High Production of Nattokinase via Fed-Batch Fermentation of the γ-PGA-Deficient Strain of Bacillus licheniformis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Plasmids
2.2. Media and Flask Culture Conditions
2.3. Construction of pgsC-Deficient Strain
2.4. Fed-Batch Fermentations
2.5. Analytic Methods
2.6. Transcriptional Analysis
3. Results
3.1. Deficiency of γ-PGA Synthesis and Its Implication on NK Production
3.2. pgsC Deficiency Enhances the Utilization of Glucose and Soybean Meals
3.3. Effects of pgsC Deficiency on the Transcriptional Levels of Genes Involved in Glycolysis and Tricarboxylic acid Cycle, and ATP Content
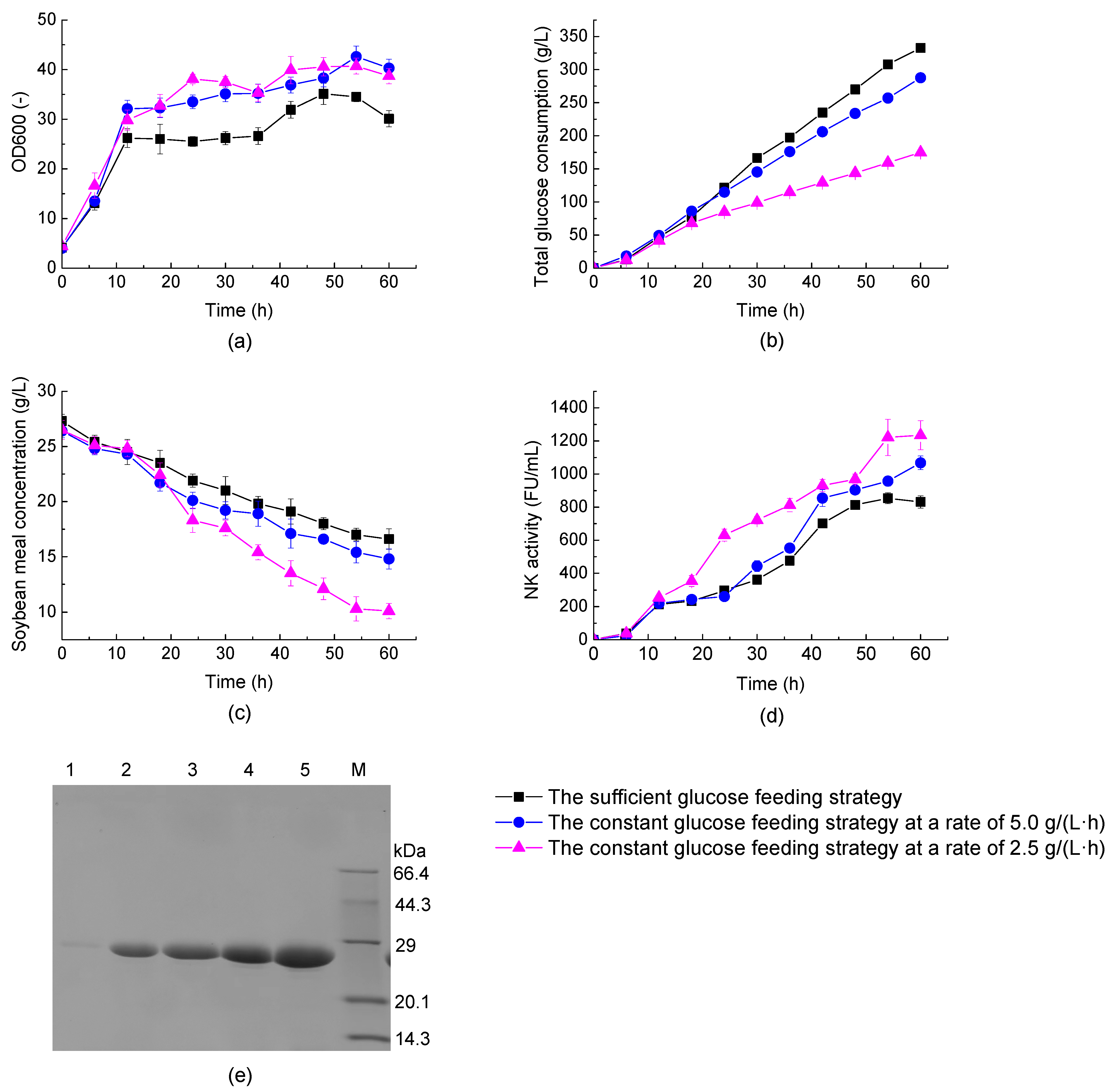
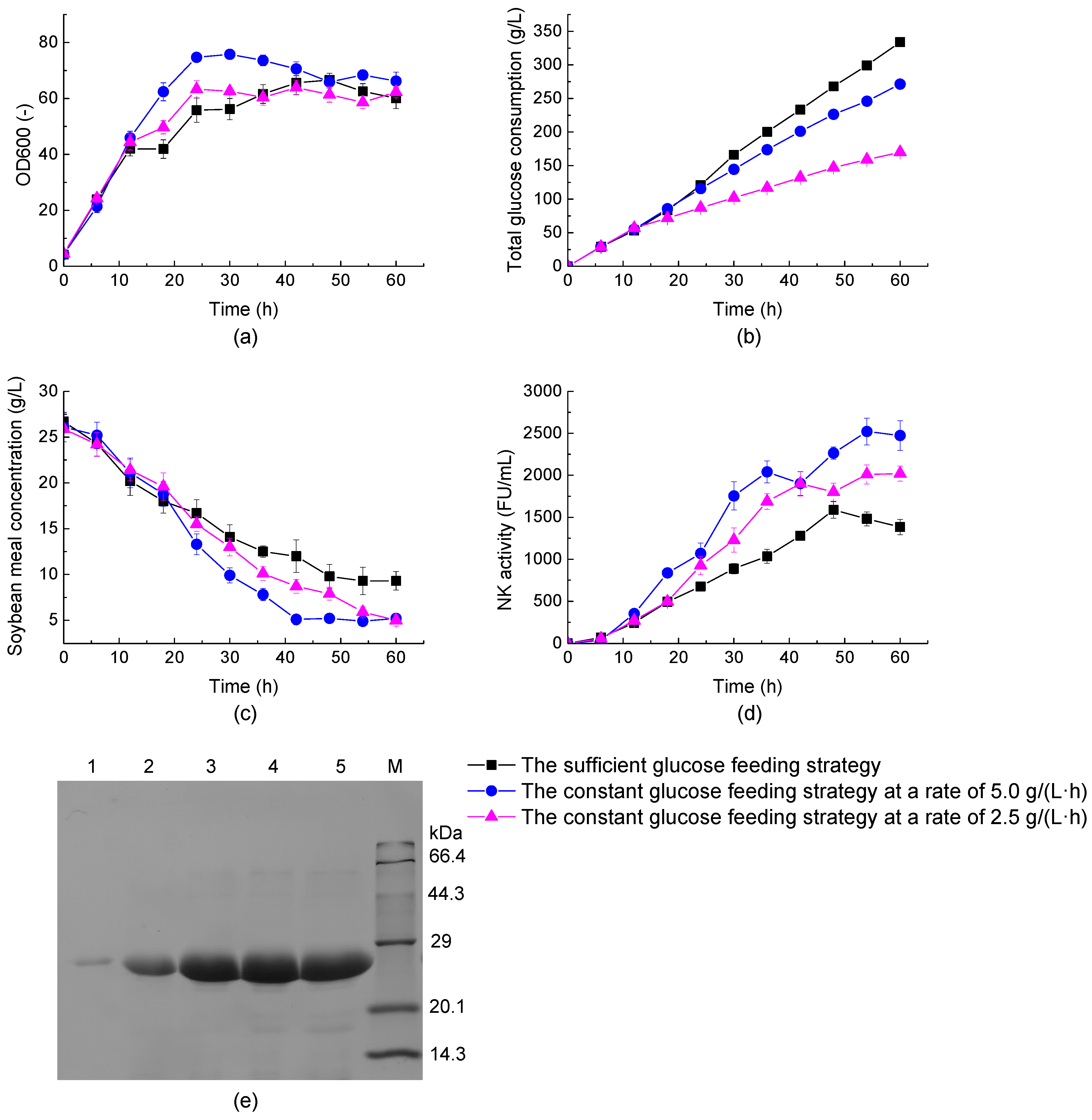
3.4. Improving NK Production from BL11 via an Optimal Glucose-Feeding Strategy under Different Oxygen Supply Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jamali, N.; Vahedi, F.; Fard, E.S.; Taheri-Anganeh, M.; Taghvimi, S.; Khatami, S.H.; Ghasemi, H.; Movahedpour, A. Nattokinase: Structure, applications and sources. Biocatal. Agric. Biotechnol. 2022, 47, 102564. [Google Scholar] [CrossRef]
- Sumi, H.; Hamada, H.; Tsushima, H.; Mihara, H.; Muraki, H. A novel fibrinolytic enzyme (nattokinase) in the vegetable cheese Natto; a typical and popular soybean food in the Japanese diet. Experientia 1987, 43, 1110–1111. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; McGowan, E.M.; Ren, N.; Lal, S.; Nassif, N.; Shad-Kaneez, F.; Qu, X.; Lin, Y. Nattokinase: A promising alternative in prevention and treatment of cardiovascular diseases. Biomark. Insights 2018, 13, 1177271918785130. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Du, M.; Yang, X.; Chen, Q.; Chen, H.; Lin, D.-H. Thrombolytic effects in vivo of nattokinase in a carrageenan-induced rat model of thrombosis. Acta Haematol. 2014, 132, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Hong, K.; Ito, Y.; Fujii, R.; Kariya, K.; Nishimuro, S. Thrombolytic effect of nattokinase on a chemically induced thrombosis model in rat. Biol. Pharm. Bull. 1995, 18, 1387–1391. [Google Scholar] [CrossRef] [PubMed]
