Oxidative Phosphorylation for Aerobic Survival, but Not for Growth: The Peculiar ‘Make-Accumulate-Consume’ Strategy in Zymomonas mobilis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid and Strain Construction
2.2. Cultivation, Starvation and Assessment of Viability
2.3. Cultivation of EGFP-Bearing Strains in the Microplate Reader and Monitoring of EGFP Fluorescence
2.4. FTIR Spectroscopy of Biomass
2.5. Preparation of Cytoplasmic Membranes and Respiratory Chain NADH Oxidase Assay
3. Results
3.1. The Impact of Respiratory Chain Activity on the Viability of Non-Growing Z. mobilis Cells
3.2. Expression of the Respiratory Chain Complexes under Ethanol Stress
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Gene/Operon Names, and Sequence i.d. | Sequences of the Amplified Chromosomal Regions, Bearing the Promoters of the Respective Genes/Operons (5’ → 3’) | Forward and Reverse Primer Sequences (5’ → 3’) |
|---|---|---|
| Cytochrome c peroxidase perC ZZ6_0192 | →ZZ6_0191 CCGATGAATGGGTAAAATAAGACCTTTTCCCTTTCTAAAATTTCAGAGGTGGCTTTATTC CTGTTAAAGCCACCTTTTATTTTATCTTTTTCTTTATTCTTTCTCAGCTTTTCTGATTAA AAAATTTTGTAAATATAGCCAATATTTCCAATGAAATATCAGGTTAAAATATCTGTAAAG AACGAGAATTTTCTTATCACTTTAATGATAACTATTGGCATCATCAAAATCATCAAGAAT TAATCAAAATTGAAATGACAGTTATGCCTTTTACTCTAAAAGGGATGTGCTTTCTGTCTT GGAATAAGGCCATATTTTTGTTAATTCACAATAAATAAAAAGGTAAATCATCTTCAATAG GGAATCGGATACTATAAATTTATAGCCGACCATCCGATCATCTCTGGCGCTGGATTAAGA TAACGAGGGGCAAGCACAGTTTTATGAATATCAAGGCTTTATT →ZZ6_0192 | F primer BamHI CCGATGAATGGGATCCATAAGACCTTTTCC |
| R primer EcoRI AGCCTTGATATTCATGAATTCGTGCTTGCC | ||
| Cytochrome bc1 (ubiquinol:cytc oxidoreductase) operon: FeS cluster (Rieske) subunit ZZ6_0349; Cyt b subunit ZZ6_0348; Cyt c1 subunit ZZ6_0347 | → ZZ6_0350 CTCGCCGCAACCGTCTTGTTCTGGCTATGAGTCCTGAGCCTAGATTTCATGATAGTCAAT ATATTAAAAAATTATATATTGTAGTAATCTTTCTTAAAAAATCAGAATAATTGATTTAAA TATCATTAATTATGGCATTTTAATTATCATTTTTAATATTTTCATGGACAATGAAGAAAA CTTATTATGTTTTTATTGTAGTAAGCGTTGCAAAAAATATTGTAAGTAATTTACAGTAGA GTAGGTTCGTCAAGGGATATGAAGAATAGCGAAGCGAA → ZZ6_0349 | F primer BamHI TTGTTCTGGCTATGGATCCTGAGCCTAGAT |
| R primer EcoRI TTCGCTATTCTTCATGAATTCTGACGAACC | ||
