1. Introduction
Malt extract is obtained by mashing malted grains and consists of the soluble part extracted during this process [
1], which includes amino acids and carbohydrates [
2,
3,
4]. This ingredient can be used as an additive in the manufacturing of bread, cookies, pasta, teas, and malted drinks for human beings [
2,
5]. It may have compounds that confer palatable characteristics and prebiotic potential [
6,
7,
8,
9].
Beta-glucans are classified as prebiotics that, within the large intestine, act as a substrate for the growth and activity of certain microorganisms that promote beneficial effects on the host’s health [
10]. Beta-glucans are polymers that constitute 75% of the barley endosperm cell wall [
11,
12,
13,
14]. Generally, barley grains contain between 2 and 11% of beta-glucans [
15].
It was observed that the concentration of beta-glucans present in the malt grain is lower than barley grain but more soluble and with lower molecular weight [
16]. The average concentration of beta-glucans in malt grains can vary from 0.1% to 1.4% of the total composition on a dry matter basis, similar to that found in malt extract [
16,
17,
18].
Most studies that evaluated the supplementation of beta-glucans to dogs used those from fungi and yeasts, which have beta-glycosidic bonds (1→3) and (1→6), different from those present in cereal beta-glucans, such as barley, (1→4) and (1→3) [
14]. Therefore, it is interesting to assess whether cereal beta-glucans present different activity in the organism or whether it is similar to fungi and yeasts.
Some sugars present in malt extract, such as maltotriose and maltotetraose, can act as a substrate for fermentation in the large intestine of some mammalian species, such as dogs, and may contribute to the increase in beneficial bacterial groups [
19,
20]. It has already been observed that maltotriose, maltotetraose and maltose may be the most abundant sugars in the wort used for malt extract production [
21].
Ingredients with prebiotic capacity also can stimulate the immune system [
22]. The functionality of these ingredients is related to their fermentation by microorganisms in the large intestine and the consequent production of SCLFA. After absorption, these metabolites can act by stimulating the immune system to produce macrophages, neutrophils, and lymphocytes, which promotes resistance to pathogenic Gram-negative bacteria [
23]. Lymphocytes are divided into three groups: T, B, and natural killer cells [
24]. T cells can play different roles in the immune response. For example: CD4
+, or helper T cells, assist in the immune response performed by other T and B cells, in addition to producing pro- and anti-inflammatory cytokines, while CD8
+ or cytotoxic T cells play the role of inducing apoptosis in infected cells by pathogens [
25]. For T cells to play their role, proliferation is an important factor, as it allows the differentiation and homeostasis of this group [
24].
Studies that evaluated the application of malt extract in dog food were not found. Based on this aspect and the increasing demand for co-products with nutraceutical characteristics, studies involving possible new additives in the dogs’ nutrition and other species have become of interest. Therefore, this study aimed to evaluate the effects of providing malt extract on apparent digestibility, fecal score, fecal microbiota, fermentation metabolites, fecal pH, and immunity in health dogs.
2. Materials and Methods
All experimental procedures were approved by the Ethics Committee on the Use of Animals (CEUA) of the Faculty of Veterinary Medicine and Animal Science of the University of São Paulo (FMVZ-USP), under protocol number 5499160221.
2.1. Location, Facilities, and Animals
This study was carried out at the Pet Nutrology Research Center (CEPEN Pet), of the Department of Nutrition and Animal Production, Faculty of Veterinary Medicine and Animal Science of University of Sao Paulo (VNP/FMVZ/USP).
Twelve dogs (six beagles and six English cocker spaniels, three males and three females of each breed) with a mean body weight (BW) of 13.45 ± 1.76 kg that were healthy, neutered, and with a mean age of two years were used. All dogs had an ideal body condition score (BCS; 5.25 ± 0.44) according to the 9-point scale from Laflamme [
26] and an ideal muscle mass score (3) according to the 4-point scale proposed by Michel et al. [
27].
The animals had their health previously evaluated by physical examination, blood count, and biochemical profile tests (urea, creatinine, alanine aminotransferase, alkaline phosphatase, cholesterol, triglycerides, and glucose). All animals that had the test results within the reference range for the species and age group were considered healthy.
The selected dogs were divided randomly in two experimental groups (n = 6 per group). They were housed in kennels with 3.42 m2 of its area covered and with a solarium of 7.21 m2, a concrete floor, and tile walls, and they were grouped according to social affinity (three animals per kennel) and had access to water ad libitum. To promote animal welfare, they were released for socialization and physical activity in grassy parks with an area of 400 m2/park, twice a day, except during the period of sample collection, in which the animals were individually housed in the kennels to avoid the ingestion of foreign bodies and released in groups to carry out a supervised physical and social activity in a concrete place.
2.2. Experimental Diets
The dogs received two experimental diets formulated for maintenance: a control diet—a diet without the addition of malt extract, and a malt diet—a diet with the addition of 1.0% of malt extract. The chemical composition of the malt extract used in this study is shown in
Table 1. The chemical composition of malt extract is shown in
Table 2, and the chemical composition and ingredient list of the experimental diets are shown in
Table 3. To reduce the variability between treatments, the ingredients were obtained from a single lot. After food formulation, the ground ingredients were weighed, homogenized, and extruded in a single-screw extruder (E-100, Ferraz Máquinas, Ribeirao Preto, Brazil). The malt extract (Dry Standard, Liotécnica Tecnologia em Alimentos S.A., Sao Paulo, Brazil) was added after the extrusion during the coating step of the kibbles, associated with swine fat, liquid palatant (SPF, Symrise Pet Food, Descalvado, Brazil) and poultry viscera oil.