- Jensen, G.S.; Lenninger, M.; Ero, M.P.; Benson, K.F. Consumption of nattokinase is associated with reduced blood pressure and von Willebrand factor, a cardiovascular risk marker: Results from a randomized, double-blind, placebo-controlled, multicenter North American clinical trial. Integr. Blood Press. Control 2016, 9, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-H.; Chen, K.-T.; Lee, T.-H.; Wang, C.-H.; Kuo, Y.-W.; Chiu, Y.-H.; Hsieh, C.-L.; Wu, C.-J.; Chang, Y.-L. Effects of natto extract on endothelial injury in a rat model. Acta Medica Okayama 2010, 64, 399–406. [Google Scholar] [PubMed]
- Ren, N.; Chen, H.; Li, Y.; Mcgowan, G.; Lin, Y. A clinical study on the effect of nattokinase on carotid artery atherosclerosis and hyperlipidaemia. Zhonghua Yi Xue Za Zhi 2017, 97, 2038–2042. [Google Scholar]
- Fujita, M.; Ohnishi, K.; Takaoka, S.; Ogasawara, K.; Fukuyama, R.; Nakamuta, H. Antihypertensive effects of continuous oral administration of nattokinase and its fragments in spontaneously hypertensive rats. Biol. Pharm. Bull. 2011, 34, 1696–1701. [Google Scholar] [CrossRef]
- Lee, B.; Lai, Y.; Wu, S. Antioxidation, angiotensin converting enzyme inhibition activity, nattokinase, and antihypertension of Bacillus subtilis (natto)-fermented pigeon pea. J. Food Drug Anal. 2015, 23, 750–757. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Yang, J.; Wang, C.; Sun, X.; Yan, L. Microbial nattokinase: From synthesis to potential application. Food Funct. 2023, 14, 2568–2585. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Liangqi, C.; Xiyu, T.; Jinyao, L. Biotechnology, bioengineering and applications of Bacillus nattokinase. Biomolecules 2022, 12, 980. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, P.M.; Nayak, S.; Lele, S.S. Fibrinolytic enzyme from newly isolated marine bacterium Bacillus subtilis ICTF-1: Media optimization, purification and characterization. J. Biosci. Bioeng. 2012, 113, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Luo, M.; Xu, L.; Zhang, Y.; Lin, X.; Kong, P.; Liu, H. Production of fibrinolytic enzyme from Bacillus amyloliquefaciens by fermentation of chickpeas, with the evaluation of the anticoagulant and antioxidant properties of chickpeas. J. Agric. Food Chem. 2011, 59, 3957–3963. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Huang, Q.; Zhang, R.; Zhang, Y. Purification and characterization of a fibrinolytic enzyme produced by Bacillus amyloliquefaciens DC-4 screened from douchi, a traditional Chinese soybean food. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2003, 134, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Choi, K.; Kim, Y.; Park, H.; Choi, J.; Lee, Y.; Oh, H.; Kwon, I.; Lee, S. Purification and characterization of a fibrinolytic enzyme produced from Bacillus sp. strain CK 11-4 screened from Chungkook-Jang. Appl. Environ. Microbiol. 1996, 62, 2482–2488. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Ning, C.; Peng, S.; Li, D.; Jin, R.; Zhang, Y.; Liu, Z.; Mou, H.; Zhu, C. High-Efficiency Fermentation of Nattokinase by Recombinant PSP2 Using Oyster Protein Hydrolysate as a Substrate. Foods 2023, 12, 1252. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Suo, F.; Cheng, J.; Han, L.; Hao, W.; Guo, J.; Zhou, Z. Stepwise modifications of genetic parts reinforce the secretory production of nattokinase in Bacillus subtilis. Microb. Biotechnol. 2018, 11, 930–942. [Google Scholar] [CrossRef]
- Chen, Y.; Cai, D.; He, P.; Mo, F.; Zhang, Q.; Ma, X.; Chen, S. Enhanced production of heterologous proteins by Bacillus licheniformis with defective d-alanylation of lipoteichoic acid. World J. Microbiol. Biotechnol. 2018, 34, 135. [Google Scholar] [CrossRef]
- Wei, X.; Zhou, Y.; Chen, J.; Cai, D.; Wang, D.; Qi, G.; Chen, S. Efficient expression of nattokinase in Bacillus licheniformis: Host strain construction and signal peptide optimization. J. Ind. Microbiol. Biotechnol. 2015, 42, 287–295. [Google Scholar] [CrossRef]
- Cai, D.; Wei, X.; Qiu, Y.; Chen, Y.; Chen, J.; Wen, Z.; Chen, S. High-level expression of nattokinase in Bacillus licheniformis by manipulating signal peptide and signal peptidase. J. Appl. Microbiol. 2016, 121, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.; Wang, H.; He, P.; Zhu, C.; Wang, Q.; Wei, X.; Nomura, C.T.; Chen, S. A novel strategy to improve protein secretion via overexpression of the SppA signal peptide peptidase in Bacillus licheniformis. Microb. Cell Factories 2017, 16, 70. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Li, W.; Shu, L.; Yi, J.; Chen, G.; Liang, Z. Non-sterilized fermentative co-production of poly (γ-glutamic acid) and fibrinolytic enzyme by a thermophilic Bacillus subtilis GXA-28. Bioresour. Technol. 2013, 142, 697–700. [Google Scholar] [CrossRef] [PubMed]
- Nie, G.; Zhu, Z.; Liu, F.; Nie, Z.; Ye, Y.; Yue, W. Co-production of nattokinase and poly (γ-Glutamic Acid) under solid-state fermentation using soybean and rice husk. Braz. Arch. Biol. Technol. 2015, 58, 718–724. [Google Scholar] [CrossRef]
