Technol. 2022, 43, 2391–2403. [Google Scholar] [CrossRef]
- Balcombe, P.; Speirs, J.F.; Brandon, N.P.; Hawkes, A.D. Methane emissions: Choosing the right climate metric and time horizon. Environ. Sci. Process. Impacts 2018, 20, 1323–1339. [Google Scholar] [CrossRef]
- Guilbert, D.; Vitale, G. Hydrogen as a clean and sustainable energy vector for global transition from fossil-based to zero-carbon. Clean Technol. 2021, 3, 881–909. [Google Scholar] [CrossRef]
- Jadhav, D.A.; Chendake, A.D.; Schievano, A.; Pant, D. Suppressing methanogens and enriching electrogens in bioelectrochemical systems. Bioresour. Technol. 2019, 277, 148–156. [Google Scholar] [CrossRef]
- Pachapur, V.L.; Kutty, P.; Pachapur, P.; Brar, S.K.; Bihan, Y.L.; Galvez-Cloutier, R.; Buelna, G. Seed pretreatment for increased hydrogen production using mixed-culture systems with advantages over pure-culture systems. Energies 2019, 12, 530. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, Y.; Wang, D.; Li, H.; Wang, Q.; Liu, Y.; Peng, L.; Yang, Q.; Li, X.; Zeng, G.; et al. Understanding the mechanisms of how poly aluminium chloride inhibits short-chain fatty acids production from anaerobic fermentation of waste activated sludge. Chem. Eng. J. 2018, 334, 1351–1360. [Google Scholar] [CrossRef]