The energy requirement for maintenance of the animals was determined by the following equation: 110 × body weight (BW)
0.75 [
28]. With the result, it was possible to determine the amount of food that was supplied daily, dividing it by the metabolizable energy (ME) of the food, which was estimated at 4000 kcal/kg based on the ingredients’ energy, which was calculated by the software used to formulate the diets (Optimal FormulaPlus, Optimal, Campinas, Brazil). The daily total amount of the food was divided into two equal portions. Food intake was monitored daily.
2.3. Experimental Design
The experimental design used was the 2 × 2 crossover, which was composed of two experimental groups (n = 6 per group), two diets, two experimental periods, and a washout period. The two experimental periods lasted 30 days, and the washout lasted 60 days. During the washout period, the dogs received a maintenance diet (Premier Formula Raças Médias, PremieRpet, Dourado, Brazil) that did not contain malt extract. Although this food contains prebiotics and fermentable fibers (dry brewer’s yeast, mannan-oligosaccharides, and yeast cell wall), this diet was chosen because it was the same diet that the animals consumed before the beginning of the study.
Following the premises of the design, each animal consumed both diets at different experimental periods. In the end, 12 experimental units were obtained for the malt treatment (MT), and 12 for the control treatment (CT).
In total, the experiment lasted 120 days. The first 23 days of periods 1 and 2 included adaptation to the diets, and the following 7 days included fecal and blood collection for analysis of apparent digestibility, fecal score, fecal microbiota, fermentation metabolites, fecal pH, and immunity.
2.4. Apparent Digestibility and Fecal Score
The apparent total tract digestibility (ATTD) of the experimental diets were determined by the total feces collection method [
29]. Food consumption was recorded daily, and feces were collected for five days. These were weighed and placed in individual plastic bags and stored in a freezer (−15 °C). At the end of the collection periods, the feces were thawed and homogenized, comprising a single sample (fecal pool) per animal. Subsequently, they were weighed and dried in a forced-air oven (Marconi MA035/2, Piracicaba, Brazil) at 55 °C for 72 h [
30]. The pre-dried feces were ground in a knife mill (Marconi MA340, Piracicaba, Brazil), with a 1 mm sieve, and afterward were ground in a micro-knife mill (Marconi MA048, Piracicaba, Brazil). Food samples were ground in an analytical mill (IKA A11 Basic Mill, Staufen, Germany).
After milling, dry matter (DM) (method 934.01), crude protein (CP) (method 935.11), acid-hydrolyzed fat (method 954.02), ash (method 942.05), and crude fiber (CF) (method 962.09) from feces and diets were determined, according to the methodologies described by the AOAC [
30]; the calcium (Ca) (method 927.02) and phosphorus (P) (method 927.02) contents of the diets were also determined [
30]. Nitrogen-free extracts (NFE) were calculated using the following equation:
Organic matter (OM) was calculated using the formula:
All analyses were performed in duplicate, exception for CF, which was performed in triplicate.
Based on the results obtained in the laboratory and after correction for DM, the ATTD of DM, OM, CP, fat, CF, and NFE of the diets were calculated with the following equation:
The gross energy (GE) of feces and diets were determined using a bomb calorimeter (IKA C200 Basic, Staufen, Germany), and the ME of the diets were calculated according to the FEDIAF [
28].
Fecal score was determined using the scale published by the Waltham Research Center [
31]. During the period of fecal collection for digestibility, values from 1 (hard, dry pellets, which are small and hard masses) to 5 (entire liquid stool) were assigned, considering values between 2.0 and 2.5 as ideal.
2.5. Fecal Microbiota
Feces were collected in a sterile way, using appropriate gloves. After collection, the samples were stored in sterile cryogenic tubes and frozen in a freezer (−80 °C).
The determination of the fecal bacteria population was performed using Illumina sequencing technology (San Diego, CA, USA). Total DNA extraction was performed using the ZR Fungal/Bacterial DNA MiniPrep™ kit (code D6005, Zymo Research, Irvine, CA, USA). PCR reactions were performed in a final volume of 20 μL, containing 10 μL of GoTaq® Colorless Master Mix 2× (USL, Promega, Madison, WI, USA), 0.3 μM of forward oligonucleotide, 0.3 μM of reverse oligonucleotide, 1 uL of genomic DNA, and 20 uL of sufficiently sterile ultrapure water.
Amplification reactions were performed in a Veriti™ Thermal Cycler (Applied Biosystems, Waltham, MA, USA) and verified by electrophoresis on 2% agarose gel stained with UniSafe Dye 0.03% (v/v) ~400 bp (amplicon size). For amplification, a universal primer (5’-ATGATACGGCGACCACCGAGATCTACACTATGGTAATTGTGTGCCAGCCTCCGCGGTAA-3’) was used. The indexing reaction was performed following the protocol of the Nextera XT Index kit (Illumina, San Diego, CA, USA).
The generated libraries were subjected to purification steps using Agencourt AMPure XP magnetic bead (Beckman Coulter, Brea, CA, USA). Quantification was performed using the Real-Time PCR methodology using Kit KAPAKK4824 (Library Quantification Kit—Illumina/Universal). An equimolar pool of DNA was generated, by normalizing all samples to 3 nM for sequencing, using the Illumina MiniSeq next-generation sequencing system (Illumina® Sequencing) and MiniSeq Reagent Output MID kit 300 cycles—reading of 2 × 150 bp.