- Li, M.; Zhang, Z.; Li, S.; Tian, Z.; Ma, X. Study on the mechanism of production of γ-PGA and nattokinase in Bacillus subtilis natto based on RNA-seq analysis. Microb. Cell Factories 2021, 20, 83. [Google Scholar] [CrossRef] [PubMed]
- Tuan, N.A.; Thuan, D.T.H.; Tam, T.T.M.; Huong, N.T. Determination the optimum fermentation in obtaining nattokinase by Bacillus subtilis natto. Int. J. Innov. Appl. Stud. 2015, 13, 663. [Google Scholar]
- Mahajan, P.M.; Gokhale, S.V.; Lele, S.S. Production of nattokinase using Bacillus natto NRRL 3666: Media optimization, scale up, and kinetic modeling. Food Sci. Biotechnol. 2010, 19, 1593–1603. [Google Scholar] [CrossRef]
- Chen, P.T.; Chiang, C.J.; Chao, Y.P. Medium optimization for the production of recombinant nattokinase by Bacillus subtilis using response surface methodology. Biotechnol. Prog. 2007, 23, 1327–1332. [Google Scholar] [CrossRef]
- Deepak, V.; Kalishwaralal, K.; Ramkumarpandian, S.; Babu, S.V.; Senthilkumar, S.; Sangiliyandi, G. Optimization of media composition for Nattokinase production by Bacillus subtilis using response surface methodology. Bioresour. Technol. 2008, 99, 8170–8174. [Google Scholar] [CrossRef]
- Kumar, D.; Rakshitha, R.; Vidhya, M.A.; Jennifer, P.S.; Prasad, S.; Kumar, M.R.; Kalaichelvan, P. Production, optimization and characterization of fibrinolytic enzyme by Bacillus subtilis RJAS19. Pak. J. Biol. Sci. 2014, 17, 529–534. [Google Scholar] [CrossRef]
- Kwon, E.Y.; Kim, K.M.; Kim, M.K.; Lee, I.Y.; Kim, B.S. Production of nattokinase by high cell density fed-batch culture of Bacillus subtilis. Bioprocess Biosyst. Eng. 2011, 34, 789–793. [Google Scholar] [CrossRef] [PubMed]
- Unrean, P.; Nguyen, N.H.; Visessanguan, W.; Kitsubun, P. Improvement of nattokinase production by Bacillus subtilis using an optimal feed strategy in fed-batch fermentation. Asia-Pac. J. Sci. Technol. 2012, 17, 769–777. [Google Scholar]
- Berenjian, A.; Mahanama, R.; Kavanagh, J.; Dehghani, F.; Ghasemi, Y. Nattokinase production: Medium components and feeding strategy studies. Chem. Ind. Chem. Eng. Q. 2014, 20, 541–547. [Google Scholar] [CrossRef]