| Cytochrome bd operon: cydA ZZ6_1532; cydB ZZ6_1531 | ZZ6_1533 ← TCAAGGGTGGAAATGGTCATGTCGCTTCAACCTTTGATTCTATGAGCGAGGAAAGACTTA GGAGATATATCTAACAAAATTCGACCCGATCTGACAGCTATTATTAAAATGCAGAATAAG GTCGCGGGCTTTTTATCAGCTAACTCTAACAAAAAACAGAATCATTCGAAAAAGAATAAT AGAATACTCTATATTTAAATTATAAATTGATCCTTTTTCATATGGCTTTTTAATCACAAC CGCCCTAAAATCGGAAAATATCTGTTCCCTTGGGTGGTCTGTTATCAGAGATAGAAAATA ATTCGATTTCAAAAATCGTTATTTTCAATATTGAGACCTAAGCCCCTTTTCCCTTGTTCC TATCGTGACTTTCTTTTGAAATCGCCCCTATTCAACGAATTTAGGGGCAAGTTTTAAAAT AAAGCCCTTAATCGCTTCATCACTATTTTATTTAAAAAATGTTATTGATCTATAAAAAGA TACTATAACTACAGCAATATACTCAAAACAATCTGACATGGTCATATTAAATTTTAAAAT TTATCAAAAAAATAATTAAAATTAAAGAAATATAAAACGATAATTAAACCCAATTGAAGA TAAATAGACTGTAGATAGAGATTATTACCCCCGAGGATCAAGAAATTCTCTATTGACACA ACACCCGATGCTATCAATACACCGATAATATATAGGGGGCATTTTACAGATTCAGGAAGC TGATCCCTGAAGAAAAGCCCGTAGACCCTAATAATTCCTAATATTATCGACGGAGCCATT TCTATGGTACCAGATGCGACCGC → ZZ6_1532 | F primer BamHI GGTGGAAATGGATCCGTCGCTTCAACCTTT |
| R primer EcoRI CGCATCTGGTACCATGAATTCGGCTCCGTC | ||
| Type II NADH: ubiquionone oxidoreductase ndh ZZ6_0213 | ZZ6_0214 ← TGATCGGTTTGTTTTTCCATAACCCATATTGACGGGAAAAGAAGCCAAAGGTCAATCAGA GAAAGCGCGCTTTAAACAAAGCCCATGAGGATGATCAAAGACAGAGACCGGATAAGGGCA AAGATAGTTTCACAGTAAGCAATAAAAACTTTCACTCTGACTTATAATTTATGTGAAACA GATTTGAATGTTCTTCGTTAGTTACAGCAAAAGGTAATTGAGGCGGCTCGCAAACCACCA CGCCAAAGGGGGCTGGGTCGCTTGGAAGAGAATAGAGGTTTCAAATGTCGAAGAATGGTA → ZZ6_0213 | F primer BamHI GGTTTGTGGATCCATAACCCATATTGACGG |
| R primer EcoRI GACATGAATTCCCTCTATTCTCTTCCAAGC | ||
| EGFP gene in plasmid pQE-EGFP egfp | → egfp ATACGGATCCACCGGTCGCCACCATGGTGAGCAAGGGCGAGGAGCTGTTCACCGGGGTGGTGCCCATCCTGGTCGAGCTGGACGGCGACGTAAACGGCCACAAGTTCAGCGTGTCCGGCGAGGGCGAGGGCGATGCCACCTACGGCAAGCTGACCCTGAAGTTCATCTGCACCACCGGCAAGCTGCCCGTGCCCTGGCCCACCCTCGTGACCACCCTGACCTACGGCGTGCAGTGCTTCAGCCGCTACCCCGACCACATGAAGCAGCACGACTTCTTCAAGTCCGCCATGCCCGAAGGCTACGTCCAGGAGCGCACCATCTTCTTCAAGGACGACGGCAACTACAAGACCCGCGCCGAGGTGAAGTTCGAGGGCGACACCCTGGTGAACCGCATCGAGCTGAAGGGCATCGACTTCAAGGAGGACGGCAACATCCTGGGGCACAAGCTGGAGTACAACTACAACAGCCACAACGTCTATATCATGGCCGACAAGCAGAAGAACGGCATCAAGGTGAACTTCAAGATCCGCCACAACATCGAGGACGGCAGCGTGCAGCTCGCCGACCACTACCAGCAGAACACCCCCATCGGCGACGGCCCCGTGCTGCTGCCCGACAACCACTACCTGAGCACCCAGTCCGCCCTGAGCAAAGACCCCAACGAGAAGCGCGATCACATGGTCCTGCTGGAGTTCGTGACCGCCGCCGGGATCACTCTCGGCATGGACGAGCTGTACAAGTAAGGAGGTTCCCGCGGA | F primer EcoRI ATACGGATCCACCGGTCGAATTCATGGTGA |
| R primer HindIII TCCGCGGGAAAGCTTTTACTTGTACAGCTC |
Appendix B
| Incubation without Substrate Addition | Carbohydrates | Nucleic Acids | Proteins | Lipids | ||||
|---|---|---|---|---|---|---|---|---|
| 0 h | 6 h | 0 h | 6 h | 0 h | 6 h | 0 h | 6 h | |
| Mean value | 12.65 | 11.97 | 17.57 | 18.45 | 57.74 | 57.71 | 2.05 | 1.87 |
| ±SEM | ±0.47 | ±0.39 | ±0.35 | ±0.61 | ±0.61 | ±0.27 | ±0.24 | ±0.17 |
| p value | 0.33 | 0.28 | 0.97 | 0.55 | ||||
| Incubation with 1 g L−1 glucose additions, once per hour | ||||||||