The bioinformatics of the fecal microbiota was performed with QIIME 2 2021.4 [
32]. The sequence data were demultiplexed and quality filtered using the q2-demux plugin, followed by denoising with DADA2 [
33]. All amplicon sequence variants were aligned with mafft [
34] and used to build a phylogeny with fasttree2 [
35]. Alpha diversity metrics (observed traits, Faith phylogenetic diversity [
36] and Pielou’s evenness index [
37]), beta diversity metrics (weighted UniFrac [
38]), unweighted UniFrac [
39], accord distance, Bray–Curtis dissimilarity [
40] and principal coordinate analysis (PCoA) were estimated using q2 diversity after the samples were rarefied (subsampled without replacement) to 40,607 sequences per sample. The taxonomy was assigned to the amplicon sequence variants through the feature classifier q2 [
41], Bayes taxonomy classifier naïve classify-sklearn against the Greengenes reference sequences 99% operational taxonomic units (OTUs) [
42].
2.6. Fecal pH and Fermentation Metabolites
Fecal pH was determined by homogenizing one gram of fresh feces with 9 mL of distilled water. The analysis was performed within 30 min after sample collection, using a benchtop digital pH meter with an autonomous electrode (STARTER 3100, PH BENCH, OHAUS Sao Paulo/SP) [
43].
To determine SLCFA and SBCFA, three grams of fresh feces sample were diluted in 9 mL of 16% formic acid. These mixtures were kept in a refrigerator (5 °C) and were homogenized daily for seven days. After this period, the samples were centrifuged (Sorvall Legend MACH 1.6 R, Waltham, MA, USA) for 15 min at 15 °C at 3075×
g force three times. The supernatant was extracted, identified, and stored in a freezer (−15 °C). The SLCFA and SBCFA determinations were performed by gas chromatography (GC HP 7890 A; Injector HP 7683 B, Agilent Technologies, Santa Clara, CA, USA) [
44]. The fatty acid concentration (mM) was calculated based on an external calibration curve with acetic, propionic, butyric, valeric, isovaleric, and isobutyric acid performed with chromatographic standards [
44]. The results of acetic, propionic, butyric and valeric acids were added to determine the total SLCFA, while isovaleric and isobutyric acids were added for the total SBCFA. Finally, the values of total SLCFA and total SBCFA were added to determine the total fatty acids.
The preparation of the samples to determine the concentration of fecal ammonia was the same as was used for SLCFA and SBCFA. The extracts were thawed at room temperature, and then 2 mL were diluted in 13 mL of distilled water and submitted for distillation in a MICRO-KJELDAHL apparatus (Marconi MA036, Piracicaba, Brazil) [
45]. These analyses were performed in duplicate.
To determine lactic acid, one gram of sample was diluted with 2 mL of distilled water (1:2
w/
v). These mixtures were kept for three days in a refrigerator (5 °C) and were mixed daily. After this period, the samples were centrifuged for 5 min at 1000×
g force (Fanem 206-R Centrifuge Excelsa Baby II, Sao Paulo, Brazil). The supernatants were extracted. The determination of this metabolite was performed by spectrophotometry at 565 nm (500 to 570 nm) [
46]. These analyzes were performed in triplicate.
2.7. Immunological Tests
Two mL of blood were collected from the jugular vein of the animals, which was stored in a lithium heparin tube. These samples were used to determine lymphocyte proliferation and immunophenotyping of TCD4+ and TCD8+ lymphocytes.
For the lymphocyte proliferation test, one mL of blood was diluted in phosphate-buffered saline (1:3 w/v). Afterward, Ficoll® Paque Plus (GE Healthcare, Chicago, IL, USA) was added to the diluted blood in a 1:2 ratio. The constituent was centrifuged for 25 min at 900× g force and 20 °C (Eppendorf Centrifuge 5804R, Hamburg, Germany). Ten mL of phosphate-buffered saline was added to the lymphocyte band that was obtained. The solution was centrifuged for five minutes at 300× g force and 8 °C. The supernatant was discarded, and another centrifugation similar to the previous one was performed. The supernatant was discarded again, and 900 µg of fetal bovine serum and 100 µg of dimethyl sulfoxide (hibri-max) were added.