- Chen, M.; Qin, Y.; Liu, Z.; Liu, K.; Wang, F.; Qu, Y. Isolation and characterization of a β-glucosidase from Penicillium decumbens and improving hydrolysis of corncob residue by using it as cellulase supplementation. Enzym. Microb. Technol. 2010, 46, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.; Zhang, B.; Rao, Y.; Li, L.; Zhu, J.; Li, J.; Ma, X.; Chen, S. Improving the utilization rate of soybean meal for efficient production of bacitracin and heterologous proteins in the aprA-deficient strain of Bacillus licheniformis. Appl. Microbiol. Biotechnol. 2019, 103, 4789–4799. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wu, D.; Ren, H. Economical production of agricultural γ-polyglutamic acid using industrial wastes by Bacillus subtilis. Biochem. Eng. J. 2019, 146, 117–123. [Google Scholar] [CrossRef]
- Kőrös, Á.; Varga, Z.; Molnár-Perl, I. Simultaneous analysis of amino acids and amines as their o-phthalaldehyde-ethanethiol-9-fluorenylmethyl chloroformate derivatives in cheese by high-performance liquid chromatography. J. Chromatogr. 2008, 1203, 146–152. [Google Scholar] [CrossRef]
- Szentirmai, V.; Wacha, A.; Németh, C.; Kitka, D.; Rácz, A.; Héberger, K.; Mihály, J.; Varga, Z. Reagent-free total protein quantification of intact extracellular vesicles by attenuated total reflection Fourier transform infrared (ATR-FTIR) spectroscopy. Anal. Bioanal. Chem. 2020, 412, 4619–4628. [Google Scholar] [CrossRef]
- Sun, S.; Wang, Y.; Chen, S.; Li, Z.; Zhao, C.; Liu, Q.; Zhang, X. Functionality, characterization and DEGs contribution by engineering isolate Pseudomonas P1 to elucidate the regulation mechanisms of p-chlorophenol-4-Chloroaniline bioremediation. Chem. Eng. J. 2023, 468, 143798. [Google Scholar] [CrossRef]
- Wang, L.; Chen, S.; Yu, B. Poly-γ-glutamic acid: Recent achievements, diverse applications and future perspectives. Trends Food Sci. Technol. 2022, 119, 1–12. [Google Scholar] [CrossRef]
- Cao, M.; Song, C.; Jin, Y.; Liu, L.; Liu, J.; Xie, H.; Guo, W.; Wang, S. Synthesis of poly (γ-glutamic acid) and heterologous expression of pgsBCA genes. J. Mol. Catal. B Enzym. 2010, 67, 111–116. [Google Scholar] [CrossRef]
- Xu, Y.; Jiang, Y.; Li, X.; Sun, B.; Teng, C.; Yang, R.; Xiong, K.; Fan, G.; Wang, W. Systematic characterization of the metabolism of acetoin and its derivative ligustrazine in Bacillus subtilis under micro-oxygen conditions. J. Agric. Food Chem. 2018, 66, 3179–3187. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Cai, D.; Chen, S. Metabolic engineering of central carbon metabolism of Bacillus licheniformis for enhanced production of poly-γ-glutamic acid. Appl. Biochem. Biotechnol. 2021, 193, 3540–3552. [Google Scholar] [CrossRef]
