Food Ingredients for the Future: In-Depth Analysis of the Effects of Lactic Acid Bacteria Fermentation on Spent Barley Rootlets
Abstract
1. Introduction
2. Materials and Methods
2.1. Raw Materials
2.2. Rootlet Preparation
2.3. Rootlet Fermentation
2.3.1. Acidification of Rootlets and Microbial Growth
2.4. Compositional Analysis of Ingredients
2.4.1. Basic Composition of Unfermented Barley Rootlets
2.4.2. Quantification of Sugars, Organic Acids and FODMAPs
Extraction and Detection of Mono-,di-, Trisaccharides and Organic Acids
Extraction and Detection of Fermentable Oligo-,di- and Monosaccharides and Polyols (FODMAPs)
2.4.3. Metabolomic Analysis
2.4.4. Alpha and Beta Amylase Activity
2.5. Scanning Electron Microscopy
2.6. Functional Properties of Rootlet Ingredients
2.6.1. Water- and Oil-Binding Capacity
2.6.2. pH and Titratable Acidity
2.6.3. Colour
2.7. Statistical Analysis
3. Results
3.1. Microbial Growth and Acidification
3.2. Composition of Ingredients
3.2.1. Compositional Analysis Results of BR
3.2.2. Mono-, di-, Trisaccharide Profile
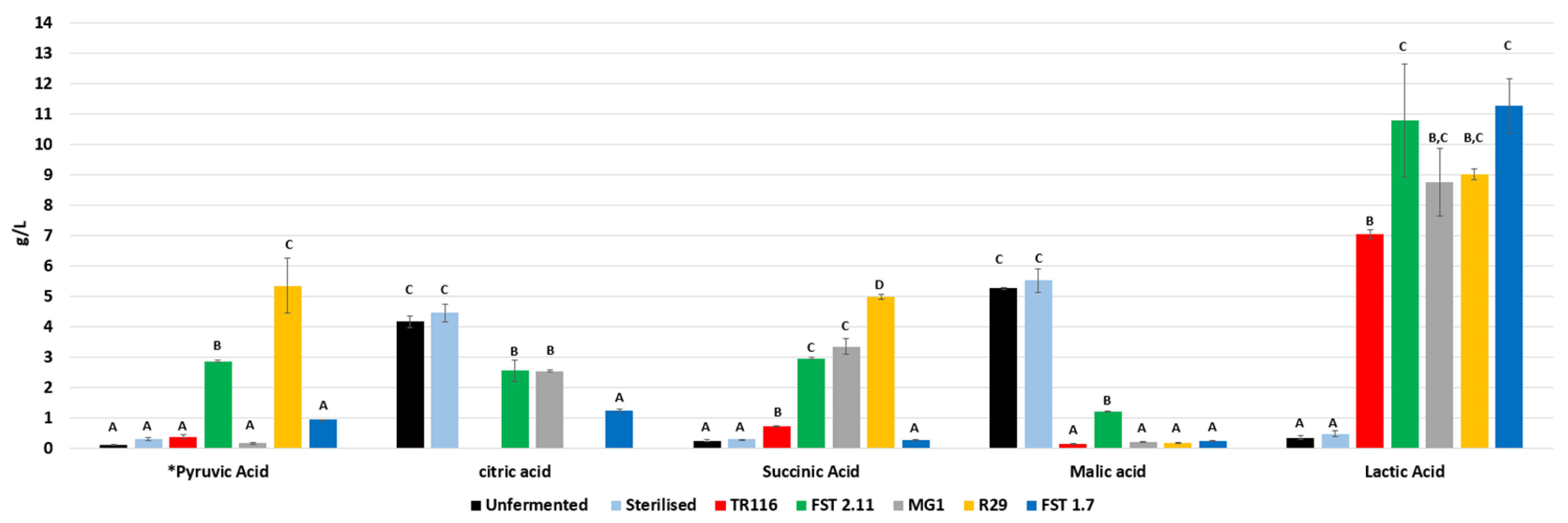
3.2.3. Organic Acids
3.2.4. FODMAPs
3.2.5. Alpha and Beta Amylase Activity
3.2.6. Metabolite Analysis
3.3. Scanning Electron Microscopy
3.4. Techno-Functional Properties of BR Ingredients
3.4.1. Water- and Oil-Binding Capacity
3.4.2. pH, TA and Colour
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Briggs, D.E. Malts and Malting, 1st ed.; Blackie Academic & Professional: London, UK, 1998. [Google Scholar]
- Kunze, W. Technology Brewing and Malting, 3rd ed.; VLB: Berlin, Germany, 2004. [Google Scholar]
- Stewart, G.G.; Russell, I.; Anstruther, A. Handbook of Brewing, 3rd ed.; Taylor & Francis Group: Boca Raton, FL, USA, 2017; ISBN 9781498751919. [Google Scholar]
- Neylon, E.; Arendt, E.K.; Lynch, K.M.; Zannini, E.; Bazzoli, P.; Monin, T.; Sahin, A.W. Rootlets, a Malting By-Product with Great Potential. Fermentation 2020, 6, 117. [Google Scholar] [CrossRef]
- Koistinen, V.M.; Tuomainen, M.; Lehtinen, P.; Peltola, P.; Auriola, S.; Jonsson, K.; Hanhineva, K. Side-stream products of malting: A neglected source of phytochemicals. npj Sci. Food 2020, 4, 21. [Google Scholar] [CrossRef] [PubMed]
- The Maltsters Association of Great Britain Malting Co-Products—Valuable Nutritional Ingredients for the Feed Industry. Available online: https://www.ukmalt.com/technical/8-food-and-feed-safety/malting-co-products/ (accessed on 11 February 2022).