| Mean value | 12.70 | 12.21 | 15.93 | 15.48 | 59.13 | 60.06 | 2.24 | 2.25 |
| ±SEM | ±0.87 | ±0.39 | ±0.19 | ±0.15 | ±0.68 | ±0.49 | ±0.36 | ±0.10 |
| p value | 0.63 | 0.11 | 0.31 | 0.98 | ||||
References
- Rogers, P.L.; Lee, K.J.; Skotnicki, M.L.; Tribe, D.E. Ethanol production by Zymomonas mobilis. In Microbial Reactions; Advances in Biochemical Engineering; Springer: Berlin/Heidelberg, Germany, 1982; Volume 23, pp. 37–84. [Google Scholar]
- Sprenger, G. Carbohydrate metabolism in Zymomonas mobilis: A catabolic highway with some scenic routes. FEMS Microbiol. Lett. 1996, 145, 301–307. [Google Scholar] [CrossRef]
- Swings, J.; De Ley, J. The biology of Zymomonas. Bacteriol. Rev. 1977, 41, 1–46. [Google Scholar] [CrossRef]
- Zhang, M.; Eddy, C.; Deanda, K.; Finkelstein, M.; Picataggio, S. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science 1995, 267, 240–243. [Google Scholar]
- Mohagheghi, A.; Linger, J.G.; Yang, S.H.; Smith, H.; Dowe, N.; Zhang, M.; Pienkos, P.T. Improving a recombinant Zymomonas mobilis strain 8b through continuous adaptation on dilute acid pretreated corn stover hydrolysate. Biotechnol. Biofuels 2015, 8, 55. [Google Scholar]
- Yang, S.; Mohagheghi, A.; Franden, M.A.; Chou, Y.C.; Chen, X.; Dowe, N.; Himmel, M.E.; Zhang, M. Metabolic engineering of Zymomonas mobilis for 2,3-butanediol production from lignocellulosic biomass sugars. Biotechnol. Biofuels 2016, 9, 189. [Google Scholar]
- Qiu, M.; Shen, W.; Yan, X.; He, Q.; Cai, D.; Chen, S.; Wei, H.; Knoshaug, E.P.; Zhang, M.; Himmel, M.E.; et al. Metabolic engineering of Zymomonas mobilis for anaerobic isobutanol production. Biotechnol. Biofuels 2020, 13, 15. [Google Scholar]
- Kalnenieks, U.; Pappas, K.M.; Bettenbrock, K. Zymomonas mobilis metabolism: Novel tools and targets for its rational engineering. Adv. Micr. Physiol. 2020, 77, 37–88. [Google Scholar]
- Brooijmans, R.J.W.; Poolman, B.; Schuurman-Wolters, G.K.; De Vos, W.M.; Hugenholtz, J. Generation of a membrane potential by Lactococcus lactis through aerobic electron transport. J. Bacteriol. 2007, 189, 5203–5209. [Google Scholar] [CrossRef]
- Kalnenieks, U.; Balodite, E.; Rutkis, R. Metabolic engineering of bacterial respiration: High vs. low P/O and the case of Zymomonas mobilis. Front. Bioeng. Biotechnol. 2019, 7, 327. [Google Scholar]
- Zelle, E.N.; Oldiges, M.; Koch-Koerfges, A.; Bott, M.; Nöh, K.; Wiechert, W. An energetic profile of Corynebacterium glutamicum underpinned by measured biomass yield on ATP. Metabol. Eng. 2021, 65, 66–78. [Google Scholar] [CrossRef]
