Historical Aspects of Restriction Endonucleases as Intelligent Scissors for Genetic Engineering
Abstract
1. Introduction
2. Diversity of Restriction Endonucleases and Their Function in Vivo
3. Nomenclature and Classification of Restriction Endonucleases
4. Genomic Organization
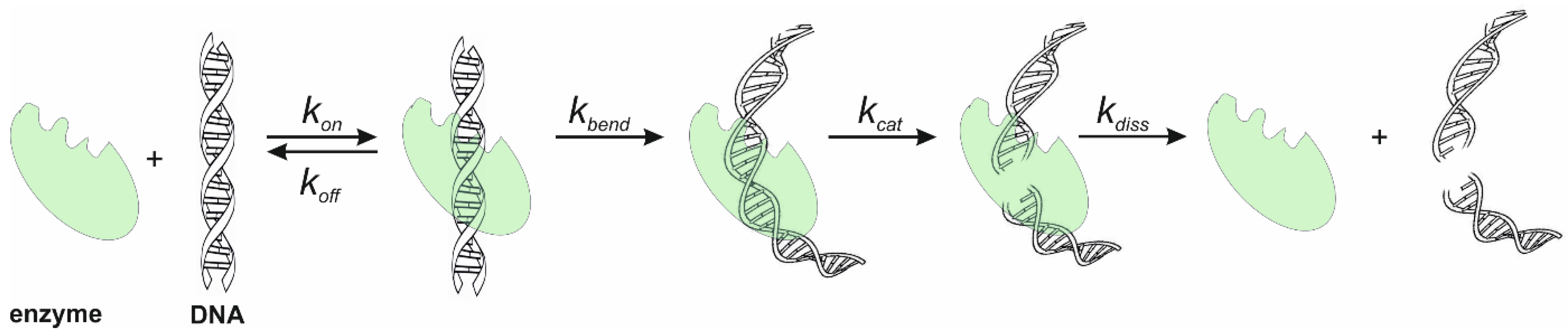
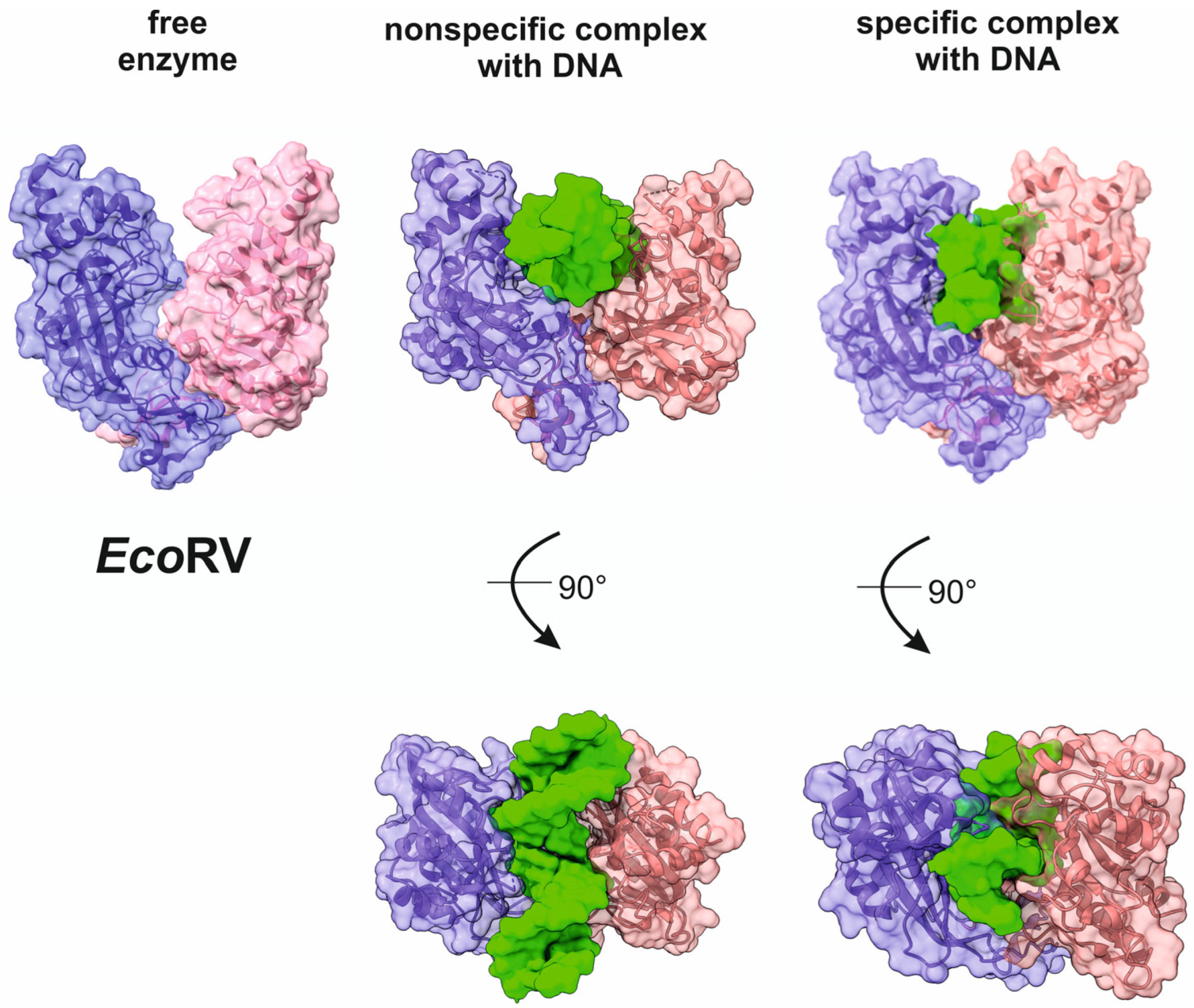
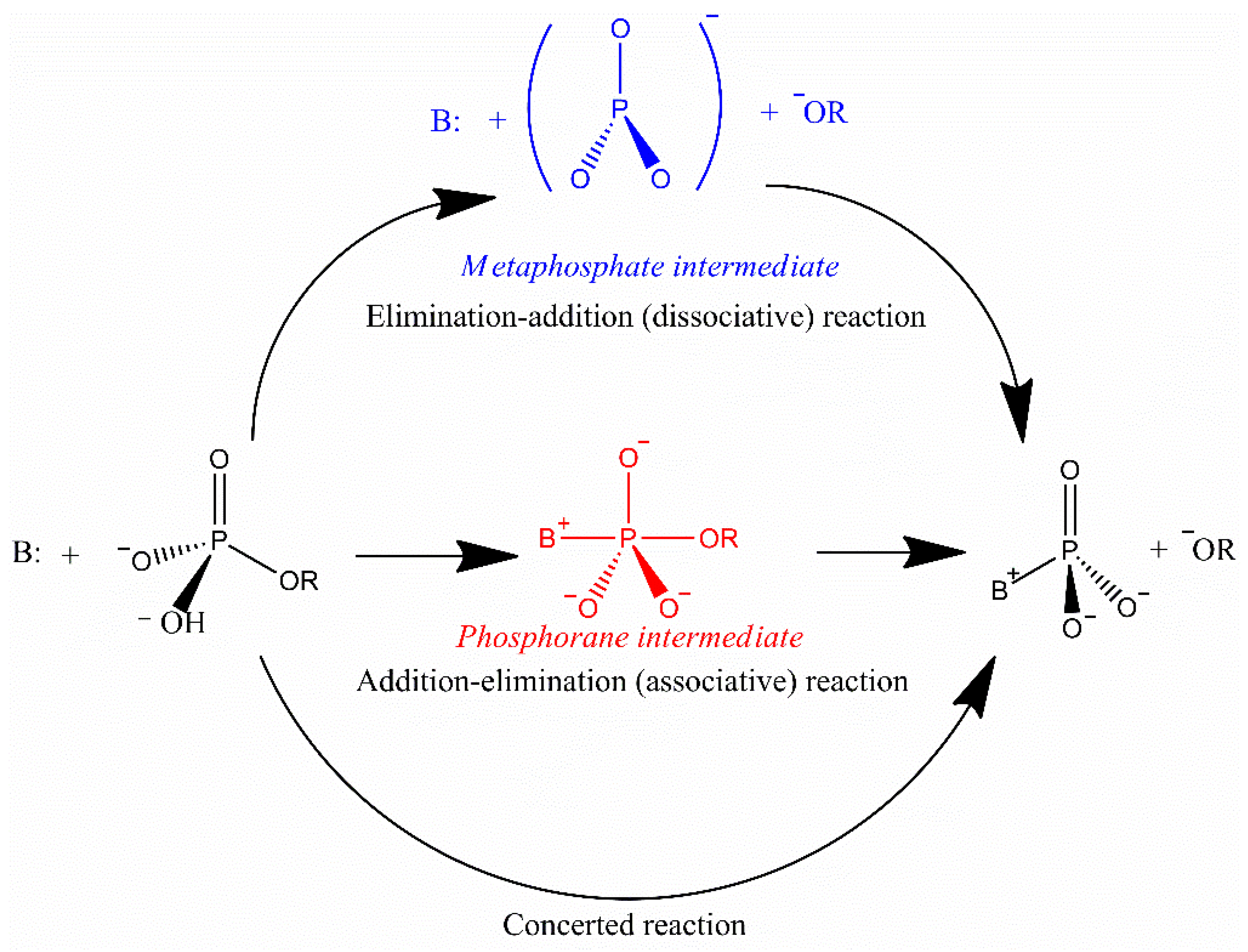
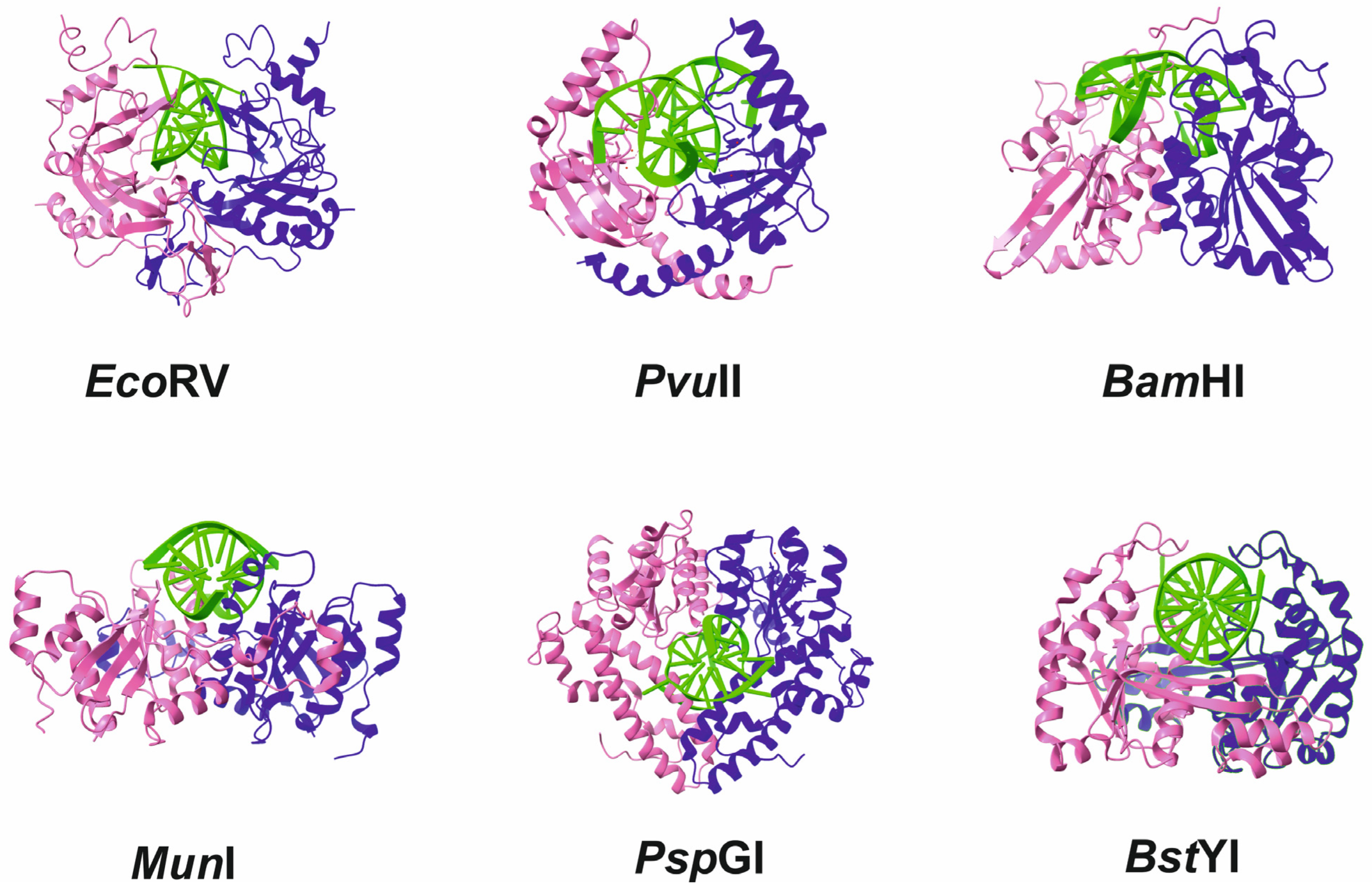
5. Mechanism of Specific-Site Recognition and Catalysis
6. Practical Application of Type II Restriction Endonucleases
6.1. Endonucleases of Subtype IIA
6.2. Endonucleases of Subtype IIB
6.3. Endonucleases of Subtype IIC
6.4. Endonucleases of Subtype IIE
6.5. Endonucleases of Subtype IIF
6.6. Endonucleases of Subtype IIG
6.7. Endonucleases of Subtype IIH
6.8. Endonucleases of Subtype IIM
6.9. Endonucleases of Subtype IIP
6.10. Endonucleases of Subtype IIS
6.11. Endonucleases of Subtype IIT
7. Off-Target Activity of Restriction Endonucleases
8. Single-Stranded DNA Cleavage
9. Changing the Specificity of Enzymes by Protein Engineering
10. Fusion Proteins
11. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. ZFN, TALEN, and CRISPR/Cas-Based Methods for Genome Engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Maguin, P.; Varble, A.; Modell, J.W.; Marraffini, L.A. Cleavage of Viral DNA by Restriction Endonucleases Stimulates the Type II CRISPR-Cas Immune Response. Mol. Cell 2022, 82, 907–919.e7. [Google Scholar] [CrossRef]
- Phan, H.T.L.; Kim, K.; Lee, H.; Seong, J.K. Progress in and Prospects of Genome Editing Tools for Human Disease Model Development and Therapeutic Applications. Genes 2023, 14, 483. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yang, Y.; Qi, H.; Cui, W.; Zhang, L.; Fu, X.; He, X.; Liu, M.; Li, P.; Yu, T. CRISPR/Cas9 Therapeutics: Progress and Prospects. Signal Transduct. Target. Ther. 2023, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Arora, L.; Narula, A. Gene Editing and Crop Improvement Using CRISPR-Cas9 System. Front. Plant Sci. 2017, 8, 1932. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Zhang, Y.; Shi, S. Development and Application of CRISPR/Cas in Microbial Biotechnology. Front. Bioeng. Biotechnol. 2020, 8, 711. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, A.; da Silva, G. CRISPR-Cas: Converting A Bacterial Defence Mechanism into A State-of-the-Art Genetic Manipulation Tool. Antibiotics 2019, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Naeem, M.; Majeed, S.; Hoque, M.Z.; Ahmad, I. Latest Developed Strategies to Minimize the Off-Target Effects in CRISPR-Cas-Mediated Genome Editing. Cells 2020, 9, 1608. [Google Scholar] [CrossRef]
- Danna, K.; Nathans, D. Specific Cleavage of Simian Virus 40 DNA by Restriction Endonuclease of Hemophilus Influenzae. Proc. Natl. Acad. Sci. USA 1971, 68, 2913–2917. [Google Scholar] [CrossRef]
- Smith, H.O.; Welcox, K.W. A Restriction Enzyme from Hemophilus Influenzae. I. Purification and General Properties. J. Mol. Biol. 1970, 51, 379–391. [Google Scholar] [CrossRef]
- Wilson, G.G.; Murray, N.E. Restriction and Modification Systems. Annu. Rev. Genet. 1991, 25, 585–627. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.J. A Nomenclature for Restriction Enzymes, DNA Methyltransferases, Homing Endonucleases and Their Genes. Nucleic Acids Res. 2003, 31, 1805–1812. [Google Scholar] [CrossRef] [PubMed]
- Thielking, V.; Alves, J.; Fliess, A.; Maass, G.; Pingoud, A. Accuracy of the EcoRI Restriction Endonuclease: Binding and Cleavage Studies with Oligodeoxynucleotide Substrates Containing Degenerate Recognition Sequences. Biochemistry 1990, 29, 4682–4691. [Google Scholar] [CrossRef] [PubMed]
- Alves, J.; Selent, U.; Wolfes, H. Accuracy of the EcoRV Restriction Endonuclease: Binding and Cleavage Studies with Oligodeoxynucleotide Substrates Containing Degenerate Recognition Sequences. Biochemistry 1995, 34, 11191–11197. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.J.; Vincze, T.; Posfai, J.; Macelis, D. REBASE—A Database for DNA Restriction and Modification: Enzymes, Genes and Genomes. Nucleic Acids Res. 2015, 43, D298–D299. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.J.; Vincze, T.; Posfai, J.; Macelis, D. REBASE: A Database for DNA Restriction and Modification: Enzymes, Genes and Genomes. Nucleic Acids Res. 2023, 51, D629–D630. [Google Scholar] [CrossRef] [PubMed]
- Di Felice, F.; Micheli, G.; Camilloni, G. Restriction Enzymes and Their Use in Molecular Biology: An Overview. J. Biosci. 2019, 44, 38. [Google Scholar] [CrossRef]
- Bickle, T.A.; Krüger, D.H. Biology of DNA Restriction. Microbiol. Rev. 1993, 57, 434–450. [Google Scholar] [CrossRef]
- Raleigh, E.A.; Brooks, J.E. Restriction Modification Systems: Where They Are and What They Do. In Bacterial Genomes; Springer US: Boston, MA, USA, 1998; pp. 78–92. [Google Scholar]