- Chiş, M.S.; Pop, A.; Păucean, A.; Socaci, S.A.; Alexa, E.; Man, S.M.; Bota, M.; Muste, S. Fatty acids, volatile and sensory profile of multigrain biscuits enriched with spent malt rootlets. Molecules 2020, 25, 442. [Google Scholar] [CrossRef] [PubMed]
- Salama, A.R.A.; El-Sahn, M.A.; Mesallam, A.S.; El-Tabey Shehata, A.M. The chemical composition, the nutritive value and the functional properties of malt sprout and its components (acrospires, rootlets and husks). J. Sci. Food Agric. 1997, 75, 50–56. [Google Scholar] [CrossRef]
- Salama, A.A.; El-Sahn, M.A.; Mesallam, A.S.; Shehata, A.M.E. Evaluation of the quality of bread, biscuit and butcher’s sausage supplemented with rootlets of malt sprouts. Nahrung 1997, 41, 228–231. [Google Scholar] [CrossRef]
- Waters, D.M.; Kingston, W.; Jacob, F.; Titze, J.; Arendt, E.K.; Zannini, E. Wheat bread biofortification with rootlets, a malting by-product. J. Sci. Food Agric. 2013, 93, 2372–2383. [Google Scholar] [CrossRef]
- Bianco, A.; Budroni, M.; Zara, S.; Mannazzu, I.; Fancello, F.; Zara, G. The role of microorganisms on biotransformation of brewers’ spent grain. Appl. Microbiol. Biotechnol. 2020, 104, 8661–8678. [Google Scholar] [CrossRef]
- Neylon, E.; Arendt, E.K.; Zannini, E.; Sahin, A.W. Fundamental study of the application of brewers spent grain and fermented brewers spent grain on the quality of pasta. Food Struct. 2021, 30, 100225. [Google Scholar] [CrossRef]
- Neylon, E.; Arendt, E.K.; Zannini, E.; Sahin, A.W. Fermentation as a tool to revitalise brewers’ spent grain and elevate techno-functional properties and nutritional value in high fibre bread. Foods 2021, 10, 1639. [Google Scholar] [CrossRef]
- Waters, D.M.; Jacob, F.; Titze, J.; Arendt, E.K.; Zannini, E. Fibre, protein and mineral fortification of wheat bread through milled and fermented brewer’s spent grain enrichment. Eur. Food Res. Technol. 2012, 235, 767–778. [Google Scholar] [CrossRef]
- Aprodu, I.; Simion, A.B.; Banu, I. Valorisation of the Brewers’ Spent Grain Through Sourdough Bread Making. Int. J. Food Eng. 2017, 13, 1–9. [Google Scholar] [CrossRef]
- Ktenioudaki, A.; Alvarez-Jubete, L.; Smyth, T.J.; Kilcawley, K.; Rai, D.K.; Gallagher, E. Application of bioprocessing techniques (sourdough fermentation and technological aids) for brewer’s spent grain breads. Food Res. Int. 2015, 73, 107–116. [Google Scholar] [CrossRef]
- Verni, M.; Pontonio, E.; Krona, A.; Jacob, S.; Pinto, D.; Rinaldi, F.; Verardo, V.; Díaz-de-Cerio, E.; Coda, R.; Rizzello, C.G. Bioprocessing of Brewers’ Spent Grain Enhances Its Antioxidant Activity: Characterization of Phenolic Compounds and Bioactive Peptides. Front. Microbiol. 2020, 11, 1831. [Google Scholar] [CrossRef]
- Schettino, R.; Verni, M.; Acin-albiac, M.; Vincentini, O.; Krona, A.; Knaapila, A.; Di Cagno, R.; Gobbetti, M.; Rizzello, C.G.; Coda, R. Bioprocessed brewers’ spent grain improves nutritional and antioxidant properties of pasta. Antioxidants 2021, 10, 742. [Google Scholar] [CrossRef]
- Pontonio, E.; Lorusso, A.; Gobbetti, M.; Rizzello, C.G. Use of fermented milling by-products as functional ingredient to develop a low-glycaemic index bread. J. Cereal Sci. 2017, 77, 235–242. [Google Scholar] [CrossRef]
- Verni, M.; Rizzello, C.G.; Coda, R. Fermentation biotechnology applied to cereal industry by-products: Nutritional and functional insights. Front. Nutr. 2019, 6, 42. [Google Scholar] [CrossRef]
- Pontonio, E.; Dingeo, C.; Gobbetti, M.; Rizzello, C.G. Maize milling by-products: From food wastes to functional ingredients through lactic acid bacteria fermentation. Front. Microbiol. 2019, 10, 561. [Google Scholar] [CrossRef]
- Schettino, R.; Pontonio, E.; Rizzello, C.G. Use of fermented hemp, chickpea and milling by-products to improve the nutritional value of semolina pasta. Foods 2019, 8, 604. [Google Scholar] [CrossRef]
- Immonen, M.; Maina, N.H.; Wang, Y.; Coda, R.; Katina, K. Waste bread recycling as a baking ingredient by tailored lactic acid fermentation. Int. J. Food Microbiol. 2020, 327, 108652. [Google Scholar] [CrossRef]
- Cantatore, V.; Filannino, P.; Gambacorta, G.; De Pasquale, I.; Pan, S.; Gobbetti, M.; Di Cagno, R. Lactic Acid Fermentation to Re-cycle Apple By-Products for Wheat Bread Fortification. Front. Microbiol. 2019, 10, 2574. [Google Scholar] [CrossRef]