- Fujii, K.; Kai, Y.; Matsunobu, S.; Sato, H.; Mikami, A. Isolation of digested sludge-assimilating fungal strains and their potential applications. J. Appl. Microbiol. 2013, 115, 718–726. [Google Scholar] [CrossRef]
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E.M.H. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 1997, 63, 3233–3241. [Google Scholar] [CrossRef]
- Zhou, M.; Hernandez-Sanabria, E.; Guan, L.L. Characterization of variation in rumen methanogenic communities under different dietary and host feed efficiency conditions, as determined by PCR-Denaturing gradient gel electrophoresis analysis. Appl. Environ. Microbiol. 2010, 76, 3776–3786. [Google Scholar] [CrossRef]
- May, L.A.; Smiley, B.; Schmidt, M.G. Comparative denaturing gradient gel electrophoresis analysis of fungal communities associated with whole plant corn silage. Can. J. Microbiol. 2001, 47, 829–841. [Google Scholar] [CrossRef]
- Tamura, T.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Silva, J.C.R.; Salgado, J.C.S.; Vici, A.C.; Ward, R.J.; Polizeli, M.L.T.W.; Guimarães, L.H.S.; Furriel, R.P.M.; Jorge, J.A. A novel Trichoderma reesei mutant RP698 with enhanced cellulase production. Braz. J. Microbiol. 2020, 51, 537–545. [Google Scholar] [CrossRef]