- Cui, W.; Han, L.; Suo, F.; Liu, Z.; Zhou, L.; Zhou, Z. Exploitation of Bacillus subtilis as a robust workhorse for production of heterologous proteins and beyond. World J. Microbiol. Biotechnol. 2018, 34, 145. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Lü, S.; Ji, Y.; Wang, Z.; Zhang, S.F.; Qi, T.; Yan, J.; Li, T.; Liu, Y.; Liu, M. A nattokinase carrier bonding with polyglutamic acid peptide dendrimer for improved thrombolysis. Polym. Adv. Technol. 2019, 30, 2353–2360. [Google Scholar] [CrossRef]
- Hsieh, C.W.; Lu, W.C.; Hsieh, W.C.; Huang, Y.P.; Lai, C.H.; Ko, W.C. Improvement of the stability of nattokinase using γ-polyglutamic acid as a coating material for microencapsulation. LWT-Food Sci. Technol. 2009, 42, 144–149. [Google Scholar] [CrossRef]
- Li, X.; Yang, H.; Zhou, M.; Zhan, Y.; Liu, J.; Yan, D.; Cai, D.; Chen, S. A novel strategy of feeding nitrate for cost-effective production of poly-γ-glutamic acid from crude glycerol by Bacillus licheniformis WX-02. Biochem. Eng. J. 2021, 176, 108156. [Google Scholar] [CrossRef]
- Wang, L.; Liu, N.; Yu, C.; Chen, J.; Hong, K.; Zang, Y.; Wang, M.; Nie, G. Inhibition of nattokinase against the production of poly (γ-glutamic Acid) in Bacillus subtilis natto. Biotechnol. Lett. 2020, 42, 2285–2291. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Hou, L.; Hu, M.; Gao, Y.; Tian, Z.; Fan, B.; Li, S.; Wang, F. Recent advances in nattokinase-enriched fermented soybean Foods: A review. Foods 2022, 11, 1867. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Shen, J.; Li, Y.; Wang, Z.; Zhao, Y.; Chen, Y.; Zhao, J. High and economical nattokinase production with acetoin as a useful byproduct from soybean milk and glucose. Probiotics Antimicrob. Proteins 2022, 14, 792–803. [Google Scholar] [CrossRef]
- Pan, S.; Chen, G.; Zeng, J.; Cao, X.; Zheng, X.; Zeng, W.; Liang, Z. Fibrinolytic enzyme production from low-cost substrates by marine Bacillus subtilis: Process optimization and kinetic modeling. Biochem. Eng. J. 2019, 141, 268–277. [Google Scholar] [CrossRef]
- Han, L.; Chen, Q.; Luo, J.; Cui, W.; Zhou, Z. Development of a glycerol-inducible expression system for high-yield heterologous protein production in Bacillus subtilis. Microbiol. Spectr. 2022, 10, e01322. [Google Scholar] [CrossRef] [PubMed]