- Sahin, A.W.; Rice, T.; Zannini, E.; Axel, C.; Coffey, A.; Lynch, K.M.; Arendt, E.K. Leuconostoc citreum TR116: In-situ production of mannitol in sourdough and its application to reduce sugar in burger buns. Int. J. Food Microbiol. 2019, 302, 80–89. [Google Scholar] [CrossRef]
- Sahin, A.W.; Rice, T.; Coffey, A. Genomic analysis of Leuconostoc citreum TR116 with metabolic reconstruction and the effects of fructose on gene expression for mannitol production. Int. J. Food Microbiol. 2021, 354, 109327. [Google Scholar] [CrossRef]
- Rice, T.; Sahin, A.W.; Heitmann, M.; Lynch, K.M.; Jacob, F.; Arendt, E.K.; Coffey, A. Application of mannitol producing Leuconostoc citreum TR116 to reduce sugar content of barley, oat and wheat malt-based worts. Food Microbiol. 2020, 90, 103464. [Google Scholar] [CrossRef]
- Hoehnel, A.; Bez, J.; Sahin, A.W.; Coffey, A.; Arendt, E.K.; Zannini, E. Leuconostoc citreum TR116 as a Microbial Cell Factory to Functionalise High-Protein Faba Bean Ingredients for Bakery Applications. Foods 2020, 9, 1706. [Google Scholar] [CrossRef]
- Boeck, T.; Ispiryan, L.; Hoehnel, A.; Sahin, A.W.; Coffey, A.; Zannini, E.; Arendt, E.K. Lentil-Based Yogurt Alternatives Fermented with Multifunctional Strains of Lactic Acid Bacteria—Techno-Functional, Microbiological, and Sensory Characteristics. Foods 2022, 11, 2013. [Google Scholar] [CrossRef]
- Ryan, L.A.M.; Zannini, E.; Dal Bello, F.; Pawlowska, A.; Koehler, P.; Arendt, E.K. Lactobacillus amylovorus DSM 19280 as a novel food-grade antifungal agent for bakery products. Int. J. Food Microbiol. 2011, 146, 276–283. [Google Scholar] [CrossRef]
- Peyer, L.C.; Zarnkow, M.; Jacob, F.; De Schutter, D.P.; Arendt, E.K. Sour brewing: Impact of Lactobacillus amylovorus FST2.11 on technological and quality attributes of acid beers. J. Am. Soc. Brew. Chem. 2017, 75, 207–216. [Google Scholar] [CrossRef]
- Peyer, L.C.; Bellut, K.; Lynch, K.M.; Zarnkow, M.; Jacob, F.; De Schutter, D.P.; Arendt, E.K. Impact of buffering capacity on the acidification of wort by brewing-relevant lactic acid bacteria. J. Inst. Brew. 2017, 123, 497–505. [Google Scholar] [CrossRef]
- Lynch, K.M.; Lucid, A.; Arendt, E.K.; Sleator, R.D.; Lucey, B.; Coffey, A. Genomics of Weissella cibaria with an examination of its metabolic traits. Microbiology 2015, 161, 914–930. [Google Scholar] [CrossRef]
- Galle, S.; Schwab, C.; Arendt, E.; Gänzle, M. Exopolysaccharide-forming weissella strains as starter cultures for sorghum and wheat sourdoughs. J. Agric. Food Chem. 2010, 58, 5834–5841. [Google Scholar] [CrossRef]
- Wolter, A.; Hager, A.S.; Zannini, E.; Arendt, E.K. Influence of sourdough on in vitro starch digestibility and predicted glycemic indices of gluten-free breads. Food Funct. 2014, 5, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Zannini, E.; Mauch, A.; Galle, S.; Gänzle, M.; Coffey, A.; Arendt, E.K.; Taylor, J.P.; Waters, D.M. Barley malt wort fermentation by exopolysaccharide-forming Weissella cibaria MG1 for the production of a novel beverage. J. Appl. Microbiol. 2013, 115, 1379–1387. [Google Scholar] [CrossRef]
- Wolter, A.; Hager, A.S.; Zannini, E.; Galle, S.; Gänzle, M.G.; Waters, D.M.; Arendt, E.K. Evaluation of exopolysaccharide producing Weissella cibaria MG1 strain for the production of sourdough from various flours. Food Microbiol. 2014, 37, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Belz, M.C.E.; Axel, C.; Arendt, E.K.; Lynch, K.M.; Brosnan, B.; Sheehan, E.M.; Coffey, A.; Zannini, E. Improvement of taste and shelf life of yeasted low-salt bread containing functional sourdoughs using Lactobacillus amylovorus DSM 19280 and Weisella cibaria MG1. Int. J. Food Microbiol. 2019, 302, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Zannini, E.; Jeske, S.; Lynch, K.; Arendt, E.K. Development of novel quinoa-based yoghurt fermented with dextran producer Weissella cibaria MG1. Int. J. Food Microbiol. 2018, 268, 19–26. [Google Scholar] [CrossRef]
- Axel, C.; Brosnan, B.; Zannini, E.; Peyer, L.C.; Furey, A.; Coffey, A.; Arendt, E.K. Antifungal activities of three different Lactobacillus species and their production of antifungal carboxylic acids in wheat sourdough. Appl. Microbiol. Biotechnol. 2016, 100, 1701–1711. [Google Scholar] [CrossRef]
- Schmidt, M.; Lynch, K.M.; Zannini, E.; Arendt, E.K. Fundamental study on the improvement of the antifungal activity of Lactobacillus reuteri R29 through increased production of phenyllactic acid and reuterin. Food Control 2018, 88, 139–148. [Google Scholar] [CrossRef]