- Zotta, T.; Guidone, A.; Ianniello, R.G.; Parente, E.; Ricciardi, A. Temperature and respiration affect the growth and stress resistance of Lactobacillus plantarum C17. J. Appl. Microbiol. 2013, 115, 848–858. [Google Scholar] [CrossRef]
- Hayashi, T.; Kato, T.; Watakabe, S.; Song, W.; Aikawa, S.; Furukawa, K. The respiratory chain provides salt stress tolerance by maintaining a low NADH/NAD+ ratio in Zymomonas mobilis. Microbiology 2015, 161, 2384–2394. [Google Scholar]
- Murata, M.; Ishii, A.; Fujimoto, H.; Nishimura, K.; Kosaka, T.; Mori, H.; Yamada, M. Update of thermotolerant genes essential for survival at a critical high temperature in Escherichia coli. PLoS ONE 2018, 13, e0189487. [Google Scholar] [CrossRef]
- Strohdeicher, M.; Neuß, B.; Bringer-Meyer, S.; Sahm, H. Electron transport chain of Zymomonas mobilis. Interaction with the membrane-bound glucose dehydrogenase and identification of ubiquinone 10. Arch. Microbiol. 1990, 154, 536–543. [Google Scholar]
- Kalnenieks, U.; Galinina, N.; Bringer-Meyer, S.; Poole, R.K. Membrane d-lactate oxidase in Zymomonas mobilis: Evidence for a branched respiratory chain. FEMS Microbiol. Lett. 1998, 168, 91–97. [Google Scholar]
- Kalnenieks, U.; Galinina, N.; Strazdina, I.; Kravale, Z.; Pickford, J.L.; Rutkis, R.; Poole, R.K. NADH dehydrogenase deficiency results in low respiration rate and improved aerobic growth of Zymomonas mobilis. Microbiology 2008, 154, 989–994. [Google Scholar] [CrossRef]
- Sootsuwan, K.; Lertwattanasakul, N.; Thanonkeo, P.; Matsushita, K.; Yamada, M. Analysis of the respiratory chain in ethanologenic Zymomonas mobilis with a cyanide-resistant bd-type ubiquinol oxidase as the only terminal oxidase and its possible physiological roles. J. Mol. Microbiol. Biotechnol. 2008, 14, 163–175. [Google Scholar] [CrossRef]
- Seo, J.-S.; Chong, H.; Park, H.S.; Yoon, K.-O.; Jung, C.; Hong, J.H.; Kim, H.; Kim, J.-H.; Kil, J.-I.; Park, C.J.; et al. The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4. Nat. Biotechnol. 2005, 23, 63–68. [Google Scholar] [CrossRef]
- Yang, S.; Pappas, K.M.; Hauser, L.J.; Land, M.L.; Chen, G.L.; Hurst, G.B.; Pan, C.; Kouvelis, V.N.; A Typas, M.; A Pelletier, D.; et al. Improved genome annotation for Zymomonas mobilis. Nat. Biotechnol. 2009, 27, 893–894. [Google Scholar] [CrossRef]