The assay was performed in 96-well microtiter plates with a U-shaped bottom. Blood lymphocytes were obtained by separating them into iron particles after purification and washing in RPMI-1640 medium. They were added to the wells at a concentration equivalent to 1 × 10
5 cells in 200 µL/well. The mitogen used was phytohemagglutinin (PHA). The plates were incubated for 72 h at 37 °C in a 5% CO
2 atmosphere. After the incubation, the cells were collected, and the proliferation evaluation was performed in a flow cytometer (FACSCalibur, Becton Dickinson, Franklin Lakes, NJ, USA). For the analysis of the fluorescence data, the values of the percentage of lymphocyte divisions and the index of cellular proliferation of the lymphocytes were considered. CellQuest
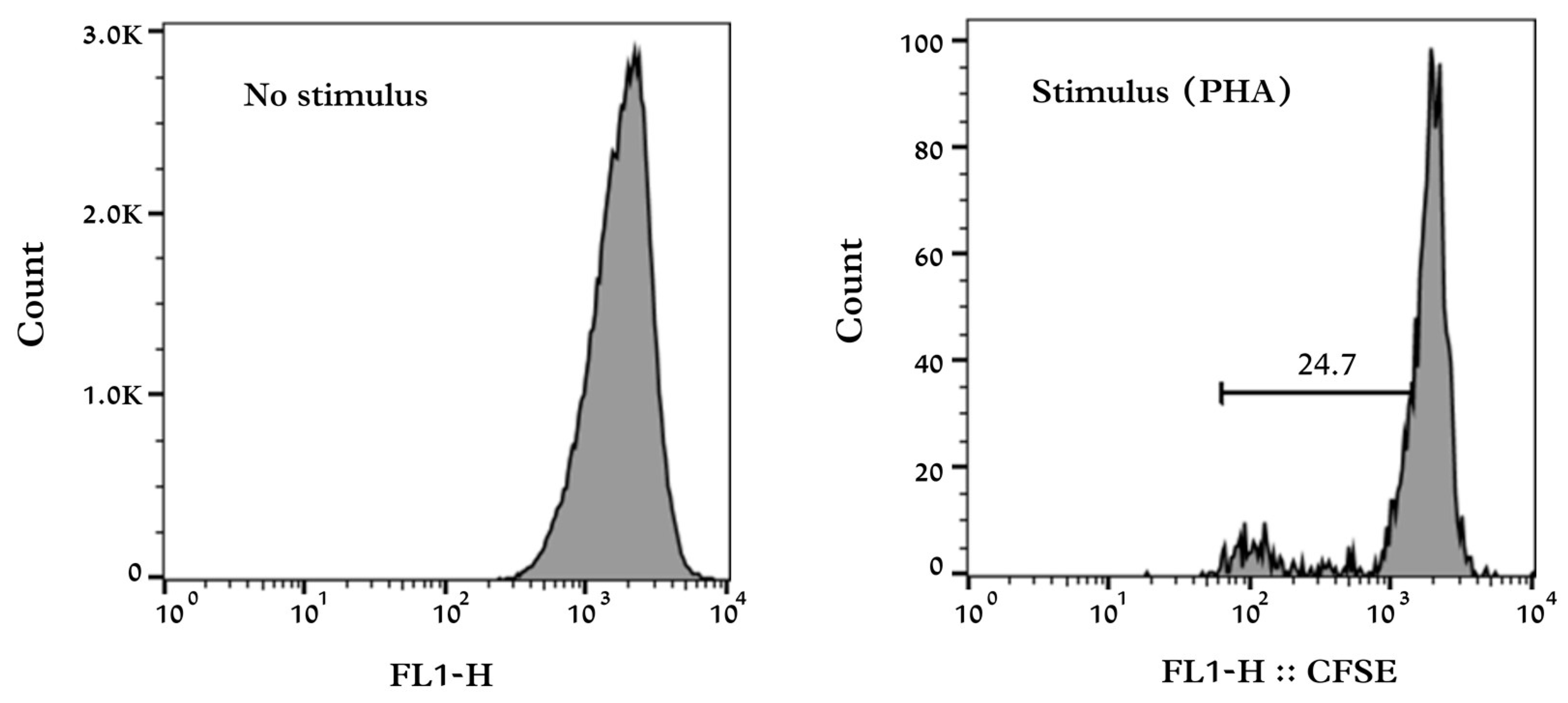
® (Becton Dickinson, Franklin Lakes, NJ, USA) was used to obtain and analyze the results (
Figure 1). To determine the proliferation index, the following equation was used:
The proliferation rate was determined by multiplying the index result by 10.
For the lymphocyte immunophenotyping test, blood samples were diluted with PBS (1:1 v/v). In a sterile 15 mL centrifuge tube, 2 mL of Ficoll® Paque Plus density gradient (GE Healthcare Life Science, Chicago, IL, USA) and 2 mL of diluted blood were placed. The constituent was centrifuged for 20 min at 400× g force and 20 °C for the separation of interface mononuclear cells. The number of naive T helper (CD4+) and cytotoxic T lymphocytes (CD8+) and the CD4+/CD8+ ratio were evaluated.
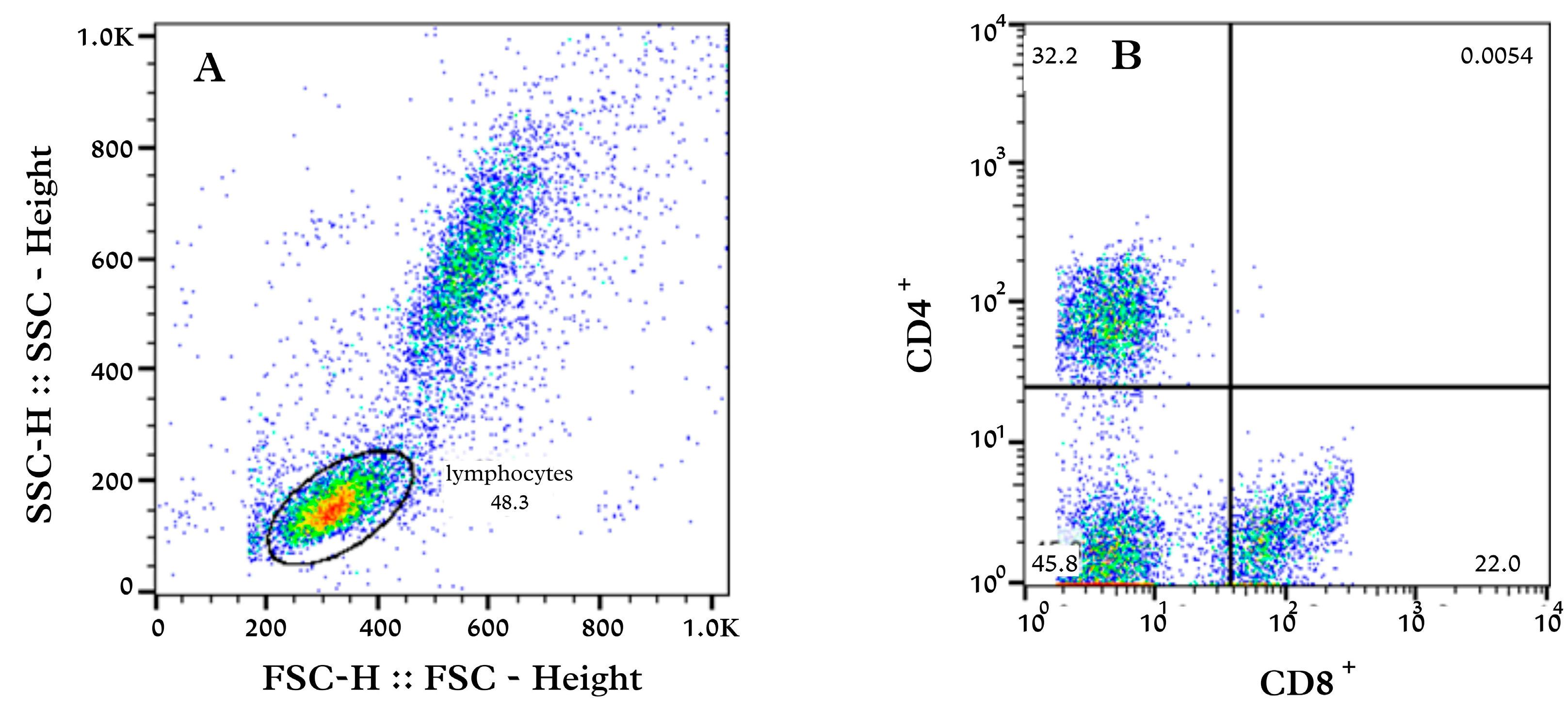
Mononuclear cells (2 × 105 cells/mL) were incubated in microtubes (1.5 mL) with CD4+ (1:10) and CD8+ (1:20) antibodies (Alexa Fluor® 647 anti Dog CD3:FITC/CD4:RPE/CD8 647, Bio-Rad, Hercules, Eugene, OR, USA) and diluted in 100 µL of cytometry buffer (PBS containing 0.5% bovine serum albumin and 0.02% sodium azide). The isotype antibodies for defining the background were included in the assay. Cells were incubated for 30 min at 4 °C, protected from light. At the end of the incubation period, the samples were washed twice with cytometry buffer in a volume of 1000 µL/microtube. Finally, the cells were resuspended in 500 µL of PBS. The population of cells with low size and low complexity, according to the delimited gate, was selected as the lymphocyte population. From this selection, the different populations of lymphocytes were determined. The acquisition and analysis of 10,000 cells were performed using a flow cytometry (FACSCalibur, Becton Dickinson, Franklin Lakes, NJ, USA) and BD CellQuest® software (Becton Dickinson, Franklin Lakes, NJ, USA).
Figure 2 illustrates some of the results obtained by the CellQuest