- Man, L.L.; Xiang, D.J.; Zhang, C.L. Strain screening from traditional fermented soybean foods and induction of nattokinase production in Bacillus subtilis MX-6. Probiotics Antimicrob. Proteins 2019, 11, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhan, C.; Guo, G.; Liu, Z.; Hao, N.; Ouyang, P. Tofu processing wastewater as a low-cost substrate for high activity nattokinase production using Bacillus subtilis. BMC Biotechnol. 2021, 21, 57. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.H.; Song, J.Y.; Kim, K.M.; Kim, M.K.; Lee, I.Y.; Kim, S.B.; Kim, H.S.; Han, N.S.; Lee, B.H.; Kim, B.S. Production of nattokinase by batch and fed-batch culture of Bacillus subtilis. N. Biotechnol. 2010, 27, 341–346. [Google Scholar] [CrossRef] [PubMed]
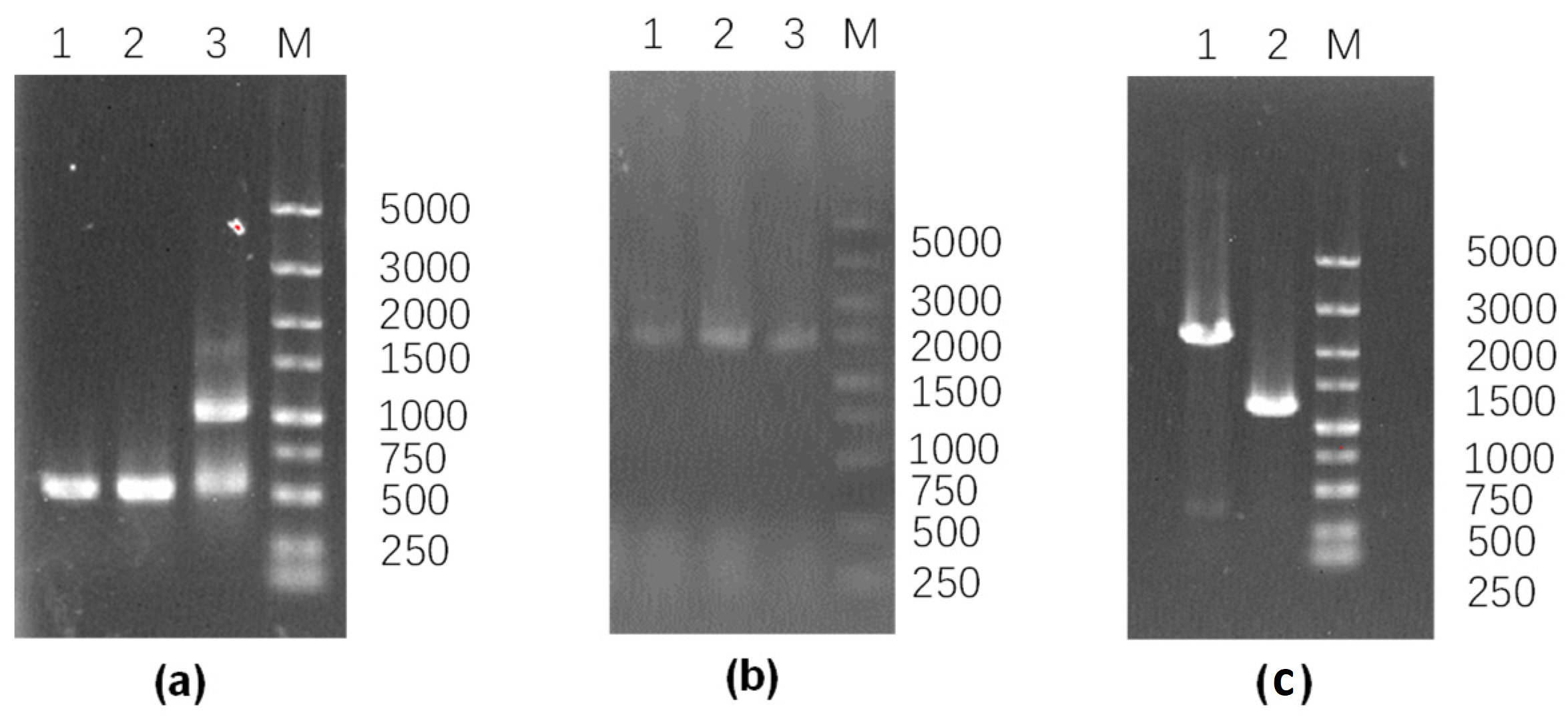
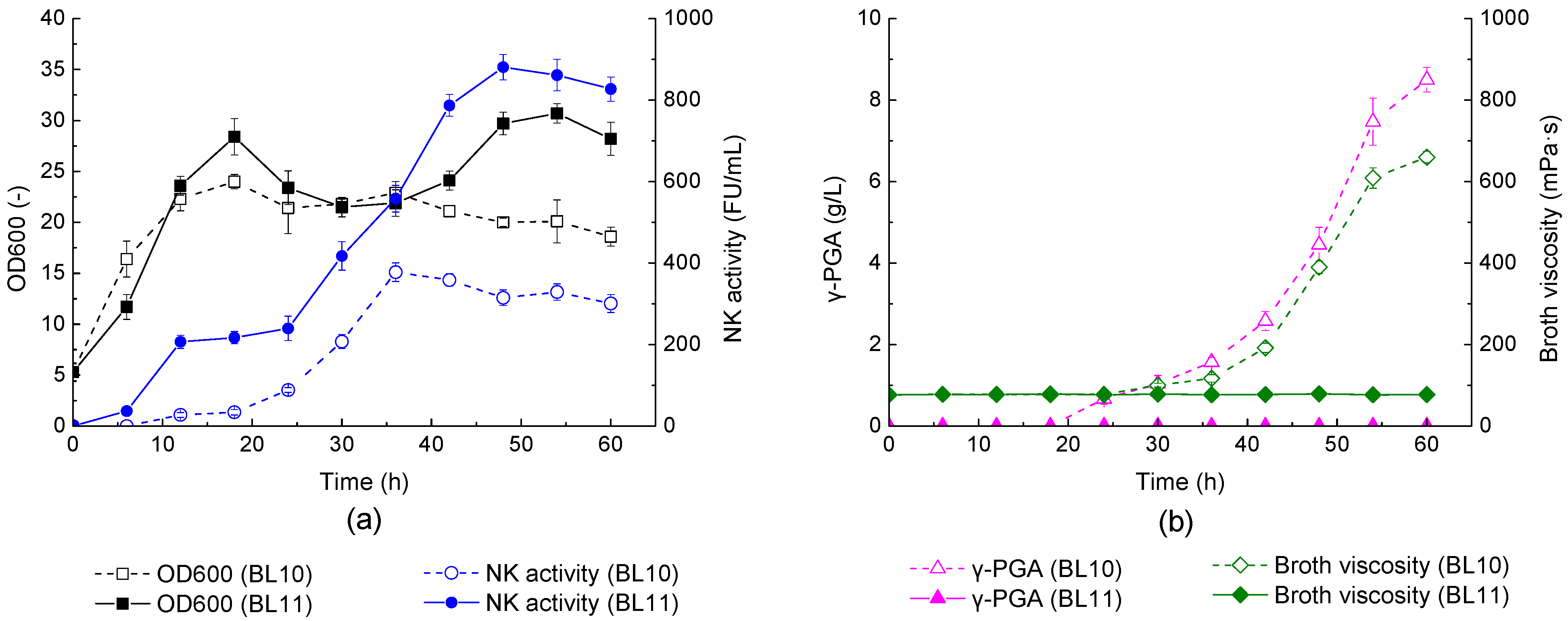
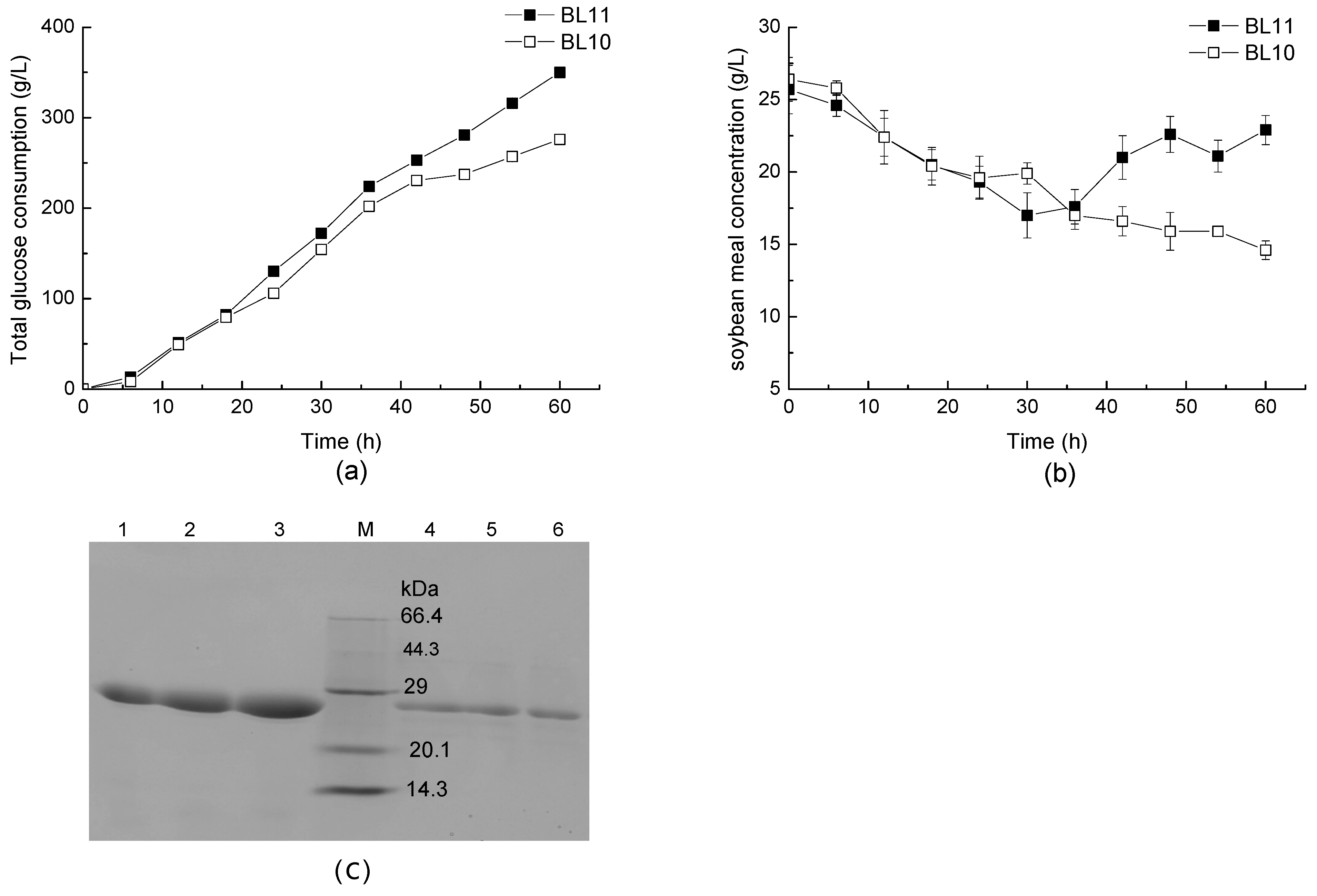

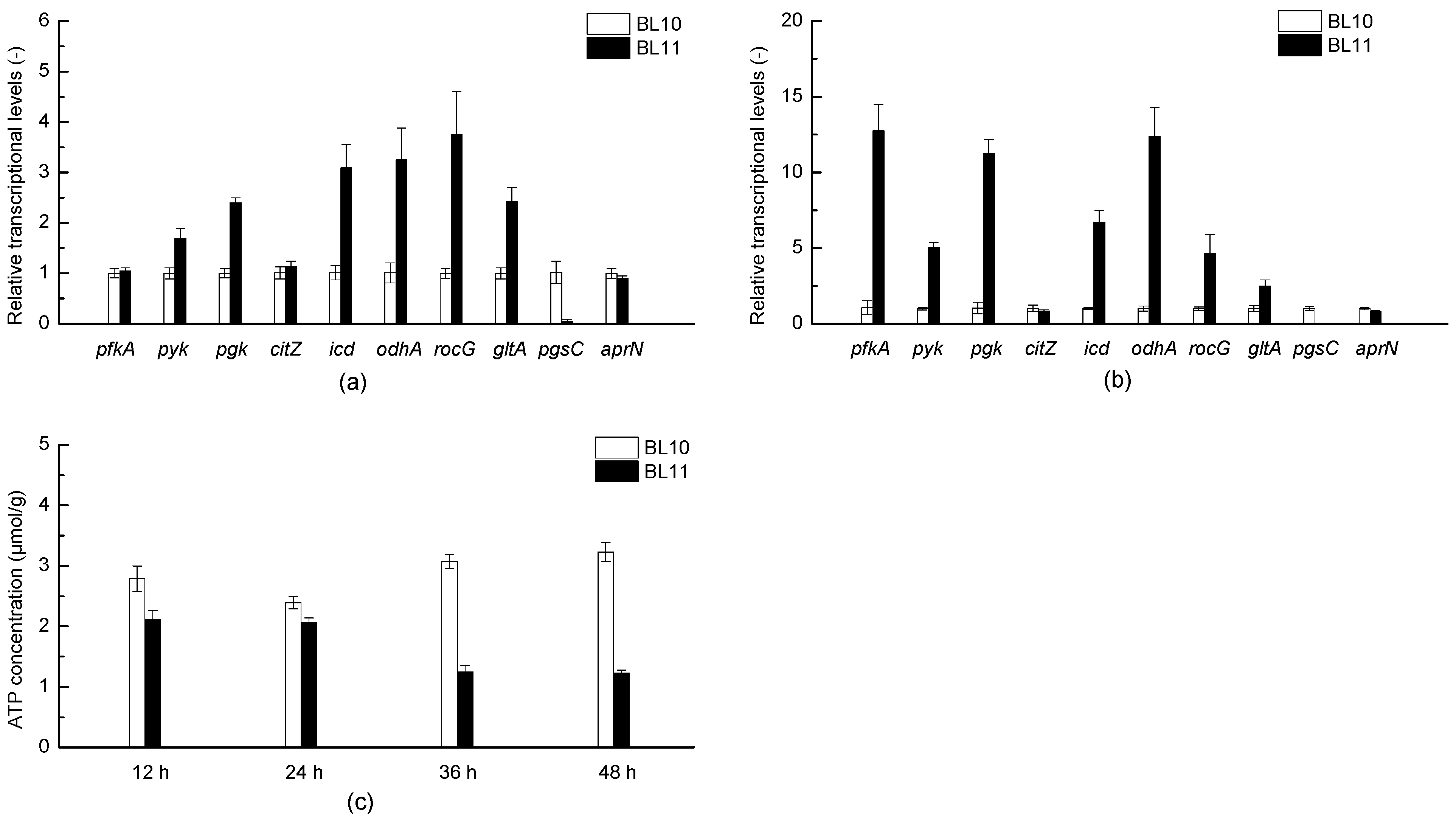
| Strains and Plasmids | Relevant Characteristics | Source of Reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | supE44 ΔlacU169 (f 80 lacZΔM15) hsdR17 recA1 gyrA96 thi1 relA1 | Lab collection |
| Bacillus licheniformis WX-02 | Wide-type host strain (CCTCC M208065) | Lab collection |
| BL10 | WX-02(Δmpr, Δvpr, ΔaprX, Δepr, Δbpr, ΔwprA, ΔaprE, ΔbprA, Δhag, ΔamyL) harboring pP43SacCNK (CCTCC M2014253) | [20] |
| BL11 | BL10(ΔpgsC) harboring pP43SacCNK | This study |
| Plasmids | ||
| pP43SacCNK | Plasmid pHY300PLK harboring P43 promoter, signal peptide of SacC, gene aprN, and amyL terminator | [20] |
| T2(2)-Ori | E. coli-B. licheniformis shuttle vector, for gene knockout | Lab collection |
| T2-pgsC | T2(2)-Ori derivative containing homologous arms of pgsC, to block poly-γ-glutamic acid synthesis | This study |
| Strains | Genetic Engineering Strategies | Culture Medium/Fermentation Substrates | Fermentation Process | Final Activity/Yield | References |
|---|---|---|---|---|---|
| Bacillus subtilis PSP2 | Heterologous expression of aprN gene | 1% trypsin, 1% oyster protein hydrolysate, 2% maltose, and 0.5% NaCl | Batch fermentation in a flask | 390.23 FU/mL | [17] |
| Bacillus subtilis WB800 | Optimization of promoters and signal peptides | 2.4% yeast extract, 2% tryptone, 0.4% glycerol, 2.3% KH2PO4, and 1.3% K2HPO4 | Batch fermentation in a flask | 292 FU/mL | [18] |