- Van Etten, J.L. Unusual Life Style of Giant Chlorella Viruses. Annu. Rev. Genet. 2003, 37, 153–195. [Google Scholar] [CrossRef]
- Pingoud, A.; Wilson, G.G.; Wende, W. Type II Restriction Endonucleases—A Historical Perspective and More. Nucleic Acids Res. 2014, 42, 7489–7527. [Google Scholar] [CrossRef]
- Jeltsch, A. Maintenance of Species Identity and Controlling Speciation of Bacteria: A New Function for Restriction/Modification Systems? Gene 2003, 317, 13–16. [Google Scholar] [CrossRef] [PubMed]
- Pingoud, A.; Fuxreiter, M.; Pingoud, V.; Wende, W. Type II Restriction Endonucleases: Structure and Mechanism. Cell. Mol. Life Sci. 2005, 62, 685–707. [Google Scholar] [CrossRef] [PubMed]
- Stein, D.C.; Gunn, J.S.; Radlinska, M.; Piekarowicz, A. Restriction and Modification Systems of Neisseria Gonorrhoeae. Gene 1995, 157, 19–22. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.F.; Posfai, J.; Roberts, R.J.; Kong, H. Comparative Genomics of the Restriction-Modification Systems in Helicobacter Pylori. Proc. Natl. Acad. Sci. USA 2001, 98, 2740–2745. [Google Scholar] [CrossRef] [PubMed]
- Arber, W. Evolution of Prokaryotic Genomes. Curr. Top. Microbiol. Immunol. 2002, 264, 1–14. [Google Scholar] [PubMed]
- Rocha, E.P.C.; Danchin, A.; Viari, A. Evolutionary Role of Restriction/Modification Systems as Revealed by Comparative Genome Analysis. Genome Res. 2001, 11, 946–958. [Google Scholar] [CrossRef] [PubMed]
- Nobusato, A.; Uchiyama, I.; Kobayashi, I. Diversity of Restriction–Modification Gene Homologues in Helicobacter Pylori. Gene 2000, 259, 89–98. [Google Scholar] [CrossRef]
- Smith, H.O.; Nathans, D. A Suggested Nomenclature for Bacterial Host Modification and Restriction Systems and Their Enzymes. J. Mol. Biol. 1973, 81, 419–423. [Google Scholar] [CrossRef]
- Loenen, W.A.M.; Dryden, D.T.F.; Raleigh, E.A.; Wilson, G.G. Type I Restriction Enzymes and Their Relatives. Nucleic Acids Res. 2014, 42, 20–44. [Google Scholar] [CrossRef]
- Pingoud, A.; Jeltsch, A. Structure and Function of Type II Restriction Endonucleases. Nucleic Acids Res. 2001, 29, 3705–3727. [Google Scholar] [CrossRef]
- Szybalski, W.; Blumenthal, R.M.; Brooks, J.E.; Hattman, S.; Raleigh, E.A. Nomenclature for Bacterial Genes Coding for Class-II Restriction Endonucleases and Modification Methyltransferases. Gene 1988, 74, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.N.; Dryden, D.T.F.; Bheemanaik, S. Type III Restriction-Modification Enzymes: A Historical Perspective. Nucleic Acids Res. 2014, 42, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Butterer, A.; Pernstich, C.; Smith, R.M.; Sobott, F.; Szczelkun, M.D.; Tóth, J. Type III Restriction Endonucleases Are Heterotrimeric: Comprising One Helicase–Nuclease Subunit and a Dimeric Methyltransferase That Binds Only One Specific DNA. Nucleic Acids Res. 2014, 42, 5139–5150. [Google Scholar] [CrossRef] [PubMed]
- Loenen, W.A.M.; Raleigh, E.A. The Other Face of Restriction: Modification-Dependent Enzymes. Nucleic Acids Res. 2014, 42, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; He, X.; Liang, J.; Li, A.; Xu, T.; Kieser, T.; Helmann, J.D.; Deng, Z. A Novel DNA Modification by Sulphur. Mol. Microbiol. 2005, 57, 1428–1438. [Google Scholar] [CrossRef] [PubMed]
- Ou, H.-Y.; He, X.; Shao, Y.; Tai, C.; Rajakumar, K.; Deng, Z. DndDB: A Database Focused on Phosphorothioation of the DNA Backbone. PLoS ONE 2009, 4, e5132. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Liang, J.; Chen, S.; Wang, L.; He, X.; You, D.; Wang, Z.; Li, A.; Xu, Z.; Zhou, X.; et al. DNA Phosphorothioation in Streptomyces Lividans: Mutational Analysis of the Dnd Locus. BMC Microbiol. 2009, 9, 41. [Google Scholar] [CrossRef]
- Gidwani, S.; Heiter, D.; Xu, S. Expression and Purification of BsaXI Restriction Endonuclease and Engineering New Specificity from BsaXI Specificity Subunit. Front. Microbiol. 2022, 13, 888435. [Google Scholar] [CrossRef]
- Tao, T.; Bourne, J.C.; Blumenthal, R.M. A Family of Regulatory Genes Associated with Type II Restriction-Modification Systems. J. Bacteriol. 1991, 173, 1367–1375. [Google Scholar] [CrossRef]
- Ives, C.L.; Sohail, A.; Brooks, J.E. The Regulatory C Proteins from Different Restriction-Modification Systems Can Cross-Complement. J. Bacteriol. 1995, 177, 6313–6315. [Google Scholar] [CrossRef][Green Version]
- Stahl, F.; Wende, W.; Jeltsch, A.; Pingoud, A. The Mechanism of DNA Cleavage by the Type II Restriction Enzyme EcoRV: Asp36 Is Not Directly Involved in DNA Cleavage but Serves to Couple Indirect Readout to Catalysis. Biol. Chem. Hoppe Seyler 1998, 379, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Thielking, V.; Selent, U.; Koehler, E.; Wolfes, H.; Pieper, U.; Geiger, R.; Urbanke, C.; Winkler, F.K.; Pingoud, A. Site-Directed Mutagenesis Studies with EcoRV Restriction Endonuclease to Identify Regions Involved in Recognition and Catalysis. Biochemistry 1991, 30, 6416–6422. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.E. Restriction Endonucleases and Modification Methylases. Curr. Opin. Struct. Biol. 1993, 3, 24–30. [Google Scholar] [CrossRef]