® software (Becton Dickinson, Franklin Lakes, NJ, USA) of the population of lymphocytes found in a sample from one dog. It also demonstrated a way of counting the number of CD4
+ and CD8
+ T lymphocytes using the software from the initial result.
2.8. Statistical Analysis
Data for digestibility, fecal production, fecal fermentation metabolites, and immunity were analyzed using the Statistical Analysis System software, version 9.4 (SAS Institute Inc., Cary, NC, USA). The normality of the residues was previously verified by the Shapiro–Wilk test (PROC UNIVARIATE), and the homogeneity of the variances by the Levine test. When necessary, logarithmic transformation (log x + 1) was applied. The analysis of variance was performed by PROC MIXED with a significance level of
p < 0.05 according to the following statistical model:
in which Yijk = dependent variable; m = overall mean; Ti = fixed treatment effect; Pj = fixed line effect; Ak = fixed column effect; and eijk = residual error.
The relative abundances for each phylum, class, family, and genus evaluated in each animal were evaluated using a Generalized Mixed Linear Model, considering the binomial distribution of the abundances of each bacterium. The logit link function was adopted to relate the observed abundances to the systematic component of the model. In case of significant effects on the treatment, despite the F Test being discriminatory, the Tukey Test was also adopted as a procedure for comparing means. All microbiota analyzes were performed using the PROC GLIMMIX from the SAS software, version 9.4. The level of significance used was p < 0.05.
4. Discussion
Dog food manufacturers’ demand for alternative ingredients has been growing. Studies have been carried out with different co-products derived from some foods to evaluate their possible effects on the nutrition of these animals [
47]. Barley has some compounds of interest, such as beta-glucans and sugars, which are also present in its co-products, including malt grains and malt extract [
48].
In this study, no differences were observed for ATTD, GE, fecal score, and fecal production. Some authors, when evaluating the supplementation of different prebiotics such as beta-glucans, fructooligosaccharides, and mannooligosaccharides in healthy dogs, also did not find changes in these variables [
49,
50,
51,
52].
It was not possible to perform the GE analysis for the calculation of ME before the beginning of this study. Therefore, the ME was calculated using the ingredients’ energy, and it was estimated for both diets at 4000 kcal/kg. It was observed that the estimated ME value was similar to that obtained after the analyses. The decrease found for ME in the MT can be explained by the reduction of 1% of swine fat from this diet to include malt extract.