- Alves, T.B.; de Oliveira Ornela, P.H.; de Oliveira, A.H.C.; Jorge, J.A.; Guimarães, L.H.S. Production and characterization of a thermostable antifungal chitinase secreted by the filamentous fungus Aspergillus niveus under submerged fermentation. 3 Biotech 2018, 8, 369. [Google Scholar] [CrossRef]
- Kumar, G.; Cho, S.K.; Sivagurunathan, P.; Anburajan, P.; Mahapatra, D.M.; Park, J.H.; Pugazhendhi, A. Insights into evolutionary trends in molecular biology tools in microbial screening for biohydrogen production through dark fermentation. Int. J. Hydrogen Energy 2018, 43, 19885–19901. [Google Scholar] [CrossRef]
- Ghassemi, N.; Poulhazan, A.; Deligey, F.; Mentink-Vigier, F.; Marcotte, I.; Wang, T. Solid-state NMR investigations of extracellular matrixes and cell walls of algae, bacteria, fungi, and plants. Chem. Rev. 2022, 122, 10036–10086. [Google Scholar] [CrossRef]
- Matsushita, O.; Okabe, A. Clostridial hydrolytic enzymes degrading extracellular components. Toxicon 2001, 39, 1769–1780. [Google Scholar] [CrossRef]
- Mercer, R.G.; Walker, B.D.; Yang, X.; McMullen, L.M.; Ganzle, M.G. The locus of heat resistance (LHR) mediates heat resistance in Salmonella enterica, Escherichia coli and Enterobacter cloacae. Food Microbiol. 2017, 64, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Li, X.M.; Zeng, G.M.; Zhou, Y. Effective hydrogen production using waste sludge and its filtrate. Energy 2010, 35, 3557–3562. [Google Scholar] [CrossRef]