- Balodite, E.; Strazdina, I.; Galinina, N.; McLean, S.; Rutkis, R.; Poole, R.K.; Kalnenieks, U. Structure of the Zymomonas mobilis respiratory chain: Oxygen affinity of electron transport and the role of cytochrome c peroxidase. Microbiology 2014, 160, 2045–2052. [Google Scholar] [CrossRef]
- Rutkis, R.; Galinina, N.; Strazdina, I.; Kalnenieks, U. The inefficient aerobic energetics of Zymomonas mobilis: Identifying the bottleneck. J. Basic Microbiol. 2014, 54, 1090–1097. [Google Scholar] [CrossRef]
- Rutkis, R.; Strazdina, I.; Balodite, E.; Lasa, Z.; Galinina, N.; Kalnenieks, U. The low energy-coupling respiration in Zymomonas mobilis accelerates flux in the Entner–Doudoroff pathway. PLoS ONE 2016, 11, e0153866. [Google Scholar]
- Kalnenieks, U.; de Graaf, A.A.; Bringer-Meyer, S.; Sahm, H. Oxidative phosphorylation in Zymomonas mobilis. Arch. Microbiol. 1993, 160, 74–79. [Google Scholar] [CrossRef]
- Belaich, J.P.; Senez, J.C. Influence of aeration and pantothenate on growth yields of Zymomonas mobilis. J. Bacteriol. 1965, 89, 1195–1200. [Google Scholar]
- Kalnenieks, U. Physiology of Zymomonas mobilis: Some unanswered questions. Adv. Microb. Physiol. 2006, 51, 73–117. [Google Scholar] [CrossRef]
- Hayashi, T.; Furuta, Y.; Furukawa, K. Respiration-deficient mutants of Zymomonas mobilis show improved growth and ethanol fermentation under aerobic and high temperature conditions. J. Biosci. Bioeng. 2011, 111, 414–419. [Google Scholar]
- Hayashi, T.; Kato, T.; Furukawa, K. Respiratory chain analysis of Zymomonas mobilis mutants producing high levels of ethanol. Appl. Environ. Microbiol. 2012, 78, 5622–5629. [Google Scholar] [CrossRef]
- Kalnenieks, U.; Balodite, E.; Strähler, S.; Strazdina, I.; Rex, J.; Pentjuss, A.; Fuchino, K.; Bruheim, P.; Rutkis, R.; Pappas, K.M.; et al. Improvement of acetaldehyde production in Zymomonas mobilis by engineering of its aerobic metabolism. Front. Microbiol. 2019, 10, 2533. [Google Scholar] [CrossRef]
- De Deken, R.H. The Crabtree effect: A regulatory system in yeast. J. Gen. Microbiol. 1966, 44, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Dashko, S.; Zhou, N.; Compagno, C.; Piskur, J. Why, when, and how did yeast evolve alcoholic fermentation? FEMS Yeast Res. 2014, 14, 826–832. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Swamy, K.B.S.; Leu, J.-Y.; McDonald, M.J.; Galafassi, S.; Compagno, C.; Piškur, J. Coevolution with bacteria drives the evolution of aerobic fermentation in Lachancea kluyveri. PLoS ONE 2017, 12, e0173318. [Google Scholar] [CrossRef]