- Peyer, L.C.; Zannini, E.; Jacob, F.; Arendt, E.K. Growth study, metabolite development, and organoleptic profile of a malt-based substrate fermented by lactic acid bacteria. J. Am. Soc. Brew. Chem. 2015, 73, 303–313. [Google Scholar] [CrossRef]
- Dal Bello, F.; Clarke, C.I.; Ryan, L.A.M.; Ulmer, H.; Schober, T.J.; Ström, K.; Sjögren, J.; van Sinderen, D.; Schnürer, J.; Arendt, E.K. Improvement of the quality and shelf life of wheat bread by fermentation with the antifungal strain Lactobacillus plantarum FST 1.7. J. Cereal Sci. 2007, 45, 309–318. [Google Scholar] [CrossRef]
- Peyer, L.C.; Axel, C.; Lynch, K.M.; Zannini, E.; Jacob, F.; Arendt, E.K. Inhibition of Fusarium culmorum by carboxylic acids released from lactic acid bacteria in a barley malt substrate. Food Control 2016, 69, 227–236. [Google Scholar] [CrossRef]
- Sahin, A.W.; Rice, T.; Zannini, E.; Lynch, K.M.; Coffey, A.; Arendt, E.K. The incorporation of sourdough in sugar-reduced biscuits: A promising strategy to improve techno-functional and sensory properties. Eur. Food Res. Technol. 2019, 245, 1841–1854. [Google Scholar] [CrossRef]
- Baldini, M.; Fabietti, F.; Giammarioli, S.; Onori, R.; Orefice, L.; Stacchini, A. Metodi di Analisi Utilizzati Per il Controllo Chimico Degli Alimenti. Rapporti ISTISAN 1996/34. 1996. Available online: https://www.iss.it/documents/20126/45616/Rapp_ISTISAN_96_34_def.pdf/e3149ce3-508d-28f5-d4d2-f30c8479213c?t=1581103178196 (accessed on 20 December 2022).
- Ispiryan, L.; Heitmann, M.; Hoehnel, A.; Zannini, E.; Arendt, E.K. Optimization and Validation of an HPAEC-PAD Method for the Quantification of FODMAPs in Cereals and Cereal-Based Products. J. Agric. Food Chem. 2019, 67, 4384–4392. [Google Scholar] [CrossRef] [PubMed]
- Smart, K.F.; Aggio, R.B.M.; Van Houtte, J.R.; Villas-Bôas, S.G. Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatization followed by gas chromatography-mass spectrometry. Nat. Protoc. 2010, 5, 1709–1729. [Google Scholar] [CrossRef] [PubMed]
- Traynham, T.L.; Myers, D.J.; Carriquiry, A.L.; Johnson, L.A. Evaluation of water-holding capacity for wheat-soy flour blends. JAOCS, J. Am. Oil Chem. Soc. 2007, 84, 151–155. [Google Scholar] [CrossRef]
- Hoehnel, A.; Axel, C.; Bez, J.; Arendt, E.K.; Zannini, E. Comparative analysis of plant-based high-protein ingredients and their impact on quality of high-protein bread. J. Cereal Sci. 2019, 89, 102816. [Google Scholar] [CrossRef]
- Hoehnel, A.; Bez, J.; Petersen, I.L.; Amarowicz, R.; Juśkiewicz, J.; Arendt, E.K.; Zannini, E. Enhancing the nutritional profile of regular wheat bread while maintaining technological quality and adequate sensory attributes. Food Funct. 2020, 11, 4732–4751. [Google Scholar] [CrossRef] [PubMed]
- Graves, T.; Narendranath, N.V.; Dawson, K.; Power, R. Effect of pH and lactic or acetic acid on ethanol productivity by Saccharomyces cerevisiae in corn mash. J. Ind. Microbiol. Biotechnol. 2006, 33, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Tirloni, E.; Bernardi, C.; Celandroni, F.; Ghelardi, E.; Stella, S. Effectiveness of lactic and acetic acids on the growth of Listeria monocytogenes and Bacillus cereus in primo sale fresh cheese. Lwt 2021, 151, 112170. [Google Scholar] [CrossRef]
- Chang, R. Physical Chemistry for the Biosciences; University Science Books: Sausalito, CA, USA, 2005. [Google Scholar]
- Von Wright, A.; Axelsson, L. Lactic Acid Bacteria—An introduction. In Lactic Acid Bacteria; Vinderola, G., Ouwehand, A.C., Salminen, S., Von Wright, A., Eds.; CRC Press: Boca Raton, FL, USA, 2019; pp. 1–17. [Google Scholar]
- Prentice, N. Invertase of Germinated Barley. J. Agric. Food Chem. 1972, 20, 764–768. [Google Scholar] [CrossRef]
- Sasaki, Y.; Laivenieks, M.; Zeikus, J.G. Lactobacillus reuteri ATCC 53608 mdh gene cloning and recombinant mannitol dehydrogenase characterization. Appl. Microbiol. Biotechnol. 2005, 68, 36–41. [Google Scholar] [CrossRef]
- Ortiz, M.E.; Fornaguera, M.J.; Raya, R.R.; Mozzi, F. Lactobacillus reuteri CRL 1101 highly produces mannitol from sugarcane molasses as carbon source. Appl. Microbiol. Biotechnol. 2012, 95, 991–999. [Google Scholar] [CrossRef]
- Ortiz, M.E.; Raya, R.R.; Mozzi, F. Efficient mannitol production by wild-type Lactobacillus reuteri CRL 1101 is attained at constant pH using a simplified culture medium. Appl. Microbiol. Biotechnol. 2015, 99, 8717–8729. [Google Scholar] [CrossRef] [PubMed]