- Oh, M.; Han, J.W.; Lee, C.; Choi, G.J.; Kim, H. Nematicidal and plant growth-promoting activity of Enterobacter asburiae HK169: Genome analysis provides insight into its biological activities. J. Microbiol. Biotechnol. 2018, 28, 968–975. [Google Scholar] [CrossRef] [PubMed]
- Gheibipour, M.; Ghiasi, S.E.; Bashtani, M.; Torbati, M.B.M.; Motamedi, H. The potential of tannin degrading bacteria isolated from rumen of Iranian Urial ram as silage additives. Bioresour. Technol. Rep. 2022, 8, 101024. [Google Scholar] [CrossRef]
- Ghimire, A.; Frunzo, L.; Pirozzi, F.; Trably, E.; Escudie, R.; Lens, P.N.L.; Esposito, G. A review on dark fermentative biohydrogen production from organic biomass: Process parameters and use of by-products. Appl. Energ. 2015, 144, 73–95. [Google Scholar] [CrossRef]
- Sato, H.; Kuribayashi, K.; Fujii, K. Possible practical utility of an enzyme cocktail produced by sludge-degrading microbes for methane and hydrogen production from digested sludge. New Biotechnol. 2016, 33, 1–6. [Google Scholar] [CrossRef]
- Kuribayashi, K.; Kobayashi, Y.; Yokoyama, K.; Fujii, K. Digested sludge-degrading and hydrogen-producing bacterial floras and their potential for biohydrogen production. Int. Biodeter. Biodegr. 2017, 120, 58–65. [Google Scholar] [CrossRef]