- He, M.X.; Wu, B.; Shui, Z.X.; Hu, Q.C.; Wang, W.G.; Tan, F.R.; Tang, X.-Y.; Zhu, Q.-L.; Pan, K.; Li, Q.; et al. Transcriptome profiling of Zymomonas mobilis under ethanol stress. Biotechnol. Biofuels 2012, 5, 75. [Google Scholar] [CrossRef]
- Yang, S.; Pan, C.; Tschaplinski, T.J.; Hurst, G.B.; Engle, N.L.; Zhou, W.; Dam, P.; Xu, Y.; Rodriguez, M.; Dice, L.; et al. Systems Biology Analysis of Zymomonas mobilis ZM4 Ethanol Stress Responses. PLoS ONE 2013, 7, e68886. [Google Scholar] [CrossRef]
- Strazdina, I.; Balodite, E.; Lasa, Z.; Rutkis, R.; Galinina, N.; Kalnenieks, U. Aerobic catabolism and respiratory lactate bypass in Ndh-negative Zymomonas mobilis. Metabol. Eng. Commun. 2018, 7, e00081. [Google Scholar]
- Zou, S.; Zhang, K.; You, L.; Zhao, X.; Jing, X.; Zhang, M. Enhanced electrotransformation of the ethanologen Zymomonas mobilis ZM4 with plasmids. Eng. Life Sci. 2012, 12, 152–161. [Google Scholar]
- Grube, M.; Bekers, M.; Upite, D.; Kaminska, E. IR-spectroscopic studies of Zymomonas mobilis and levan precipitate. Vibr. Spectrosc. 2002, 28, 277–285. [Google Scholar] [CrossRef]
- DeLey, J.; Swings, J. Phenotypic description, numerical analysis, and proposal for an improved taxonomy and nomeclature of the genus Zymomonas Kluyver and van Niel 1936. Int. J. Syst. Bacteriol. 1976, 26, 146–157. [Google Scholar]
- Dawes, E.A.; Large, P.J. Effect of starvation on the viability and cellular constituents of Zymomonas anaerobia and Zymomonas mobilis. J. Gen. Microbiol. 1970, 60, 31–62. [Google Scholar]
- Jones-Burrage, S.E.; Kremer, T.A.; McKinlay, J.B. Cell aggregation and aerobic respiration are important for Zymomonas mobilis ZM4 survival in an aerobic minimal medium. Appl. Environ. Microbiol. 2019, 85, e00193-19. [Google Scholar] [CrossRef]
- Hong, Y.; Zeng, J.; Wang, X.; Drlica, K.; Zhao, X. Post-stress bacterial cell death mediated by reactive oxygen species. Proc. Natl. Acad. Sci. USA 2019, 116, 10064–10071. [Google Scholar] [CrossRef]
- Cao, H.; Wei, D.; Yang, Y.; Shang, Y.; Li, G.; Zhou, Y.; Ma, Q.; Xu, Y. Systems-level understanding of ethanol-induced stresses and adaptation in E. coli. Sci. Rep. 2017, 7, 44150. [Google Scholar] [CrossRef]
- Perez-Gallardo, R.V.; Briones, L.S.; Diaz-Perez, A.L.; Gutierrez, S.; Rodriguez-Zavala, J.S.; Campos-Garcia, J. Reactive oxygen species production induced by ethanol in Saccharomyces cerevisiae increases because of a dysfunctional mitochondrial iron–sulfur cluster assembly system. FEMS Yeast Res. 2013, 13, 804–819. [Google Scholar] [CrossRef]