- Smith, H.O.; Annau, T.M.; Chandrasegaran, S. Finding Sequence Motifs in Groups of Functionally Related Proteins. Proc. Natl. Acad. Sci. USA 1990, 87, 826–830. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch, A.; Pingoud, A. Kinetic Characterization of Linear Diffusion of the Restriction Endonuclease EcoRV on DNA. Biochemistry 1998, 37, 2160–2169. [Google Scholar] [CrossRef] [PubMed]
- Engler, L.E.; Sapienza, P.; Dorner, L.F.; Kucera, R.; Schildkraut, I.; Jen-Jacobson, L. The Energetics of the Interaction of BamHI Endonuclease with Its Recognition Site GGATCC11Edited by R. Ebright. J. Mol. Biol. 2001, 307, 619–636. [Google Scholar] [CrossRef] [PubMed]
- Horton, N.C.; Perona, J.J. Crystallographic Snapshots along a Protein-Induced DNA-Bending Pathway. Proc. Natl. Acad. Sci. USA 2000, 97, 5729–5734. [Google Scholar] [CrossRef]
- Schildkraut, I.; Banner, C.D.B.; Rhodes, C.S.; Parekh, S. The Cleavage Site for the Restriction Endonuclease EcoRV Is 5′-GAT/ATC-3′. Gene 1984, 27, 327–329. [Google Scholar] [CrossRef]
- Winkler, F.K.; Banner, D.W.; Oefner, C.; Tsernoglou, D.; Brown, R.S.; Heathman, S.P.; Bryan, R.K.; Martin, P.D.; Petratos, K.; Wilson, K.S. The Crystal Structure of EcoRV Endonuclease and of Its Complexes with Cognate and Non-Cognate DNA Fragments. EMBO J. 1993, 12, 1781–1795. [Google Scholar] [CrossRef]
- Kostrewa, D.; Winkler, F.K. Mg2+ Binding to the Active Site of EcoRV Endonuclease: A Crystallographic Study of Complexes with Substrate and Product DNA at 2-.ANG. Resolution. Biochemistry 1995, 34, 683–696. [Google Scholar] [CrossRef]
- Perona, J.J.; Martin, A.M. Conformational Transitions and Structural Deformability of EcoRV Endonuclease Revealed by Crystallographic Analysis. J. Mol. Biol. 1997, 273, 207–225. [Google Scholar] [CrossRef] [PubMed]
- Lassila, J.K.; Zalatan, J.G.; Herschlag, D. Biological Phosphoryl-Transfer Reactions: Understanding Mechanism and Catalysis. Annu. Rev. Biochem. 2011, 80, 669–702. [Google Scholar] [CrossRef] [PubMed]
- Kovall, R.A.; Matthews, B.W. Type II Restriction Endonucleases: Structural, Functional and Evolutionary Relationships. Curr. Opin. Chem. Biol. 1999, 3, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Kovall, R.A.; Matthews, B.W. Structural, Functional, and Evolutionary Relationships between λ-Exonuclease and the Type II Restriction Endonucleases. Proc. Natl. Acad. Sci. USA 1998, 95, 7893–7897. [Google Scholar] [CrossRef] [PubMed]
- Kubareva, E.A.; Thole, H.; Karyagina, A.S.; Oretskaya, T.S.; Pingoud, A.; Pingoud, V. Identification of a Base-Specific Contact between the Restriction Endonuclease Ssoll and Its Recognition Sequence by Photocross-Linking. Nucleic Acids Res. 2000, 28, 1085–1091. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mucke, M. Diversity of Type II Restriction Endonucleases That Require Two DNA Recognition Sites. Nucleic Acids Res. 2003, 31, 6079–6084. [Google Scholar] [CrossRef] [PubMed]
- Huai, Q. Crystal Structure of NaeI–an Evolutionary Bridge between DNA Endonuclease and Topoisomerase. EMBO J. 2000, 19, 3110–3118. [Google Scholar] [CrossRef] [PubMed]
- Lukacs, C.M.; Kucera, R.; Schildkraut, I.; Aggarwal, A.K. Understanding the Immutability of Restriction Enzymes: Crystal Structure of Bg/II and Its DNA Substrate at 1.5 Å Resolution. Nat. Struct. Biol. 2000, 7, 134–140. [Google Scholar] [CrossRef]
- Wah, D.A.; Bitinaite, J.; Schildkraut, I.; Aggarwal, A.K. Structure of FokI Has Implications for DNA Cleavage. Proc. Natl. Acad. Sci. USA 1998, 95, 10564–10569. [Google Scholar] [CrossRef]
- Bitinaite, J.; Wah, D.A.; Aggarwal, A.K.; Schildkraut, I. Foki Dimerization Is Required for Dna Cleavage. Proc. Natl. Acad. Sci. USA 1998, 95, 10570–10575. [Google Scholar] [CrossRef]
- Marshall, J.J.T.; Halford, S.E. The Type IIB Restriction Endonucleases. Biochem. Soc. Trans. 2010, 38, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Kong, H. Analyzing the Functional Organization of a Novel Restriction Modification System, the BcgI System. J. Mol. Biol. 1998, 279, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.J.T.; Smith, R.M.; Ganguly, S.; Halford, S.E. Concerted Action at Eight Phosphodiester Bonds by the BcgI Restriction Endonuclease. Nucleic Acids Res. 2011, 39, 7630–7640. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.J.T.; Gowers, D.M.; Halford, S.E. Restriction Endonucleases That Bridge and Excise Two Recognition Sites from DNA. J. Mol. Biol. 2007, 367, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Roemer, S.E.; Waite-Rees, P.A.; Benner, J.S.; Wilson, G.G.; Nwankwo, D.O. Characterization of BcgI, a New Kind of Restriction-Modification System. J. Biol. Chem. 1994, 269, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Morgan, R.D.; Maunus, R.E.; Schildkraut, I. A Unique Restriction Endonuclease, Bcgl, from Bacillus Coagulans. Nucleic Acids Res. 1993, 21, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Piekarowicz, A.; Golaszewska, M.; Sunday, A.O.; Siwińska, M.; Stein, D.C. The HaeIV Restriction Modification System of Haemophilus Aegyptius Is Encoded by a Single Polypeptide. J. Mol. Biol. 1999, 293, 1055–1065. [Google Scholar] [CrossRef] [PubMed]