The consumption of prebiotics modulates the composition of the intestinal microbiome. They serve as a fermentative substrate for some bacteria, which causes an increase in their population [
53,
54]. Some ways to evaluate fecal microbiota data is through alpha and beta diversity.
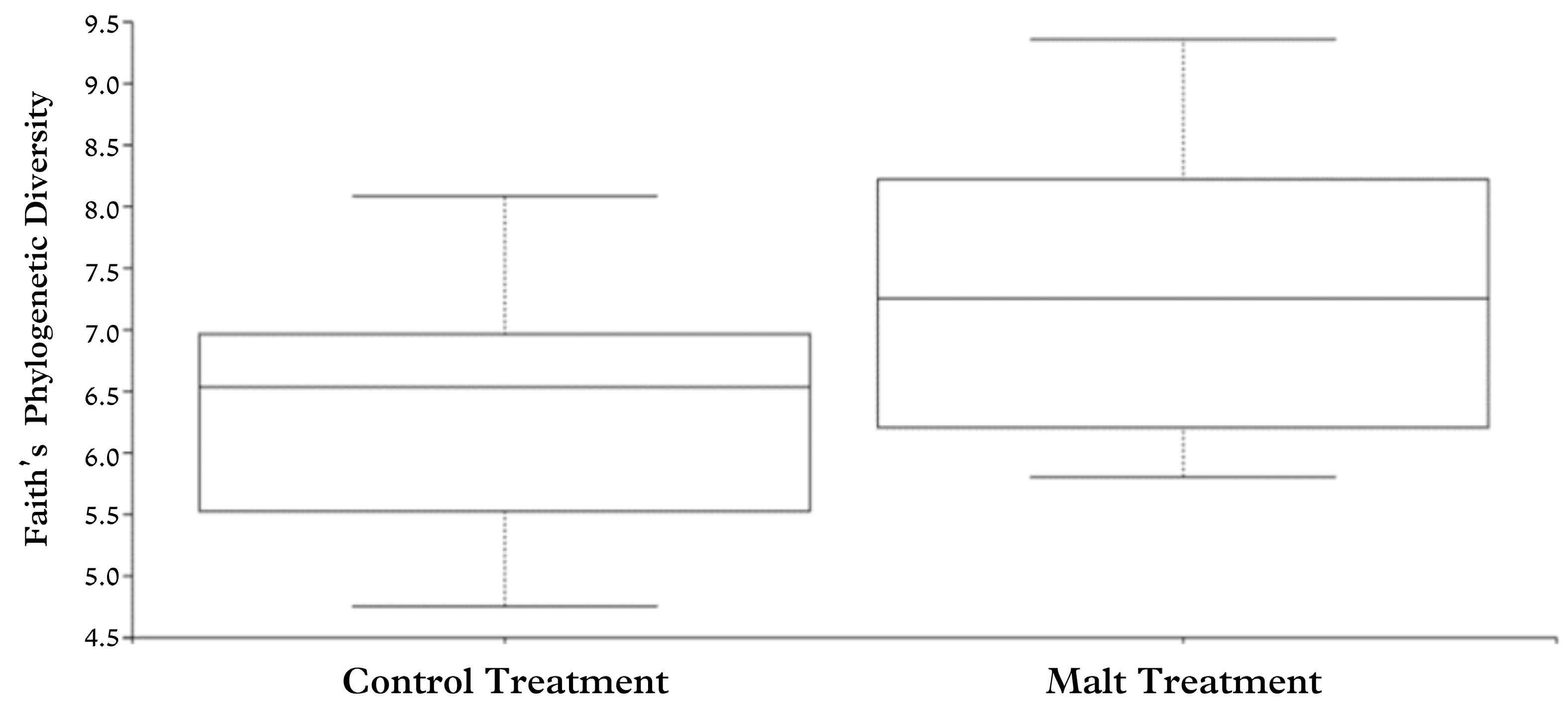
Alpha diversity refers to methods that evaluate the richness, uniformity, and diversity of samples from the same treatment. The Faith index is one of these methods, which calculates the diversity of the treatment based on the number of species and their proportion in the sample [
36]. In this index, the higher the calculated value, the higher the diversity. The Pielou’s index is another method, which shows the uniformity of the distribution of the bacterial groups found in the sample within each treatment [
37]. This index ranges from 0 (minimal uniformity) to 1 (maximum uniformity). In this study, diversity and uniformity indices showed similar results between CT and MT. However, this does not indicate that bacterial groups did not differ between treatments, as the indices were compared and not the relative values.