- Ghosh, S. Assessment and update of status of pilot scale fermentative biohydrogen production with focus on candidate bioprocesses and decisive key parameters. Int. J. Hydrogen Energy 2022, 47, 17161–17183. [Google Scholar] [CrossRef]
- Kainthola, J.; Kalamdhada, A.S.; Gouda, V.V. A review on enhanced biogas production from anaerobic digestion of lignocellulosic biomass by different enhancement techniques. Process Biochem. 2019, 84, 81–90. [Google Scholar] [CrossRef]
- Almomani, F.; Bhosale, R.R. Enhancing the production of biogas through anaerobic co-digestion of agricultural waste and chemical pre-treatments. Chemosphere 2020, 255, 126805. [Google Scholar] [CrossRef]
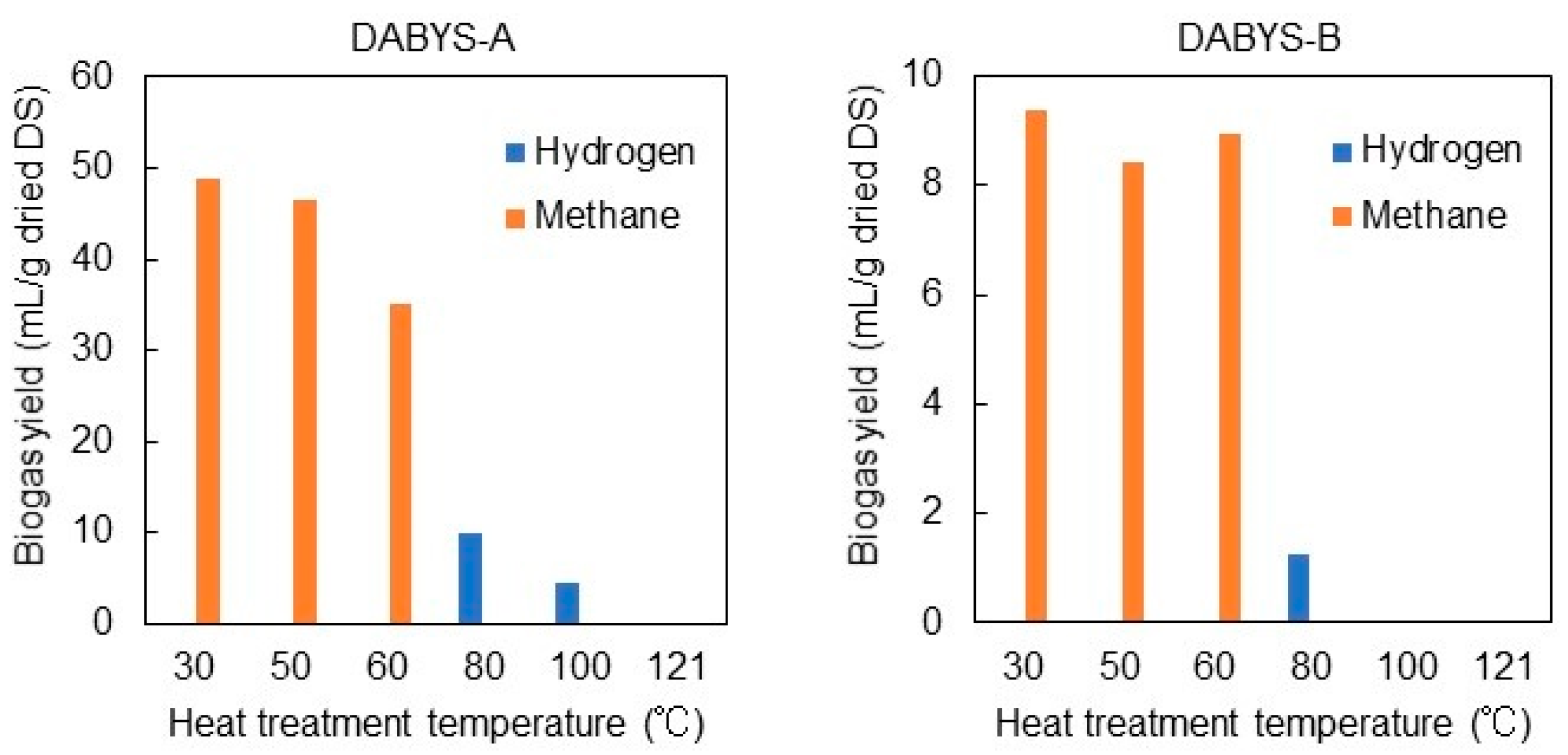
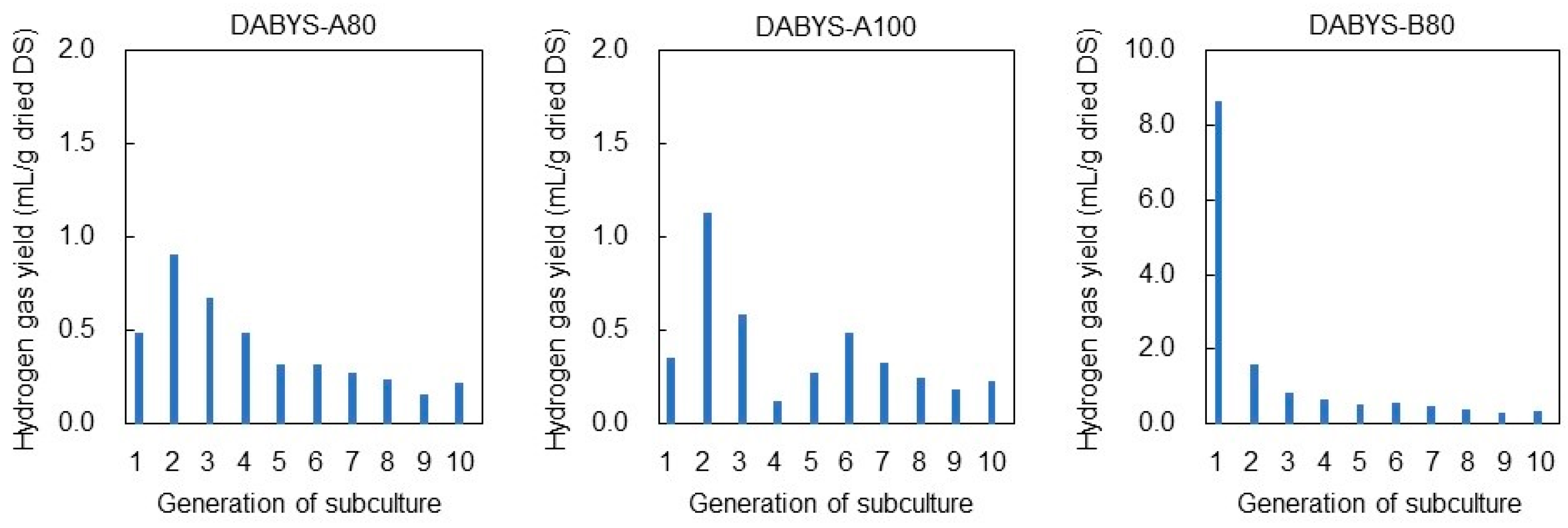
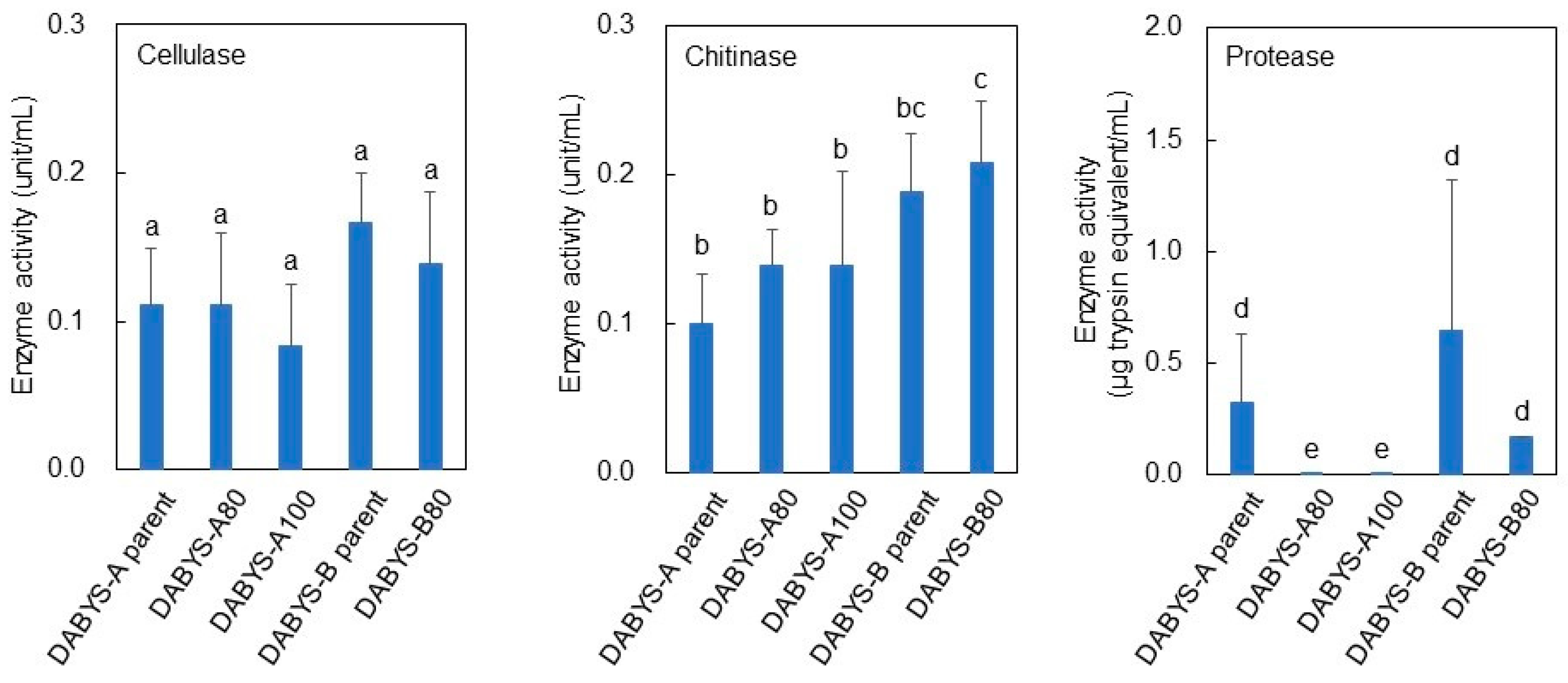
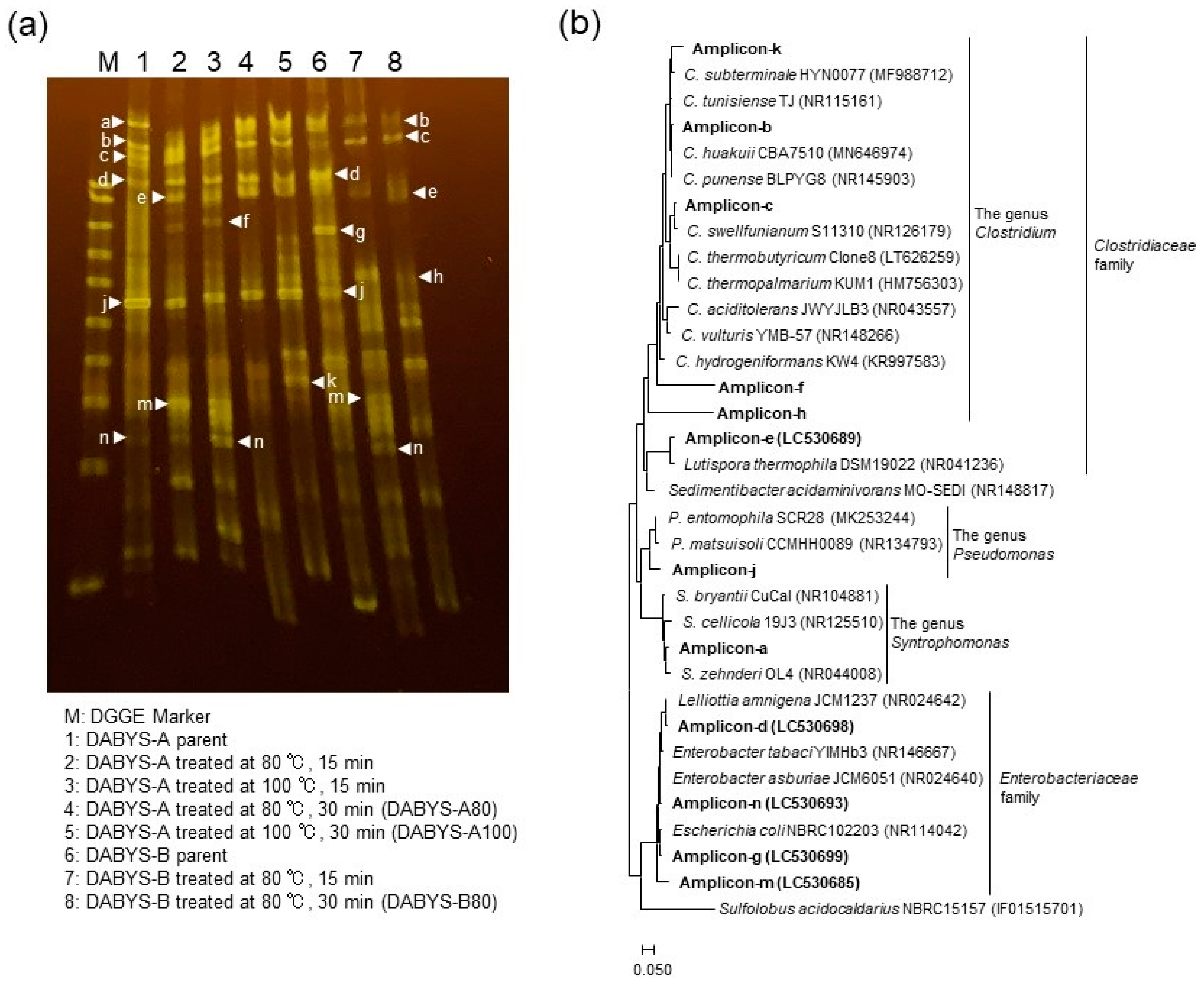
| Procedure | Parent Bacterial Flora | Temperature and Time for Heat Treatment | Gas Produced in the Subculture | Designation of the Heat-Treated Bacterial Flora |
|---|---|---|---|---|
| 1 | DABYS-A | 30 °C for 15 min | Methane | — |
| 2 | 50 °C for 15 min | Methane | — | |
| 3 | 60 °C for 15 min | Methane | — | |
| 4 | 80 °C for 15 min | Hydrogen | — | |
| 5 | 100 °C for 15 min | Hydrogen | — | |
| 6 | 121 °C for 15 min | None | — | |
| 7 | DABYS-B | 30 °C for 15 min | Methane | — |
| 8 | 50 °C for 15 min | Methane | — | |