- Česnavičiene, E.; Petrušyte, M.; Kazlauskiene, R.; Maneliene, Z.; Timinskas, A.; Lubys, A.; Janulaitis, A. Characterization of AloI, a Restriction-Modification System of a New Type. J. Mol. Biol. 2001, 314, 205–216. [Google Scholar] [CrossRef]
- Reuter, M.; Kupper, D.; Meisel, A.; Schroeder, C.; Krüger, D.H. Cooperative Binding Properties of Restriction Endonuclease EcoRII with DNA Recognition Sites. J. Biol. Chem. 1998, 273, 8294–8300. [Google Scholar] [CrossRef]
- Oiler, A.R.; Vanden Broek, W.; Conrad, M.; Topal, M.D. Ability of DNA and Spermidine to Affect the Activity of Restriction Endonucleases from Several Bacterial Species. Biochemistry 1991, 30, 2543–2549. [Google Scholar] [CrossRef]
- Pein, C.-d.; Reuter, M.; Meisel, A.; Cech, D.; Krüger, D.H. Activation of Restriction Endonuclease EcoRII Does Not Depend on the Cleavage of Stimulator DNA. Nucleic Acids Res. 1991, 19, 5139–5142. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Topal, M.D. Modified DNA Fragments Activate Nael Cleavage of Refractory DNA Sites. Nucleic Acids Res. 1992, 20, 5127–5130. [Google Scholar] [CrossRef] [PubMed]
- Embleton, M.L.; Siksnys, V.; Halford, S.E. DNA Cleavage Reactions by Type II Restriction Enzymes That Require Two Copies of Their Recognition Sites. J. Mol. Biol. 2001, 311, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Jo, K.; Topal, M.D. Step-Wise DNA Relaxation and Decatenation by NaeI-43K. Nucleic Acids Res. 1998, 26, 2380–2384. [Google Scholar] [CrossRef] [PubMed]
- Jo, K.; Topal, M.D. DNA Topoisomerase and Recombinase Activities in Nae I Restriction Endonuclease. Science 1995, 67, 1817–1820. [Google Scholar] [CrossRef] [PubMed]
- Wentzell, L.M.; Nobbs, T.J.; Halford, S.E. The SfiI Restriction Endonuclease Makes a Four-Strand DNA Break at Two Copies of Its Recognition Sequence. J. Mol. Biol. 1995, 248, 581–595. [Google Scholar] [CrossRef] [PubMed]
- Nwankwo, D.O.; Maunus, R.E.; Xu, S. Cloning and Expression of AatII Restriction-Modification System in Escherichia coli. Gene 1997, 185, 105–109. [Google Scholar] [CrossRef]
- Deibert, M.; Grazulis, S.; Sasnauskas, G.; Siksnys, V.; Huber, R. Structure of the Tetrameric Restriction Endonuclease NgoMIV in Complex with Cleaved DNA. Nat. Struct. Biol. 2000, 7, 792–799. [Google Scholar] [CrossRef]
- Eckstein, F. Phosphorothioate Oligodeoxynucleotides: What Is Their Origin and What Is Unique about Them? Antisense Nucleic Acid. Drug Dev. 2000, 10, 117–121. [Google Scholar] [CrossRef]
- Nobbs, T.J.; Halford, S.E. DNA Cleavage at Two Recognition Sites by the SfiI Restriction Endonuclease: Salt Dependence of Cis and Trans Interactions between Distant DNA Sites. J. Mol. Biol. 1995, 252, 399–411. [Google Scholar] [CrossRef]
- Williams, S.A.; Halford, S.E. Sfil Endonuclease Activity Is Strongly Influenced by the Non-Specific Sequence in the Middle of Its Recognition Site. Nucleic Acids Res. 2001, 29, 1476–1483. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pingoud, A.; Jeltsch, A. Recognition and Cleavage of DNA by Type-II Restriction Endonucleases. Eur. J. Biochem. 1997, 246, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Janulaitis, A.; Petrusyte, M.; Maneliene, Z.; Klimasauskas, S.; Butkus, V. Purification and Properties of the Eco57lrestriction Endonuclease and Methylas—Prototypes of a New Class (Type IV). Nucleic Acids Res. 1992, 20, 6043–6049. [Google Scholar] [CrossRef] [PubMed]
- Siwek, W.; Czapinska, H.; Bochtler, M.; Bujnicki, J.M.; Skowronek, K. Crystal Structure and Mechanism of Action of the N6-Methyladenine-Dependent Type IIM Restriction Endonuclease R.DpnI. Nucleic Acids Res. 2012, 40, 7563–7572. [Google Scholar] [CrossRef] [PubMed]
- Gowers, D.M. One Recognition Sequence, Seven Restriction Enzymes, Five Reaction Mechanisms. Nucleic Acids Res. 2004, 32, 3469–3479. [Google Scholar] [CrossRef][Green Version]
- Wah, D.A.; Hirsch, J.A.; Dorner, L.F.; Schildkraut, I.; Aggarwal, A.K. Structure of the multimodular endonuclease Fok I bound to DNA. Nature 1997, 388, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Vanamee, É.S.; Santagata, S.; Aggarwal, A.K. FokI Requires Two Specific DNA Sites for Cleavage. J. Mol. Biol. 2001, 309, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.A.; Hosford, C.J.; Azumaya, C.M.; Luyten, Y.A.; Chen, M.; Morgan, R.D.; Stoddard, B.L. Structures, Activity and Mechanism of the Type IIS Restriction Endonuclease PaqCI. Nucleic Acids Res. 2023, 51, 4467–4487. [Google Scholar] [CrossRef]