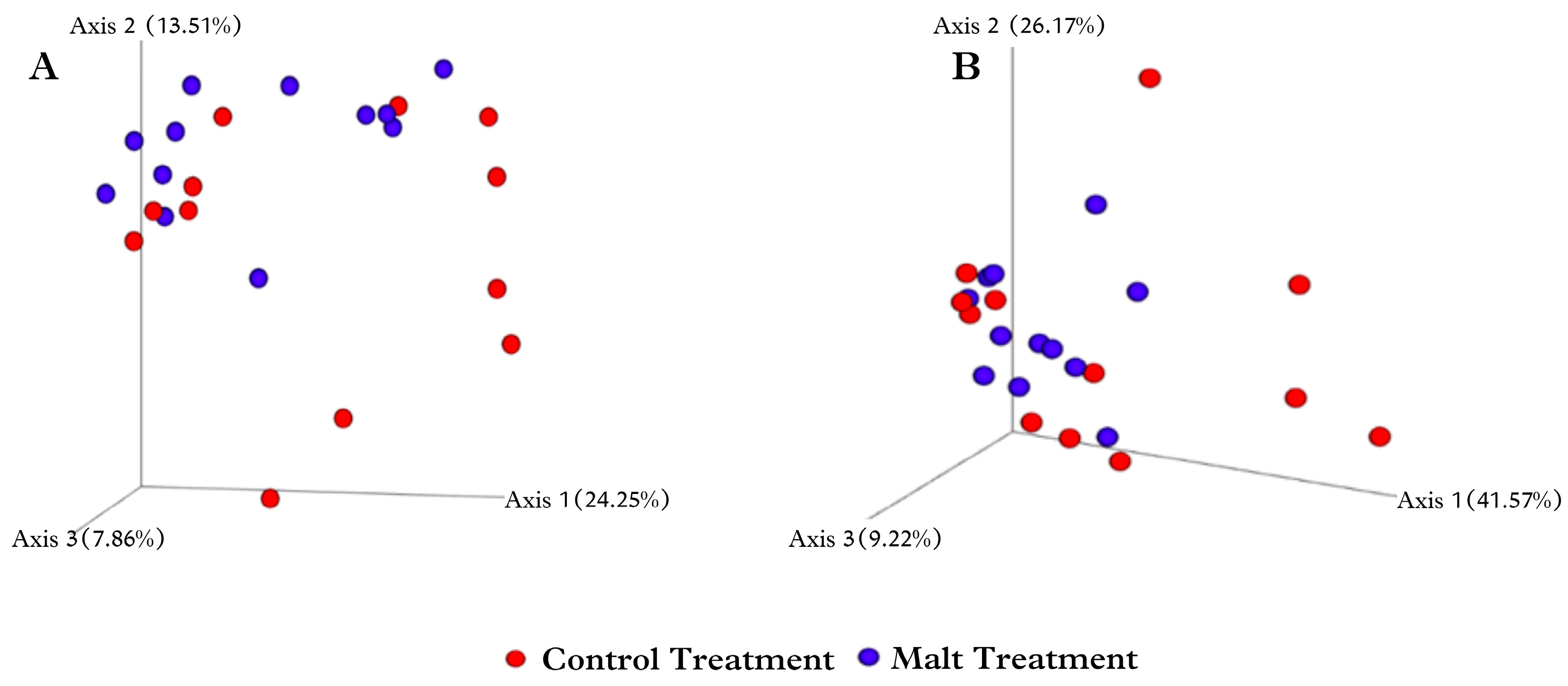
The beta-diversity analysis aims to compare results between treatments. One of the methods used is the Bray–Curtis method [
40], which evaluates the dissimilarity between the species found. In this study, a trend was observed (
p < 0.10) for it when analyzed by PCoA with three axes. However, differences between some groups were found when analyzing phyla, classes, families, and genera. Other authors have also reported similar results [
55,
56]. A possible explanation is that beta-analysis considers all species present in both treatments, but evaluating them individually or in groups can identify differences.
The phylum Firmicutes had the highest relative abundance for both treatments and was higher in MT. The Clostridia class belongs to this phylum, which is known to benefit mucosal health as it produces butyric acid through fermentation [
57]. The genus
Peptostreptococcacea belongs to this class. In this study, it was found in greater abundance than the other genera in both experimental treatments. The role of this bacterial group in the gut is unclear; however, in cats, it has been associated with protein fermentation [
58].
Generally, the phylum Bacteroidetes is the second most abundant in dog feces [
59,
60]. In this study, this phylum was observed in greater abundance in the feces of the animals from the MT. The CT presented Fusobacteria as the predominant second phylum, followed by Bacteroidetes. The other most abundant phyla, which are usually found in the feces of these animals, were also observed in this study: Actinobacteria and Proteobacteria [
53,
60,
61].
Among the genera that compose the phylum Bacteroidetes, Prevotella and Bacteroides showed greater abundance in the MT. In a study conducted by Jackson et al. [
20] with healthy dogs, a positive correlation was observed between these genera and the presence in the diet of maltotriose and maltotetraose. Prevotella is associated with high carbohydrate diets, and it is involved in the process of carbohydrate fermentation to produce SLCFA [
62,
63,
64]. Bacteroides are associated with a healthy microbiome, as they are reduced in unhealthy animals [
65,
66].
Regarding the Actinobacteria phylum, the results are consistent with other studies that observed low abundance for this phylum in dog feces [
59,
67]. In this study, the CT presented a greater relative abundance of this phylum than that of the MT. Middelbos et al. [
67] observed a difference in dog feces for the Actinobacteria phylum, and the treatment that received a diet with 7.5% of beet pulp showed a reduction in it when compared to CT. In rats, this phylum is associated with the production of SLCFA from polysaccharide fermentation [
68]. The relative abundance reduction of this phylum in MT may have occurred due to the increase in other fermenting groups, such as Firmicutes [
57].
Handl et al. [
59] observed that the predominant bacteria class in the feces of healthy dogs is Clostridia, which agrees with the results observed in this study for both treatments, and for this class, the MT presented the highest mean. Handl et al. [
59] also observed a higher abundance of
Clostridium and
Ruminococcus, members of the Clostridia class; for these genera, the averages in the MT group were also higher [
67,
69]. The increase in these bacterial groups in the MT group when compared to the CT group may have been caused by the presence of sugars such as maltotetraose, and fermentable fibers such as beta-glucans, present in malt extract, which serve as a substrate for fermentation by these groups [
70,
71].