- Martien, J.I.; Hebert, A.S.; Stevenson, D.M.; Regner, M.R.; Khana, D.B.; Coon, J.J.; Amador-Noguez, D. Systems-level analysis of oxygen exposure in Zymomonas mobilis: Implications for isoprenoid production. mSystems 2019, 4, e00284-18. [Google Scholar] [CrossRef]
- Charoensuk, K.; Irie, A.; Lertwattanasakul, N.; Sootsuwan, K.; Thanonkeo, P.; Yamada, M. Physiological importance of cytochrome c peroxidase in ethanologenic thermotolerant Zymomonas mobilis. J. Mol. Microbiol. Biotechnol. 2011, 20, 70–82. [Google Scholar] [CrossRef]
- Messner, K.R.; Imlay, J.A. The Identification of Primary Sites of Superoxide and Hydrogen Peroxide Formation in the Aerobic Respiratory Chain and Sulfite Reductase Complex of Escherichia coli. J. Biol. Chem. 1999, 274, 10119–10128. [Google Scholar]
- Jarmuszkiewicz, W.; Dominiak, K.; Budzinska, A.; Wojcicki, K.; Galganski, L. Mitochondrial Coenzyme Q Redox Homeostasis and Reactive Oxygen Species Production. Front. Biosci. 2023, 28, 61. [Google Scholar] [CrossRef]
- Felczak, M.M.; TerAvest, M.A. Respiration is essential for aerobic growth of Zymomonas mobilis ZM4. bioRxiv 2023. [Google Scholar] [CrossRef]
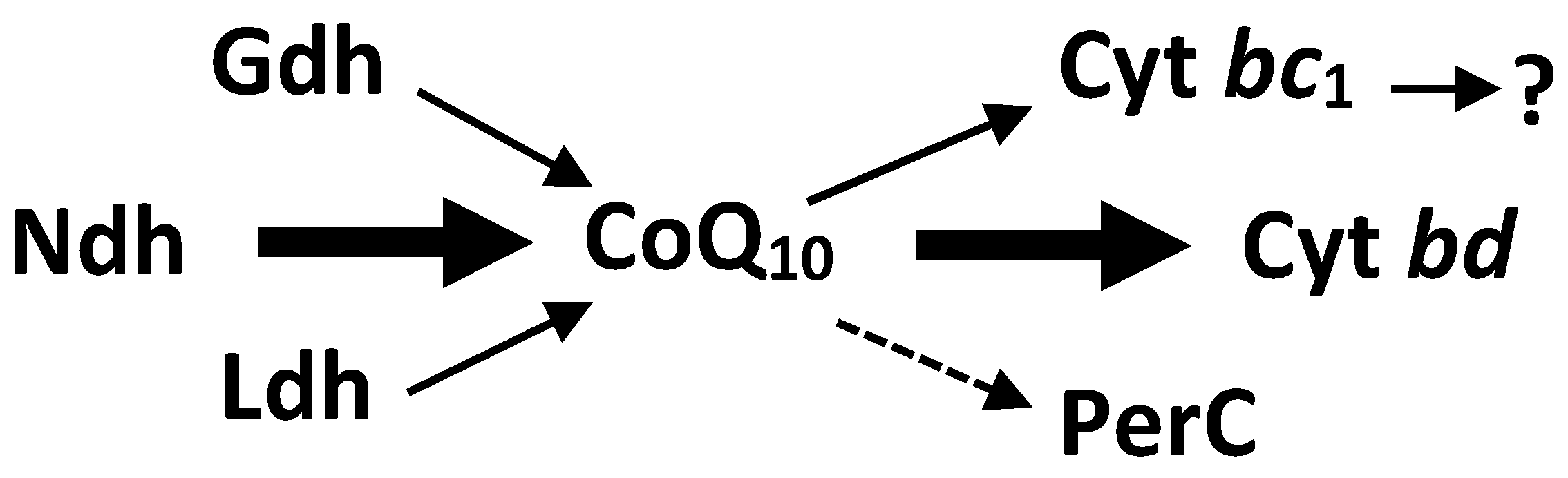
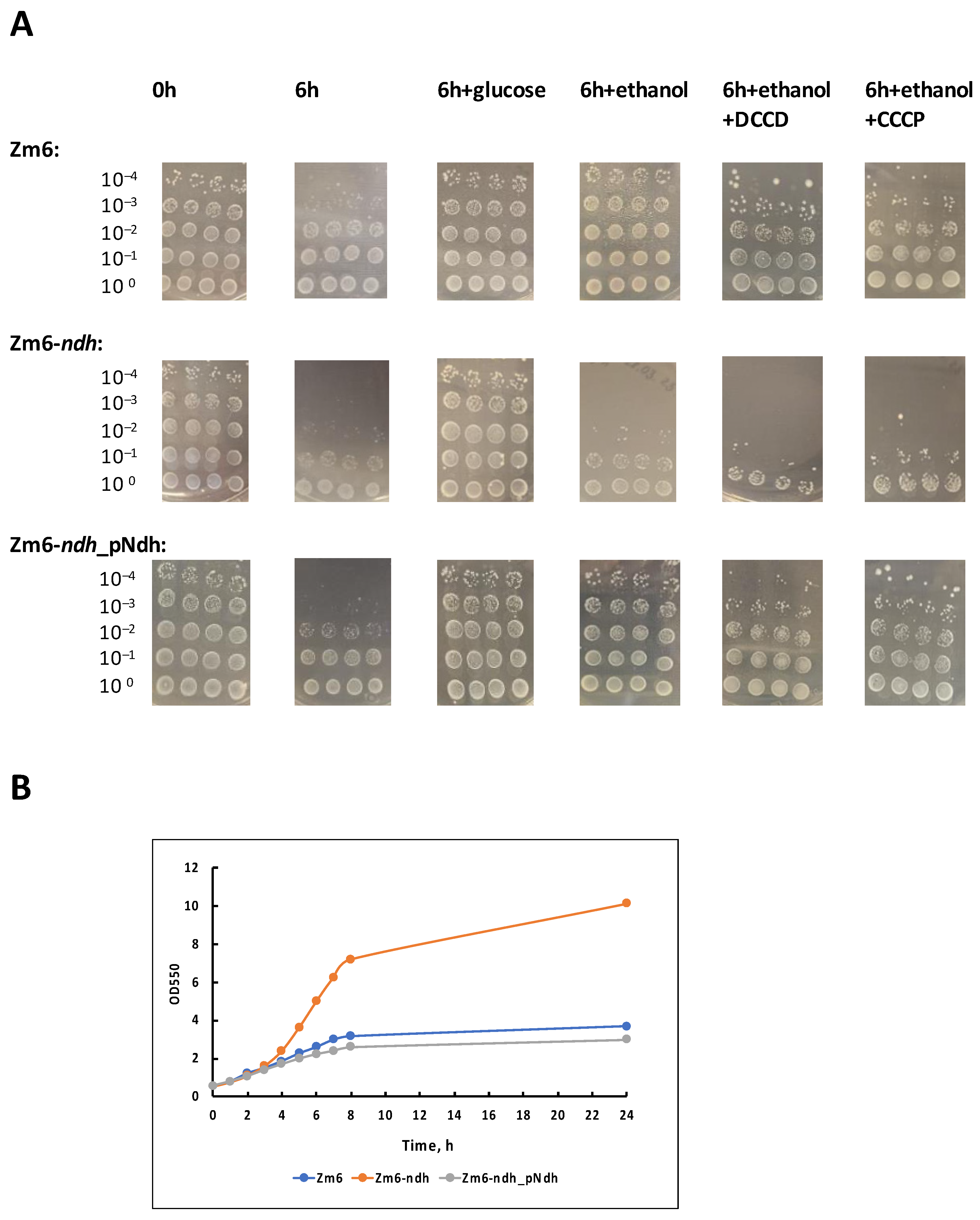

| Plasmid/Strain | Characteristics | Source |
|---|---|---|
| pBBR1MCS-2 | Plasmid vector, kanr | Addgene |
| pNdh | pBBR1MCS-2 derivative with a 1.55 kb insert between the HindIII and BamHI sites of its MCS, containing the ORF of the type II respiratory NADH dehydrogenase (ndh, gene ZZ6_0213) together with its native promoter | [23] |
| pQE-EGFP | pQE-30 plasmid with a 0.72 kb insert of the EGFP coding sequence in the MCS | Addgene |
| pBBR-EGFP | pBBR1MCS-2 derivative with a 0.72 kb insert of the EGFP coding sequence in the MCS between EcoRI and HindIII restriction sites | Present work |
| pBBR:Pndh-EGFP | pBBR-EGFP derivative with a 0.26 kb insert, containing the promoter region of the type II respiratory NADH dehydrogenase (ndh, gene ZZ6_0213), upstream of the EGFP coding sequence in the MCS between BamHI and EcoRI restriction sites | Present work |
| pBBR:Pbc1-EGFP | pBBR-EGFP derivative with a 0.23 kb insert, containing the promoter region of the cytochrome bc1 operon (genes ZZ6_0349, ZZ6_0348, ZZ6_0347), upstream of the EGFP coding sequence in the MCS between BamHI and EcoRI restriction sites | Present work |