| 9 | 60 °C for 15 min | Methane | — | |
| 10 | 80 °C for 15 min | Hydrogen | — | |
| 11 | 100 °C for 15 min | None | — | |
| 12 | 121 °C for 15 min | None | — | |
| 13 | DABYS-A treated at 80 °C for 15 min * | 80 °C for 30 min | Hydrogen | DABYS-A80 |
| 14 | DABYS-A treated at 100 °C for 15 min ** | 100 °C for 30 min | Hydrogen | DABYS-A100 |
| 15 | DABYS-B treated at 80 °C for 15 min *** | 80 °C for 30 min | Hydrogen | DABYS-B80 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hayakawa, Y.; Aburai, N.; Fujii, K. Development of Digested Sludge-Assimilating and Biohydrogen-Yielding Microflorae. Fermentation 2023, 9, 175. https://doi.org/10.3390/fermentation9020175
Hayakawa Y, Aburai N, Fujii K. Development of Digested Sludge-Assimilating and Biohydrogen-Yielding Microflorae. Fermentation. 2023; 9(2):175. https://doi.org/10.3390/fermentation9020175
Chicago/Turabian StyleHayakawa, Yuhei, Nobuhiro Aburai, and Katsuhiko Fujii. 2023. "Development of Digested Sludge-Assimilating and Biohydrogen-Yielding Microflorae" Fermentation 9, no. 2: 175. https://doi.org/10.3390/fermentation9020175
APA StyleHayakawa, Y., Aburai, N., & Fujii, K. (2023). Development of Digested Sludge-Assimilating and Biohydrogen-Yielding Microflorae. Fermentation, 9(2), 175. https://doi.org/10.3390/fermentation9020175