- Ispiryan, L.; Zannini, E.; Arendt, E.K. Characterization of the FODMAP-profile in cereal-product ingredients. J. Cereal Sci. 2020, 92, 102916. [Google Scholar] [CrossRef]
- Gibson, P.R.; Halmos, E.P.; Muir, J.G. Review article: FODMAPS, prebiotics and gut health-the FODMAP hypothesis revisited. Aliment. Pharmacol. Ther. 2020, 52, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, M.E.; Bleckwedel, J.; Fadda, S.; Picariello, G.; Hebert, E.M.; Raya, R.R.; Mozzi, F. Global analysis of mannitol 2-dehydrogenase in Lactobacillus reuteri crl 1101 during mannitol production through enzymatic, genetic and proteomic approaches. PLoS ONE 2017, 12, e0169441. [Google Scholar] [CrossRef] [PubMed]
- Gänzle, M.G. Lactic metabolism revisited: Metabolism of lactic acid bacteria in food fermentations and food spoilage. Curr. Opin. Food Sci. 2015, 2, 106–117. [Google Scholar] [CrossRef]
- Sahin, A.W.; Axel, C.; Zannini, E.; Arendt, E.K. Xylitol, mannitol and maltitol as potential sucrose replacers in burger buns. Food Funct. 2018, 9, 2201–2212. [Google Scholar] [CrossRef]
- Ispiryan, L.; Kuktaite, R.; Zannini, E.; Arendt, E.K. Fundamental study on changes in the FODMAP profile of cereals, pseudo-cereals, and pulses during the malting process. Food Chem. 2021, 343, 128549. [Google Scholar] [CrossRef]
- Van Hijum, S.A.F.T.; Van Geel-Schutten, G.H.; Rahaoui, H.; Van der Maarel, M.J.E.C.; Dijkhuizen, L. Characterization of a novel fructosyltransferase from Lactobacillus reuteri that synthesizes high-molecular-weight inulin and inulin oligosaccharides. Appl. Environ. Microbiol. 2002, 68, 4390–4398. [Google Scholar] [CrossRef] [PubMed]
- Van Geel-Schutten, G.H.; Faber, E.J.; Smit, E.; Bonting, K.; Smith, M.R.; Ten Brink, B.; Kamerling, J.P.; Vliegenthart, J.F.G.; Dijkhuizen, L. Biochemical and structural characterization of the glucan and fructan exopolysaccharides synthesized by the Lactobacillus reuteri wild-type strain and by mutant strains. Appl. Environ. Microbiol. 1999, 65, 3008–3014. [Google Scholar] [CrossRef]
- Van Hijum, S.A.F.T.; Bonting, K.; Van Der Maarel, M.J.E.C.; Dijkhuizen, L. Purification of a novel fructosyltransferase from Lactobacillus reuteri strain 121 and characterization of the levan produced. FEMS Microbiol. Lett. 2001, 205, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Ozimek, L.K.; Kralj, S.; van der Maarel, M.J.E.C.; Dijhuizen, L. The levansucrase and inulosucrase enzymes of Lactobacillus reuteri 121 catalyse processive and non-processive transglycosylation reactions. Microbiology 2006, 152, 1187–1196. [Google Scholar] [CrossRef] [PubMed]
- Verspreet, J.; Hemdane, S.; Dornez, E.; Cuyvers, S.; Delcour, J.A.; Courtin, C.M. Maximizing the concentrations of wheat grain fructans in bread by exploring strategies to prevent their yeast (Saccharomyces cerevisiae)-mediated degradation. J. Agric. Food Chem. 2013, 61, 1397–1404. [Google Scholar] [CrossRef]
- Ispiryan, L.; Zannini, E.; Arendt, E.K. FODMAP modulation as a dietary therapy for IBS: Scientific and market perspective. Compr. Rev. Food Sci. Food Saf. 2022, 21, 1491–1516. [Google Scholar] [CrossRef]
- Fedewa, A.; Rao, S.S.C. Dietary fructose intolerance, fructan intolerance and FODMAPs. Curr. Gastroenterol. Rep. 2014, 16, 370. [Google Scholar] [CrossRef]
- Laëtitia, G.; Pascal, D.; Yann, D. The Citrate Metabolism in Homo- and Heterofermentative LAB: A Selective Means of Becoming Dominant over Other Microorganisms in Complex Ecosystems. Food Nutr. Sci. 2014, 05, 953–969. [Google Scholar] [CrossRef]
- Mauro, C.S.I.; Garcia, S. Coconut milk beverage fermented by Lactobacillus reuteri: Optimization process and stability during refrigerated storage. J. Food Sci. Technol. 2019, 56, 854–864. [Google Scholar] [CrossRef]
- Saulnier, D.M.; Santos, F.; Roos, S.; Mistretta, T.A.; Spinler, J.K.; Molenaar, D.; Teusink, B.; Versalovic, J. Exploring metabolic pathway reconstruction and genome-wide expression profiling in Lactobacillus reuteri to define functional probiotic features. PLoS ONE 2011, 6, e18783. [Google Scholar] [CrossRef]
- Østlie, H.M.; Helland, M.H.; Narvhus, J.A. Growth and metabolism of selected strains of probiotic bacteria in milk. Int. J. Food Microbiol. 2003, 87, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, S.M.B.; Mousavi Khaneghah, A.; Barba, F.J.; Nemati, Z.; Sohrabi Shokofti, S.; Alizadeh, F. Fermented sweet lemon juice (Citrus limetta) using Lactobacillus plantarum LS5: Chemical composition, antioxidant and antibacterial activities. J. Funct. Foods 2017, 38, 409–414. [Google Scholar] [CrossRef]