- Santoshi, M.; Engleng, B.; Eligar, S.M.; Ratnakar, I.S.; Nagamalleshwari, E.; Nagaraja, V. Identification and Characterization of a New HNH Restriction Endonuclease with Unusual Properties. Appl. Microbiol. Biotechnol. 2023. [Google Scholar] [CrossRef]
- Hsieh, P.C.; Xiao, J.P.; O’Loane, D.; Xu, S.Y. Cloning, Expression, and Purification of a Thermostable Nonhomodimeric Restriction Enzyme, BslI. J. Bacteriol. 2000, 182, 949–955. [Google Scholar] [CrossRef]
- Stankevicius, K.; Lubys, A.; Timinskas, A.; Vaitkevicius, D.; Janulaitis, A. Cloning and Analysis of the Four Genes Coding for Bpu10I Restriction-Modification Enzymes. Nucleic Acids Res. 1998, 26, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Lesser, D.R.; Kurpiewski, M.R.; Jen-Jacobson, L. The Energetic Basis of Specificity in the Eco RI Endonuclease—DNA Interaction. Science 1990, 250, 776–780. [Google Scholar] [CrossRef] [PubMed]
- Sidorova, N.Y.; Rau, D.C. Linkage of EcoRI Dissociation from Its Specific DNA Recognition Site to Water Activity, Salt Concentration, and PH: Separating Their Roles in Specific and Non-Specific Binding. J. Mol. Biol. 2001, 310, 801–816. [Google Scholar] [CrossRef] [PubMed]
- Szybalski, W.; Kim, S.C.; Hasan, N.; Podhajska, A.J. Class-IIS Restriction Enzymes—A Review. Gene 1991, 100, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Vermote, C.L.M.; Halford, S.E. EcoRV Restriction Endonuclease: Communication between Catalytic Metal Ions and DNA Recognition. Biochemistry 1992, 31, 6082–6089. [Google Scholar] [CrossRef] [PubMed]
- Yoo, O.J.; Agarwal, K.L. Cleavage of Single Strand Oligonucleotides and Bacteriophage ΦX174 DNA by Msp I Endonuclease. J. Biol. Chem. 1980, 255, 10559–10562. [Google Scholar] [CrossRef] [PubMed]
- Reckmann, B.; Krauss, G. The Cleavage of Single-Stranded DNA by the Isoschizomeric Restriction Endonuclease HhaI and CfoI. Biochim. Biophys. Acta Gene Struct. Expr. 1987, 908, 90–96. [Google Scholar] [CrossRef]
- Raskó, T.; Dér, A.; Klement, É.; Ślaska-Kiss, K.; Pósfai, E.; Medzihradszky, K.F.; Marshak, D.R.; Roberts, R.J.; Kiss, A. BspRI Restriction Endonuclease: Cloning, Expression in Escherichia Coli and Sequential Cleavage Mechanism. Nucleic Acids Res. 2010, 38, 7155–7166. [Google Scholar] [CrossRef][Green Version]
- Kriukiene, E.; Lubiene, J.; Lagunavicius, A.; Lubys, A. MnlI—The Member of H-N-H Subtype of Type IIS Restriction Endonucleases. Biochim. Biophys. Acta Proteins Proteom. 2005, 1751, 194–204. [Google Scholar] [CrossRef]
- Zheleznaya, L.A.; Kachalova, G.S.; Artyukh, R.I.; Yunusova, A.K.; Perevyazova, T.A.; Matvienko, N.I. Nicking Endonucleases. Biochemistry 2009, 74, 1457–1466. [Google Scholar] [CrossRef]
- Xu, S. Sequence-Specific DNA Nicking Endonucleases. Biomol. Concepts 2015, 6, 253–267. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.-H.; Stoddard, B.L.; Xu, S.-Y. Natural and Engineered Nicking Endonucleases—From Cleavage Mechanism to Engineering of Strand-Specificity. Nucleic Acids Res. 2011, 39, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Tang, X.; Chen, T.; Chen, G. Types and Applications of Nicking Enzyme-Combined Isothermal Amplification. Int. J. Mol. Sci. 2022, 23, 4620. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Liu, Q.; Zhou, M.; Zhao, H.; Yang, T.; Ma, C. Nicking Endonuclease-Mediated Isothermal Exponential Amplification for Double-Stranded DNA Detection. Sens. Actuators B Chem. 2016, 222, 221–225. [Google Scholar] [CrossRef]
- Kiesling, T.; Cox, K.; Davidson, E.A.; Dretchen, K.; Grater, G.; Hibbard, S.; Lasken, R.S.; Leshin, J.; Skowronski, E.; Danielsen, M. Sequence Specific Detection of DNA Using Nicking Endonuclease Signal Amplification (NESA). Nucleic Acids Res. 2007, 35, e117. [Google Scholar] [CrossRef]
- Zheng, G.; Dai, J.; Wang, H.; Li, L.; Yuan, D.; Bai, S.; Song, X.; Zhao, Y. A Hairpin-Mediated Nicking Enzymatic Signal Amplification for Nucleic Acids Detection. Talanta 2021, 225, 121991. [Google Scholar] [CrossRef]
- Xu, S. Engineering Infrequent DNA Nicking Endonuclease by Fusion of a BamHI Cleavage-Deficient Mutant and a DNA Nicking Domain. Front. Microbiol. 2022, 12, 787073. [Google Scholar] [CrossRef]
- Xu, S.; Gupta, Y.K. Natural Zinc Ribbon HNH Endonucleases and Engineered Zinc Finger Nicking Endonuclease. Nucleic Acids Res. 2013, 41, 378–390. [Google Scholar] [CrossRef]
- Zhang, P.; Too, P.H.-M.; Samuelson, J.C.; Chan, S.-H.; Vincze, T.; Doucette, S.; Bäckström, S.; Potamousis, K.D.; Schramm, T.M.; Forrest, D.; et al. Engineering BspQI Nicking Enzymes and Application of N.BspQI in DNA Labeling and Production of Single-Strand DNA. Protein Expr. Purif. 2010, 69, 226–234. [Google Scholar] [CrossRef]