Bacteria belonging to the Clostridium cluster XIVa are more abundant in the large intestine of dogs, and this group comprises several genera within the Lachnospiraceae family [
72]. These microorganisms, through fermentation, produce, mainly, SLCFAs (acetate, propionate and butirate) that are beneficial to the intestinal epithelium and host immune system and which can cause an increase in T cell concentration [
69]. Some bacteria of the genus
Faecalibacterium can also produce SLCFA (acetate, propionate and butirate) [
69].
In this study, the MT resulted in a higher relative abundance of the
Faecalibacterium genus and the Lachnospiraceae family. In addition, it also resulted in higher abundances of three genera that compose the Clostridium cluster XIVa:
Clostridium, Ruminococcus, and
Lachnospiraceae [
70,
71]. Jackson et al. [
20] observed that the family
Lachnospiraceae is correlated with the presence of maltose, maltotriose, maltotetraose, and glucose in the diet of healthy dogs.
Bacteria responsible for lactic acid production, such as the genera
Lactobacillus and
Bifidobacterium, are microorganisms of interest because, like carbohydrates, lactic acid can also be used as a substrate by some bacterial groups for SLCFA production [
49,
59]. In this study, bacteria of both genera were identified; however, only nine animals after ingestion of the CT diet and three after ingestion of the MT diet had Lactobacillus in their feces. The results found for this genus agree with other studies, as it is usually identified only in samples from some animals [
61,
73]. Regarding Bifidobacterium, a greater abundance was observed in the CT group. Further studies are needed to understand the reduction in these lactic acid bacteria in the feces of animals that consume malt extract and to understand its possible relationship with the increase in other bacterial groups also observed in this study.
The adaptation period for the diets must be considered when analyzing fecal microbiota and fermentation metabolites. It is necessary to collect samples after this period so that the diet modulates the large intestine microbiome and thereby changes the fermentation metabolite concentration. In this study, there was 29 days of adaptation for the diets, a period similar to that used by other authors [
50,
52], and there was 60 days of washout, a longer time than usual, to ensure that one treatment did not interfere with the other. A study conducted by Perini et al. [
51] with healthy dogs showed that a period of 30 or 60 days of food intake with prebiotics did not change most of the fermentation metabolites analyzed.
SLCFAs are fermentation metabolites that cause a reduction in intestinal pH [
69]. In this study, we expected to find a difference for these metabolites due to the increase in the relative abundance of bacterial groups belonging to the Clostridium cluster XIVa and the
Faecalibacterium genus; however, this did not occur. A possible explanation for these results is the rapid absorption of these metabolites by colonocytes because SLCFAs are an energy source for these cells [
52,
74,
75,
76,
77]. Due to this rapid absorption, fecal and large intestine pH differ, as has been observed in rats and dogs [
74,
78,
79]. So, even if there is a reduction in pH in the large intestine, this difference may not be observed in the feces [
74,
78,
79]. In addition, the level of prebiotic inclusion in the diet can influence the fecal concentration of SLCFAs (acetate, propionate, and butyrate). Fecal concentration changes only when the production of metabolites exceeds the absorption capacity [
52,
74,
75,
76,
77]. In this study, supplementing the diet with 1.0% malt extract was insufficient to alter the fecal concentration of SLCFAs and, therefore, the fecal pH. Other authors that evaluated the supplementation of various prebiotics to dog food also did not observe differences in fecal pH and SLCFA concentration between treatments, but they found differences in other variables, such as fecal microbiota and immunity [
50,
52,
61,
74,
80].
Another objective of adding prebiotics to dog food is to promote animal health [
50,
52]. Some studies have shown that different prebiotics can alter dogs’ immunity due to their ability to modulate the gut microbiome [
52,
61,
74,
76].
For the analysis of lymphocyte proliferation, PHA, which is a T cell-stimulating lectin, was used [
81]. This mitogen, as well as concanavalin A, have been used in several studies on immunity and have shown better results when compared to other mitogens and antigens [
82,
83,
84,
85]. T cells play an important role in regulating the immune system and defending against invading pathogens. Proliferation is essential for lymphocyte differentiation, homeostasis, and immune response [
24].
The lymphocyte proliferation index and rate were higher for the MT. It demonstrates a possible increase in the lymphocyte proliferation response in the face of the identification by the immune system of some antigens or pathogen. These results may be related to the increase in SLCFA-producing bacteria, which were also found in higher concentrations for the MT. The increase in the number of bacterial groups belonging to the Clostridium IV and XIVa clusters has been associated with a rise in T cells in mice [
86]. As reported by other authors, one of the ways SLCFA stimulates the immune system is through the increase in the T cell population [
69]. In this study, there was no difference between the effects of the MT and CT diets on SLCFA. Further research is required to fully comprehend the mechanism of action of malt extract on immunity, although it appears to be related to an increased production of SLCFA in the large intestine.
Currently, there are no established reference values for the rate and index of lymphocyte proliferation in dogs. The literature shows significant variation in results for these variables among healthy adult dogs, making it difficult to compare studies [
85,
87]. Bruin et al. [
88] observed that this index for healthy dogs varied between 0.5 and 4.8 over 24 months.
CD4
+ and CD8
+ cells are T lymphocytes that have T helper and cytotoxic T cell functions, respectively. It is important that during the invasion of a microorganism, the concentration of circulating CD8
+ T cells increases, and after the control of the pathogens, it decreases [
89]. This increase cannot be long-lasting, as it can result in severe inflammatory responses [
90]. The CD4
+:CD8
+ ratio of this study was similar to that observed by other authors [
91]. However, there is no reference value for the CD4
+:CD8
+ ratio in dogs, similar to other immunity variables evaluated in this study.