- Hashemi, S.M.B.; Jafarpour, D. Fermentation of bergamot juice with Lactobacillus plantarum strains in pure and mixed fermentations: Chemical composition, antioxidant activity and sensorial properties. Lwt 2020, 131, 109803. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.; Lv, M.; Shao, Z.; Hungwe, M.; Wang, J.; Bai, X.; Xie, J.; Wang, Y.; Geng, W. Metabolism Characteristics of Lactic Acid Bacteria and the Expanding Applications in Food Industry. Front. Bioeng. Biotechnol. 2021, 9, 612285. [Google Scholar] [CrossRef] [PubMed]
- Kaneuchi, C.; Seki, M.; Komagata, K. Production of Succinic Acid from Citric Acid and Related Acids by Lactobacillus Strains. Appl. Environ. Microbiol. 1988, 54, 3053–3056. [Google Scholar] [CrossRef] [PubMed]
- Bartowsky, E.J.; Costello, P.J.; Chambers, P.J. Emerging trends in the application of malolactic fermentation. Aust. J. Grape Wine Res. 2015, 21, 663–669. [Google Scholar] [CrossRef]
- Ferreira, A.M.; Mendes-Faia, A. The role of yeasts and lactic acid bacteria on the metabolism of organic acids during winemaking. Foods 2019, 9, 1231. [Google Scholar] [CrossRef]
- Garcerci Garcera, M.A.J.; Campos, M.A.; Zuniga, M.A.; Uruburu, F.E. Growth and Metabolism of L-malic Acid by Lactobacillus plantarum CECT 220 in a Defined Medium. J. Food Sci. 1992, 57, 778–780. [Google Scholar] [CrossRef]
- Arendt, E.K.; Hammes, W.P. Isolation and characterization of Leuconostoc oenos phages from German wines. Appl. Microbiol. Biotechnol. 1992, 37, 643–646. [Google Scholar] [CrossRef]
- Rosell, C..; Rojas, J.A.; Benedito de Barber, C. Influence of hydrocolloids on dough rheology and bread quality. Food Hydrocoll. 2001, 15, 75–81. [Google Scholar] [CrossRef]
- Sudha, M.L.; Vetrimani, R.; Leelavathi, K. Influence of fibre from different cereals on the rheological characteristics of wheat flour dough and on biscuit quality. Food Chem. 2007, 100, 1365–1370. [Google Scholar] [CrossRef]
- Shi, Y.; Singh, A.; Kitts, D.D.; Pratap-Singh, A. Lactic acid fermentation: A novel approach to eliminate unpleasant aroma in pea protein isolates. Lwt 2021, 150, 111927. [Google Scholar] [CrossRef]
- Mirhosseini, H.; Amid, B.T. Effect of different drying techniques on flowability characteristics and chemical properties of natural carbohydrate-protein Gum from durian fruit seed. Chem. Cent. J. 2013, 7, 1. [Google Scholar] [CrossRef]
- Olaerts, H.; Vandekerckhove, L.; Courtin, C.M. A closer look at the bread making process and the quality of bread as a function of the degree of preharvest sprouting of wheat (Triticum aestivum). J. Cereal Sci. 2018, 80, 188–197. [Google Scholar] [CrossRef]
- Nindo, C.I.; Sun, T.; Wang, S.W.; Tang, J.; Powers, J.R. Evaluation of drying technologies for retention of physical quality and antioxidants in asparagus (Asparagus officinalis, L.). LWT—Food Sci. Technol. 2003, 36, 507–516. [Google Scholar] [CrossRef]
- Argyropoulos, D.; Heindl, A.; Müller, J. Assessment of convection, hot-air combined with microwave-vacuum and freeze-drying methods for mushrooms with regard to product quality. Int. J. Food Sci. Technol. 2011, 46, 333–342. [Google Scholar] [CrossRef]
- Voda, A.; Homan, N.; Witek, M.; Duijster, A.; van Dalen, G.; van der Sman, R.; Nijsse, J.; van Vliet, L.; Van As, H.; van Duynhoven, J. The impact of freeze-drying on microstructure and rehydration properties of carrot. Food Res. Int. 2012, 49, 687–693. [Google Scholar] [CrossRef]
- Rizvi, S.M.H.; Beattie, A.D.; Rossnagel, B.; Scoles, G. Thermostability of barley malt proteases in Western Canadian two-row malting barley. Cereal Chem. 2011, 88, 609–613. [Google Scholar] [CrossRef]
 ), Lactobacillus amylovorus FST2.11 (FST2.11) (
), Lactobacillus amylovorus FST2.11 (FST2.11) ( ), Weissella cibaria MG1 (MG1) (
), Weissella cibaria MG1 (MG1) ( ), Limosilactobacillus reuteri R29 (R29) (
), Limosilactobacillus reuteri R29 (R29) ( ) and Lactiplantibacillus plantarum FST1.7 (FST1.7) (
) and Lactiplantibacillus plantarum FST1.7 (FST1.7) ( ). (A) Development of total titratable acids (TTA) (solid line) and pH (dotted line); (B) cell count (CFU/mL) during fermentation. Points which share the same letter do not differ significantly.