- Jeffet, J.; Margalit, S.; Michaeli, Y.; Ebenstein, Y. Single-Molecule Optical Genome Mapping in Nanochannels: Multidisciplinarity at the Nanoscale. Essays Biochem. 2021, 65, 51–66. [Google Scholar] [CrossRef]
- English, A.C.; Salerno, W.J.; Hampton, O.A.; Gonzaga-Jauregui, C.; Ambreth, S.; Ritter, D.I.; Beck, C.R.; Davis, C.F.; Dahdouli, M.; Ma, S.; et al. Assessing Structural Variation in a Personal Genome—Towards a Human Reference Diploid Genome. BMC Genom. 2015, 16, 286. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Mukama, O.; Lu, X.; Cao, S.; Lin, D. Strand Displacement Amplification for Multiplex Detection of Nucleic Acids. In Modulating Gene Expression—Abridging the RNAi and CRISPR-Cas9 Technologies; IntechOpen: London, UK, 2019. [Google Scholar]
- Qian, J.; Ferguson, T.M.; Shinde, D.N.; Ramírez-Borrero, A.J.; Hintze, A.; Adami, C.; Niemz, A. Sequence Dependence of Isothermal DNA Amplification via EXPAR. Nucleic Acids Res. 2012, 40, e87. [Google Scholar] [CrossRef] [PubMed]
- Van Ness, J.; Van Ness, L.K.; Galas, D.J. Isothermal Reactions for the Amplification of Oligonucleotides. Proc. Natl. Acad. Sci. USA 2003, 100, 4504–4509. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Wang, R.; Wu, H.; Ji, F.; Wu, J. Nicking Enzyme-Assisted Amplification (NEAA) Technology and Its Applications: A Review. Anal. Chim. Acta 2019, 1050, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.-H. Cloning of CviPII Nicking and Modification System from Chlorella Virus NYs-1 and Application of Nt.CviPII in Random DNA Amplification. Nucleic Acids Res. 2004, 32, 6187–6199. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ponnaluri, V.K.C.; Zhang, G.; Estève, P.-O.; Spracklin, G.; Sian, S.; Xu, S.; Benoukraf, T.; Pradhan, S. NicE-Seq: High Resolution Open Chromatin Profiling. Genome Biol. 2017, 18, 122. [Google Scholar] [CrossRef] [PubMed]
- James, A.; Alawneh, J. COVID-19 Infection Diagnosis: Potential Impact of Isothermal Amplification Technology to Reduce Community Transmission of SARS-CoV-2. Diagnostics 2020, 10, 399. [Google Scholar] [CrossRef]
- Blakesley, R.W.; Dodgson, J.B.; Nes, I.F.; Wells, R.D. Duplex Regions in “single-Stranded” PhiX174 DNA Are Cleaved by a Restriction Endonuclease from Haemophilus Aegyptius. J. Biol. Chem. 1977, 252, 7300–7306. [Google Scholar] [CrossRef]
- Smith, M.; Smith, K.; Olstein, A.; Oleinikov, A.; Ghindilis, A. Restriction Endonuclease-Based Assays for DNA Detection and Isothermal Exponential Signal Amplification. Sensors 2020, 20, 3873. [Google Scholar] [CrossRef]
- Ghindilis, A.L.; Smith, M.W.; Simon, H.M.; Seoudi, I.A.; Yazvenko, N.S.; Murray, I.A.; Fu, X.; Smith, K.; Jen-Jacobson, L.; Xu, S. Restriction Cascade Exponential Amplification (RCEA) Assay with an Attomolar Detection Limit: A Novel, Highly Specific, Isothermal Alternative to QPCR. Sci. Rep. 2015, 5, 7737. [Google Scholar] [CrossRef]
- Ghindilis, A.L.; Chesnokov, O.; Ngasala, B.; Smith, M.W.; Smith, K.; Mårtensson, A.; Oleinikov, A.V. Detection of Sub-Microscopic Blood Levels of Plasmodium Falciparum Using Tandem Oligonucleotide Repeat Cascade Amplification (TORCA) Assay with an Attomolar Detection Limit. Sci. Rep. 2019, 9, 2901. [Google Scholar] [CrossRef] [PubMed]
- Pang-Chui, S.; Yu-Keung, M. XcmI as a Universal Restriction Enzyme for Single-Stranded DNA. Gene 1993, 133, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Wang, K.; Mayo, S.L. Genome Manipulation by Guide-Directed Argonaute Cleavage. Nucleic Acids Res. 2023, 51, 4078–4085. [Google Scholar] [CrossRef] [PubMed]
- Kropocheva, E.V.; Lisitskaya, L.A.; Agapov, A.A.; Musabirov, A.A.; Kulbachinskiy, A.V.; Esyunina, D.M. Prokaryotic Argonaute Proteins as a Tool for Biotechnology. Mol. Biol. 2022, 56, 854–873. [Google Scholar] [CrossRef] [PubMed]
- Ryazansky, S.; Kulbachinskiy, A.; Aravin, A.A. The Expanded Universe of Prokaryotic Argonaute Proteins. mBio 2018, 9, e01935-18. [Google Scholar] [CrossRef] [PubMed]
- Esyunina, D.; Okhtienko, A.; Olina, A.; Panteleev, V.; Prostova, M.; Aravin, A.A.; Kulbachinskiy, A. Specific Targeting of Plasmids with Argonaute Enables Genome Editing. Nucleic Acids Res. 2023, 51, 4086–4099. [Google Scholar] [CrossRef] [PubMed]
- Lisitskaya, L.; Shin, Y.; Agapov, A.; Olina, A.; Kropocheva, E.; Ryazansky, S.; Aravin, A.A.; Esyunina, D.; Murakami, K.S.; Kulbachinskiy, A. Programmable RNA Targeting by Bacterial Argonaute Nucleases with Unconventional Guide Binding and Cleavage Specificity. Nat. Commun. 2022, 13, 4624. [Google Scholar] [CrossRef]
- Kuzmenko, A.; Yudin, D.; Ryazansky, S.; Kulbachinskiy, A.; Aravin, A.A. Programmable DNA Cleavage by Ago Nucleases from Mesophilic Bacteria Clostridium Butyricum and Limnothrix Rosea. Nucleic Acids Res. 2019, 47, 5822–5836. [Google Scholar] [CrossRef]
- Zander, A.; Willkomm, S.; Ofer, S.; van Wolferen, M.; Egert, L.; Buchmeier, S.; Stöckl, S.; Tinnefeld, P.; Schneider, S.; Klingl, A.; et al. Guide-Independent DNA Cleavage by Archaeal Argonaute from Methanocaldococcus Jannaschii. Nat. Microbiol. 2017, 2, 17034. [Google Scholar] [CrossRef]