| Bacillus licheniformis DW2 | Deficiency of dltABCD, and expression of plasmid pP43SacCNK | 2% glucose, 1% peptone, 1% soy peptone, 1.5% yeast extract, 0.5% corn steep liquor, 0.6% (NH4)2SO4, 0.3% K2HPO4·3H2O, and 1% NaCl | Batch fermentation in a flask | 38.02 FU/mL | [19] |
| Bacillus licheniformis BL10 | Deletion of protease genes, and expression of plasmid pP43SNT | Same as above | Batch fermentation in a flask | 33.83 FU/mL | [20] |
| Bacillus licheniformis BL10 | Overexpression of sppA in BL10 | Same as above | Batch fermentation in a flask | 45.05 FU/mL | [22] |
| Bacillus licheniformis DW2 | Deficiency of bacA and aprA, and expression of plasmid pP43SacCNK | 2% glucose, 1% peptone, 1% soy peptone, 1.5% yeast extract, 0.5% corn steep liquor, 0.3% K2HPO4, 0.6% (NH4)2SO4, and 1% NaCl | Batch fermentation in a flask | 38.43 FU/mL | [35] |
| Bacillus licheniformis BL11 | Deficiency of pgsC in BL10 | 6% glucose, 3% soybean meal, 0.5% corn steep liquor, 1% (NH4)2SO4, 1% NaNO3, 0.15% K2HPO4·3H2O, 0.05% MgSO4·7H2O, and 0.05% CaCl2·2H2O | Fed-batch fermentation in a 5 L fermenter | 2522.2 FU/mL | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Yang, J.; Liu, J.; Zhang, X.; Wu, W.; Yan, D.; Miao, L.; Cai, D.; Ma, X.; Chen, S. High Production of Nattokinase via Fed-Batch Fermentation of the γ-PGA-Deficient Strain of Bacillus licheniformis. Fermentation 2023, 9, 1018. https://doi.org/10.3390/fermentation9121018
Li X, Yang J, Liu J, Zhang X, Wu W, Yan D, Miao L, Cai D, Ma X, Chen S. High Production of Nattokinase via Fed-Batch Fermentation of the γ-PGA-Deficient Strain of Bacillus licheniformis. Fermentation. 2023; 9(12):1018. https://doi.org/10.3390/fermentation9121018
Chicago/Turabian StyleLi, Xin, Jing Yang, Jun Liu, Xiaohui Zhang, Wei Wu, Dazhong Yan, Lihong Miao, Dongbo Cai, Xin Ma, and Shouwen Chen. 2023. "High Production of Nattokinase via Fed-Batch Fermentation of the γ-PGA-Deficient Strain of Bacillus licheniformis" Fermentation 9, no. 12: 1018. https://doi.org/10.3390/fermentation9121018
APA StyleLi, X., Yang, J., Liu, J., Zhang, X., Wu, W., Yan, D., Miao, L., Cai, D., Ma, X., & Chen, S. (2023). High Production of Nattokinase via Fed-Batch Fermentation of the γ-PGA-Deficient Strain of Bacillus licheniformis. Fermentation, 9(12), 1018. https://doi.org/10.3390/fermentation9121018