). (A) Development of total titratable acids (TTA) (solid line) and pH (dotted line); (B) cell count (CFU/mL) during fermentation. Points which share the same letter do not differ significantly.
 ), Lactobacillus amylovorus FST2.11 (FST2.11) (
), Lactobacillus amylovorus FST2.11 (FST2.11) ( ), Weissella cibaria MG1 (MG1) (
), Weissella cibaria MG1 (MG1) ( ), Limosilactobacillus reuteri R29 (R29) (
), Limosilactobacillus reuteri R29 (R29) ( ) and Lactiplantibacillus plantarum FST1.7 (FST1.7) (
) and Lactiplantibacillus plantarum FST1.7 (FST1.7) ( ). (A) Development of total titratable acids (TTA) (solid line) and pH (dotted line); (B) cell count (CFU/mL) during fermentation. Points which share the same letter do not differ significantly.
). (A) Development of total titratable acids (TTA) (solid line) and pH (dotted line); (B) cell count (CFU/mL) during fermentation. Points which share the same letter do not differ significantly.

| Species | Leuconostoc citreum | Lactobacillus amylovorus | Weissella cibaria | Limosilactobacillus reuteri | Lactiplantibacillus plantarum |
|---|---|---|---|---|---|
| Strain | TR116 | FST 2.11 | MG1 | R29 | FST 1.7 |
| Metabolism | Heterofermentative | Homofermentative | Heterofermentative | Heterofermentative | Heterofermentative |
| Fermentation substrate | Fructose | Sucrose | Sucrose | Sucrose | Sucrose |
| Source | Yellow pea sourdough | Brewing environment | Sourdough | Human intestine | Malted barley |
| Special traits | Mannitol producer, antifungal producer | Antimicrobial producer, high acid producer | Dextran exopolysaccharide producer | Mannitol producer, antifungal producer | Antifungal producer, high acid producer |
| References | [25,26,27,28,29] | [30,31,32] | [33,34,35,36,37,38,39] | [40,41,42] | [32,35,42,43,44] |
| BR-UnF | BR-Ster | BR-TR116 | BR-MG1 | BR-FST2.11 | BR-R29 | BR-FST1.7 | |
|---|---|---|---|---|---|---|---|
| Sugars a | |||||||
| Glucose | 0.261 ± 0.021 A | 0.521 ± 0.009 B | n.d. | 8.094 ± 0.147 F | 2.176 ± 0.029 C | 3.190 ± 0.040 D | 4.581 ± 0.050 E |
| Fructose | 1.149 ± 0.076 C | 1.249 ± 0.023 C | 0.347 ± 0.006 A | 13.234 ± 0.229 F | 11.870 ± 0.167 E | 0.495 ± 0.004 B | 9.991 ± 0.134 D |
| Sucrose | 0.063 ± 0.002 C | 0.016 ± 0.000 A | n.d. | 0.586 ± 0.033 D | 2.690 ± 0.069 F | 0.042 ± 0.000 B | 2.060 ± 0.036 E |
| Maltose | 0.006 ± 0.000 A | 0.137 ± 0.001 D | n.d. | n.d. | 0.041 ± 0.001 C | 0.012 ± 0.001 B | 0.007 ± 0.001 A |
| Maltotriose | n.d. | 0.026 ± 0.000 B | 0.010 ± 0.000 A | 0.036 ± 0.001 D | n.d. | 0.046 ± 0.001 E | 0.034 ± 0.001 C |
| Acids a | |||||||
| Lactic | n.d. | n.d. | 2.822 ± 0.035 A | 3.306 ± 0.050 B | 11.743 ± 0.441 D | 5.929 ± 0.036 C | 12.498 ± 0.029 D |
| Aceticb | 0.573 ± 0.019 B | 3.980 ± 0.012 E | 5.523 ± 0.045 G | 1.681 ± 0.024 C | 0.428 ± 0.020 A | 2.060 ± 0.008 D | 4.272 ± 0.031 F |
| FODMAPs a,c | |||||||
| Excess Fructose d | 0.944 ± 0.035 B | 0.764 ± 0.004 B | 0.400 ± 0.004 A | 6.109 ± 0.112 D | 11.780 ± 0.127 E | 0.000 ± 0.000 | 5.648 ± 0.035 C |
| Sorbitol | 0.377 ± 0.017 E,F | 0.340 ± 0.002 F | 0.241 ± 0.001 A | 0.283 ± 0.002 D | 0.278 ± 0.002 C | 0.258 ± 0.002 B | 0.292 ± 0.002 E |