- Kropocheva, E.; Kuzmenko, A.; Aravin, A.A.; Esyunina, D.; Kulbachinskiy, A. A Programmable PAgo Nuclease with Universal Guide and Target Specificity from the Mesophilic Bacterium Kurthia massiliensis. Nucleic Acids Res. 2021, 49, 4054–4065. [Google Scholar] [CrossRef]
- Liu, Y.; Li, W.; Jiang, X.; Wang, Y.; Zhang, Z.; Liu, Q.; He, R.; Chen, Q.; Yang, J.; Wang, L.; et al. A Programmable Omnipotent Argonaute Nuclease from Mesophilic Bacteria Kurthia massiliensis. Nucleic Acids Res. 2021, 49, 1597–1608. [Google Scholar] [CrossRef]
- Hegge, J.W.; Swarts, D.C.; Chandradoss, S.D.; Cui, T.J.; Kneppers, J.; Jinek, M.; Joo, C.; van der Oost, J. DNA-Guided DNA Cleavage at Moderate Temperatures by Clostridium butyricum Argonaute. Nucleic Acids Res. 2019, 47, 5809–5821. [Google Scholar] [CrossRef] [PubMed]
- Swarts, D.C.; Hegge, J.W.; Hinojo, I.; Shiimori, M.; Ellis, M.A.; Dumrongkulraksa, J.; Terns, R.M.; Terns, M.P.; van der Oost, J. Argonaute of the Archaeon Pyrococcus furiosus Is a DNA-Guided Nuclease That Targets Cognate DNA. Nucleic Acids Res. 2015, 43, 5120–5129. [Google Scholar] [CrossRef]
- Olina, A.; Kuzmenko, A.; Ninova, M.; Aravin, A.A.; Kulbachinskiy, A.; Esyunina, D. Genome-Wide DNA Sampling by Ago Nuclease from the Cyanobacterium Synechococcus elongatus. RNA Biol. 2020, 17, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Swarts, D.C.; Jore, M.M.; Westra, E.R.; Zhu, Y.; Janssen, J.H.; Snijders, A.P.; Wang, Y.; Patel, D.J.; Berenguer, J.; Brouns, S.J.J.; et al. DNA-Guided DNA Interference by a Prokaryotic Argonaute. Nature 2014, 507, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Pingoud, V.; Sudina, A.; Geyer, H.; Bujnicki, J.M.; Lurz, R.; Lüder, G.; Morgan, R.; Kubareva, E.; Pingoud, A. Specificity Changes in the Evolution of Type II Restriction Endonucleases. J. Biol. Chem. 2005, 280, 4289–4298. [Google Scholar] [CrossRef]
- Chan, S.-H.; Bao, Y.; Ciszak, E.; Laget, S.; Xu, S.-Y. Catalytic Domain of Restriction Endonuclease BmrI as a Cleavage Module for Engineering Endonucleases with Novel Substrate Specificities. Nucleic Acids Res. 2007, 35, 6238–6248. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Samuelson, J.C.; Xu, S. Directed Evolution of Restriction Endonuclease BstYI to Achieve Increased Substrate Specificity. J. Mol. Biol. 2002, 319, 673–683. [Google Scholar] [CrossRef]
- Lanio, T.; Jeltsch, A.; Pingoud, A. On the Possibilities and Limitations of Rational Protein Design to Expand the Specificity of Restriction Enzymes: A Case Study Employing EcoRV as the Target. Protein Eng. Des. Sel. 2000, 13, 275–281. [Google Scholar] [CrossRef][Green Version]
- Schottler, S.; Wenz, C.; Lanio, T.; Jeltsch, A.; Pingoud, A. Protein Engineering of the Restriction Endonuclease EcoRV. Structure-Guided Design of Enzyme Variants That Recognize the Base Pairs Flanking the Recognition Site. Eur. J. Biochem. 1998, 258, 184–191. [Google Scholar] [CrossRef]
- Skowronek, K.; Boniecki, M.J.; Kluge, B.; Bujnicki, J.M. Rational Engineering of Sequence Specificity in R.MwoI Restriction Endonuclease. Nucleic Acids Res. 2012, 40, 8579–8592. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lanio, T.; Jeltsch, A.; Pingoud, A. Towards the Design of Rare Cutting Restriction Endonucleases: Using Directed Evolution to Generate Variants of EcoRV Differing in Their Substrate Specificity by Two Orders of Magnitude. J. Mol. Biol. 1998, 283, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Wenz, C.; Selent, U.; Wende, W.; Jeltsch, A.; Wolfes, H.; Pingoud, A. Protein Engineering of the Restriction Endonuclease EcoRV. Replacement of an Amino Acid Residue in the DNA Binding Site Leads to an Altered Selectivity towards Unmodified and Modified Substrates. Biochim. Biophys. Acta Gene Struct. Expr. 1994, 1219, 73–80. [Google Scholar] [CrossRef]
- Lanio, T.; Selent, U.; Wenz, C.; Wende, W.; Schulz, A.; Adiraj, M.; Katti, S.B.; Pingoud, A. EcoRV-T94V: A Mutant Restriction Endonuclease with an Altered Substrate Specificity towards Modified Oligodeoxynucleotides. Protein Eng. 1996, 9, 1005–1010. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stahl, F.; Wende, W.; Jeltsch, A.; Pingoud, A. Introduction of Asymmetry in the Naturally Symmetric Restriction Endonuclease EcoRV to Investigate Intersubunit Communication in the Homodimeric Protein. Proc. Natl. Acad. Sci. USA 1996, 93, 6175–6180. [Google Scholar] [CrossRef] [PubMed]
- Dorner, L.F.; Bitinaite, J.; Whitaker, R.D.; Schildkraut, I. Genetic Analysis of the Base-Specific Contacts of Bam HI Restriction Endonuclease 1 1Edited by J. A. Wells. J. Mol. Biol. 1999, 285, 1515–1523. [Google Scholar] [CrossRef] [PubMed]