| pBBR:Pbd-EGFP | pBBR-EGFP derivative with a 0.76 kb insert, containing the promoter region of the cytochrome bd operon (cydAB; genes ZZ6_1532, ZZ6_1531), upstream of the EGFP coding sequence in the MCS between BamHI and EcoRI restriction sites | Present work |
| pBBR:PperC-EGFP | pBBR-EGFP derivative with a 0.42 kb insert, containing the promoter region of the cytochrome c peroxidase (perC, gene ZZ6_0192), upstream of the EGFP coding sequence in the MCS between BamHI and EcoRI restriction sites | Present work |
| Zm6 | Wild type, parent strain | ATCC 29191 |
| Zm6-ndh | Zm6 with a Cmr (chloramphenicol acetyltransferase gene) inserted in the AgeI site of the chromosomal ORF of its ndh | [17] |
| Zm6-ndh_pNdh | Zm6-ndh transformed with plasmid pNdh | [23] |
| Zm6_pBBR-EGFP | Zm6 transformed with plasmid pBBR-EGFP | Present work |
| Zm6_pBBR:Pndh-EGFP | Zm6 transformed with plasmid pBBR:Pndh-EGFP | Present work |
| Zm6_pBBR:Pbc1-EGFP | Zm6 transformed with plasmid pBBR:Pbc1-EGFP | Present work |
| Zm6_pBBR:Pbd-EGFP | Zm6 transformed with plasmid pBBR:Pbd-EGFP | Present work |
| Zm6_pBBR:PperC-EGFP | Zm6 transformed with plasmid pBBR:PperC-EGFP | Present work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Strazdina, I.; Bikerniece, M.; Paegle, E.R.; Shvirksts, K.; Grube, M.; Lasa, Z.; Rutkis, R.; Kalnenieks, U. Oxidative Phosphorylation for Aerobic Survival, but Not for Growth: The Peculiar ‘Make-Accumulate-Consume’ Strategy in Zymomonas mobilis. Fermentation 2023, 9, 951. https://doi.org/10.3390/fermentation9110951
Strazdina I, Bikerniece M, Paegle ER, Shvirksts K, Grube M, Lasa Z, Rutkis R, Kalnenieks U. Oxidative Phosphorylation for Aerobic Survival, but Not for Growth: The Peculiar ‘Make-Accumulate-Consume’ Strategy in Zymomonas mobilis. Fermentation. 2023; 9(11):951. https://doi.org/10.3390/fermentation9110951
Chicago/Turabian StyleStrazdina, Inese, Mara Bikerniece, Evelina Rezija Paegle, Karlis Shvirksts, Mara Grube, Zane Lasa, Reinis Rutkis, and Uldis Kalnenieks. 2023. "Oxidative Phosphorylation for Aerobic Survival, but Not for Growth: The Peculiar ‘Make-Accumulate-Consume’ Strategy in Zymomonas mobilis" Fermentation 9, no. 11: 951. https://doi.org/10.3390/fermentation9110951
APA StyleStrazdina, I., Bikerniece, M., Paegle, E. R., Shvirksts, K., Grube, M., Lasa, Z., Rutkis, R., & Kalnenieks, U. (2023). Oxidative Phosphorylation for Aerobic Survival, but Not for Growth: The Peculiar ‘Make-Accumulate-Consume’ Strategy in Zymomonas mobilis. Fermentation, 9(11), 951. https://doi.org/10.3390/fermentation9110951