| Mannitol | 0.025 ± 0.000 A | 0.024 ± 0.000 A | 20.074 ± 0.065 D | n.d. | n.d. | 16.779 ± 0.292 C | 0.149 ± 0.001 B |
| Σ Polyols | 0.402 ± 0.003 B,C | 0.364 ± 0.000 B | 20.315 ± 0.066 E | 0.283 ± 0.002 A | 0.278 ± 0.002 A | 17.037 ± 0.291 D | 0.441 ± 0.003 C |
| Raffinose/Stachyose | 0.012 ± 0.000 A | 0.035 ± 0.000 B | n.d. | 0.101 ± 0.001 C | n.d. | n.d. | n.d. |
| Verbascose | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| Σ GOS e | 0.012 ± 0.000 A | 0.035 ± 0.000 B | - | 0.101 ± 0.001 C | - | - | - |
| Fructans | 1.711 ± 0.083 C | 1.786 ± 0.042 C,D | 0.895 ± 0.022 A | 1.277 ± 0.123 B | 1.277 ± 0.099 B | 1.925 ± 0.014 D | 1.090 ± 0.041 A,B |
| Properties | BR-UnF | BR- Ster | BR-TR116 | BR-MG1 | BR-FST2.11 | BR-R29 | BR-FST1.7 |
|---|---|---|---|---|---|---|---|
| Water-Binding Capacity (g H2O/100 g sample) | 500.29 ±11.02 C | 523.90 ± 25.78 C | 427.33 ± 17.20 A,B | 377.76 ± 18.61 A | 459.25 ± 39.09 B,C | 384.07 ± 23.88 A | 418.09 ± 25.25 A,B |
| Oil-Binding Capacity (g oil/100 g sample) | 237.20 ± 22.74 A | 367.93 ± 21.24 C | 409.39 ± 12.98 D | 287.87 ± 9.62 B | 305.61 ± 3.50 B | 319.90 ± 5.27 B | 290.23 ± 3.84 B |
| pH | 5.20 ± 0.01 D | 5.16 ± 0.01 C,D | 4.09 ± 0.02 B,C | 4.09 ± 0.01 B,C,D | 3.37 ± 0.01 A | 3.69 ± 0.01 A,B,C | 3.38 ± 0.01 A,B |
| Total Titratable Acidity (mL 0.1 M NaOH/g) | 4.5 ± 0.13 A | 6.08 ± 0.06 B | 10.55 ± 0.21 D | 8.42 ± 0.0.11 C | 16.82 ± 0.38 F | 17.01 ± 0.25 F | 14.31 ± 0.06 E |
| Colour (ΔE − BR-UnF) | - | 8.15 ± 0.77 B | 9.13 ± 0.57 B | 10.14 ± 1.15 B | 3.65 ± 0.47 A | 8.32 ± 0.85 B | 9.02 ± 0.25 B |
| Alpha amylase (cu/g d.m) | 5.120 ± 0.302 B | 0.026 ± 0.000 A | 0.000 ± 0.000 A | 0.025 ± 0.005 A | 0.030 ± 0.000 A | 0.025 ± 0.015 A | 0.041 ± 0.016 A |
| Beta amylase (cu/g d.m) | 0.772 ± 0.185 B | 0.010 ± 0.014 A | 0.000 ± 0.000 A | 0.006 ± 0.000 A | 0.124 ± 0.000 A | 0.045 ± 0.016 A | 0.035 ± 0.017 A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neylon, E.; Nyhan, L.; Zannini, E.; Monin, T.; Münch, S.; Sahin, A.W.; Arendt, E.K. Food Ingredients for the Future: In-Depth Analysis of the Effects of Lactic Acid Bacteria Fermentation on Spent Barley Rootlets. Fermentation 2023, 9, 78. https://doi.org/10.3390/fermentation9010078
Neylon E, Nyhan L, Zannini E, Monin T, Münch S, Sahin AW, Arendt EK. Food Ingredients for the Future: In-Depth Analysis of the Effects of Lactic Acid Bacteria Fermentation on Spent Barley Rootlets. Fermentation. 2023; 9(1):78. https://doi.org/10.3390/fermentation9010078
Chicago/Turabian StyleNeylon, Emma, Laura Nyhan, Emanuele Zannini, Thomas Monin, Steffen Münch, Aylin W. Sahin, and Elke K. Arendt. 2023. "Food Ingredients for the Future: In-Depth Analysis of the Effects of Lactic Acid Bacteria Fermentation on Spent Barley Rootlets" Fermentation 9, no. 1: 78. https://doi.org/10.3390/fermentation9010078
APA StyleNeylon, E., Nyhan, L., Zannini, E., Monin, T., Münch, S., Sahin, A. W., & Arendt, E. K. (2023). Food Ingredients for the Future: In-Depth Analysis of the Effects of Lactic Acid Bacteria Fermentation on Spent Barley Rootlets. Fermentation, 9(1), 78. https://doi.org/10.3390/fermentation9010078