- Rimseliene, R.; Maneliene, Z.; Lubys, A.; Janulaitis, A. Engineering of Restriction Endonucleases: Using Methylation Activity of the Bifunctional Endonuclease Eco57I to Select the Mutant with a Novel Sequence Specificity. J. Mol. Biol. 2003, 327, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.D.; Dwinell, E.A.; Bhatia, T.K.; Lang, E.M.; Luyten, Y.A. The MmeI Family: Type II Restriction–Modification Enzymes That Employ Single-Strand Modification for Host Protection. Nucleic Acids Res. 2009, 37, 5208–5221. [Google Scholar] [CrossRef][Green Version]
- Morgan, R.D.; Luyten, Y.A. Rational Engineering of Type II Restriction Endonuclease DNA Binding and Cleavage Specificity. Nucleic Acids Res. 2009, 37, 5222–5233. [Google Scholar] [CrossRef][Green Version]
- Czapinska, H.; Siwek, W.; Szczepanowski, R.H.; Bujnicki, J.M.; Bochtler, M.; Skowronek, K.J. Crystal Structure and Directed Evolution of Specificity of NlaIV Restriction Endonuclease. J. Mol. Biol. 2019, 431, 2082–2094. [Google Scholar] [CrossRef]
- Xu, Y.; Lunnen, K.D.; Kong, H. Engineering a Nicking Endonuclease N. AlwI by Domain Swapping. Proc. Natl. Acad. Sci. USA 2001, 98, 12990–12995. [Google Scholar] [CrossRef] [PubMed]
- Chandrasegaran, S. Chimeric Restriction Endonuclease. Proc. Natl. Acad. Sci. USA 1994, 91, 883–887. [Google Scholar] [CrossRef]
- Kim, Y.G.; Shi, Y.; Berg, J.M.; Chandrasegaran, S. Site-Specific Cleavage of DNA-RNA Hybrids by Zinc Finger/FokI Cleavage Domain Fusions. Gene 1997, 203, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.G.; Durgesha, M.; Chandrasegaran, S.; Smith, J. Chimeric Restriction Enzyme: Gal4 Fusion to Fok Cleavage Domain. Biol. Chem. Hoppe Seyler 1998, 379, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Jurėnaitė-Urbanavičienė, S.; Šerkšnaitė, J.; Kriukienė, E.; Giedrienė, J.; Venclovas, Č.; Lubys, A. Generation of DNA Cleavage Specificities of Type II Restriction Endonucleases by Reassortment of Target Recognition Domains. Proc. Natl. Acad. Sci. USA 2007, 104, 10358–10363. [Google Scholar] [CrossRef] [PubMed]
- Ain, Q.U.; Chung, J.Y.; Kim, Y.-H. Current and Future Delivery Systems for Engineered Nucleases: ZFN, TALEN and RGEN. J. Control. Release 2015, 205, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Wani, A.K.; Akhtar, N.; Singh, R.; Prakash, A.; Raza, S.H.A.; Cavalu, S.; Chopra, C.; Madkour, M.; Elolimy, A.; Hashem, N.M. Genome Centric Engineering Using ZFNs, TALENs and CRISPR-Cas9 Systems for Trait Improvement and Disease Control in Animals. Vet. Res. Commun. 2023, 47, 1–16. [Google Scholar] [CrossRef]
- Yanik, M.; Alzubi, J.; Lahaye, T.; Cathomen, T.; Pingoud, A.; Wende, W. TALE-PvuII Fusion Proteins—Novel Tools for Gene Targeting. PLoS ONE 2013, 8, e82539. [Google Scholar] [CrossRef]

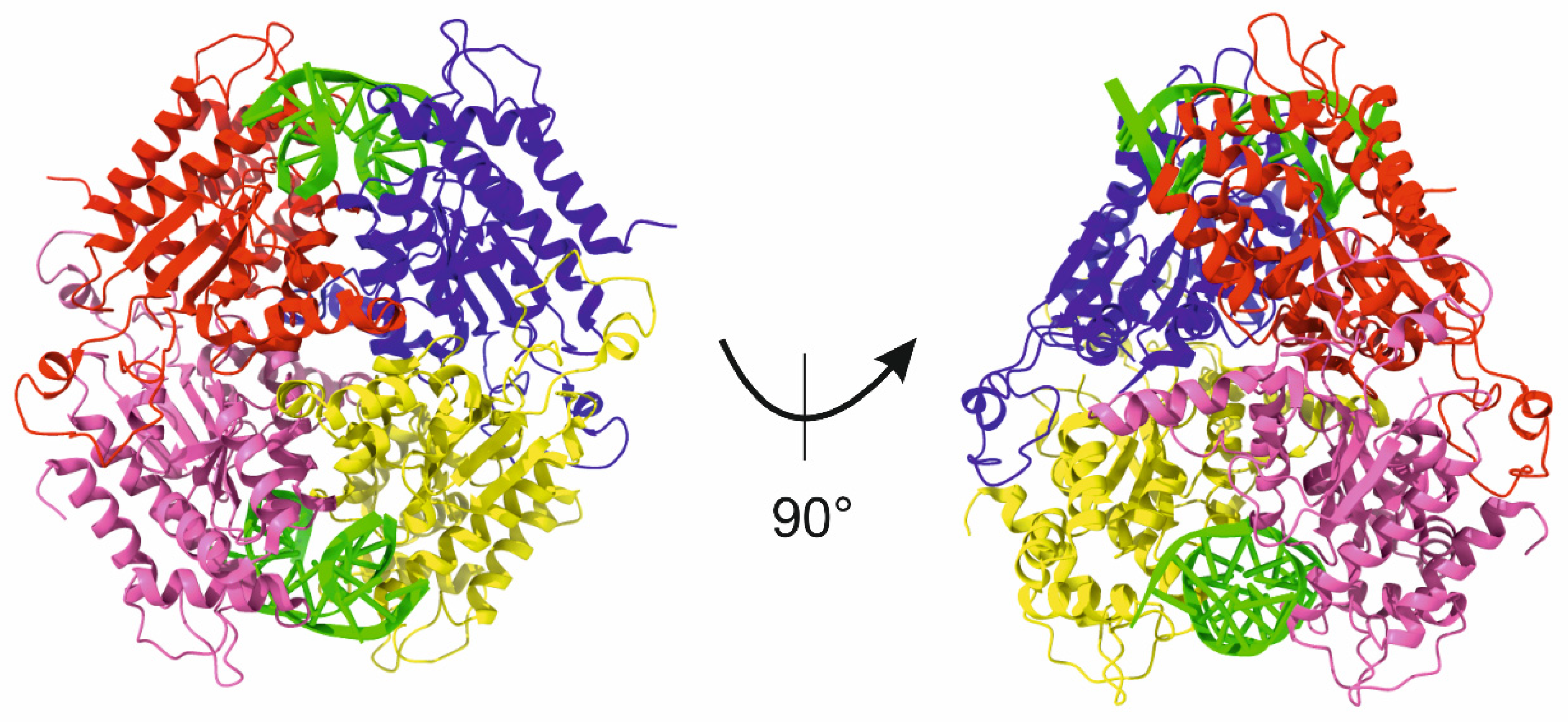

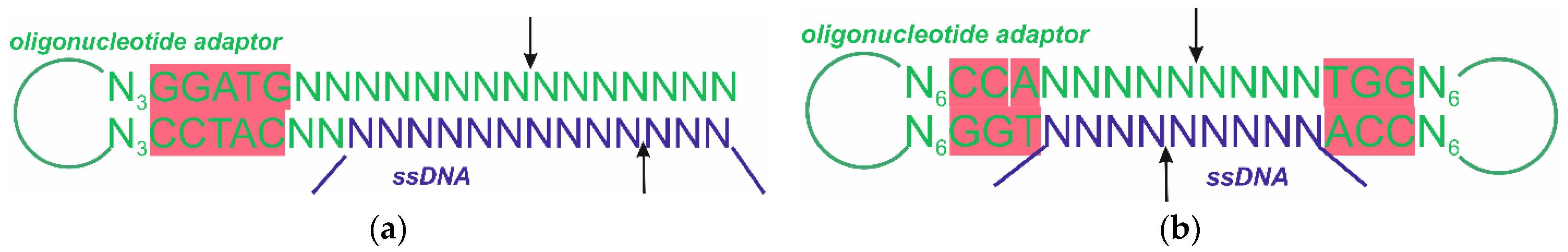
| Subtype | Defining Feature |
|---|---|
| A | Asymmetric recognition sequence. |
| B | Cleaves both sides of the recognition site on both strands. |
| C | Symmetric or asymmetric recognition site. Functions of endonuclease and methyltransferase in one polypeptide. |
| E | Two recognition sites: one cleavable, one effector site. |
| F | Two recognition sites: coordinated cleavage of both sites. |
| G | Symmetric or asymmetric recognition site. Stimulation of the activity by AdoMet. |
| H | Symmetric or asymmetric recognition site. Gene structure similar to that of type I restriction enzymes |
| M | Subtype IIP or IIA, but the recognition site must be methylated. |
| P | Symmetric recognition sequence and cleavage product. |
| S | Asymmetric recognition sequence and cleavage product. |
| T | Symmetric or asymmetric recognition site. The restriction enzyme functions as a heterodimer or heterotetramer. |
| Any Nucleotide | Three of the Four | Two of the Four |
|---|---|---|
| N = A, C, G, or T | B = not A (C, G, or T) D = not C (A, G, or T) V = not T (A, C, or G) H = not G (A, C, or T) | Y = C or T S = G or C M = A or C W = A or T R = A or G K = G or T |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alekseeva, I.V.; Kuznetsov, N.A. Historical Aspects of Restriction Endonucleases as Intelligent Scissors for Genetic Engineering. Fermentation 2023, 9, 874. https://doi.org/10.3390/fermentation9100874
Alekseeva IV, Kuznetsov NA. Historical Aspects of Restriction Endonucleases as Intelligent Scissors for Genetic Engineering. Fermentation. 2023; 9(10):874. https://doi.org/10.3390/fermentation9100874
Chicago/Turabian StyleAlekseeva, Irina V., and Nikita A. Kuznetsov. 2023. "Historical Aspects of Restriction Endonucleases as Intelligent Scissors for Genetic Engineering" Fermentation 9, no. 10: 874. https://doi.org/10.3390/fermentation9100874
APA StyleAlekseeva, I. V., & Kuznetsov, N. A. (2023). Historical Aspects of Restriction Endonucleases as Intelligent Scissors for Genetic Engineering. Fermentation, 9(10), 874. https://doi.org/10.3390/fermentation9100874






