Abstract
Since milk whey is an abundant dairy by-product and a significant threat to the environment, its utilization is of great interest. The study compares valorization of lactose and lactates—the main carbon sources of whey—by fermentation—an environmentally friendly process. Antimicrobials released during fermentation by food-grade bacteria can help increase the microbiological safety of food. Propionic acid—a strong antimicrobial—is obtained mainly by the petrochemical route, yet there is increasing interest in its synthesis in biotechnological pathway. Five strains of propionic acid bacteria (Acidipropionibacterium acidipropionici, Propionibacterium cyclohexanicum, Propionibacterium freudenreichii, Acidipropionibacterium jensenii and Acidipropionibacterium thoenii) were investigated for their ability to produce organic acids and biomass using Na lactate or lactose as carbon sources. Selected fermentates were investigated for their antimicrobial efficacy during in vitro studies with foodborne pathogens: Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Bacillus subtilis and Staphylococcus aureus. Results confirm that the production of acids and biomass is considerably influenced by the added carbon source. The tested fermentates have strong and specific antimicrobial activity against Pseudomonas aeruginosa, Bacillus subtilis and Staphylococcus aureus. In addition, inhibition of Staphylococcus aureus and Klebsiella pneumonia depends on the activity of produced bacteriocins. The article also discusses the possibility of increasing the antimicrobial activity of fermentates by acidification.
1. Introduction
The dairy industry records a continuous increase in the amount of whey [1]. Due to whey and its permeates’ abundance and its high lactose content and chemical and biological oxygen demand, a significant threat to the environment is posed; therefore, its conversion into new products is of great interest [2,3,4,5]. Sweet whey permeate, containing lactose (~4.7%), is derived from the production of cheese or rennet casein; its pH is around 6–7. Acid or sour whey (pH < 5), containing higher levels of calcium, phosphorus and lactic acid, but a lower level of lactose (~3.8%), is a by-product of curd, acid casein and fresh cheese production [1,3]. Whey, as well as other similar dairy by-products (e.g., mozzarella stretching water generated from mozzarella cheese production), are inexpensive raw materials for biotechnological production of various nutraceuticals (e.g., bioactive lipids, B group vitamins, probiotics and bioactive peptides), antimicrobials (e.g., organic acids and bacteriocins) and other value-added products [1,2,3,4,5]. Food preservation by fermentation is widely used in traditional techniques to prevent the growth of undesirable microorganisms [6,7]. Nowadays, consumer interest and demand for naturally preserved food products are increasing [8,9]. Unfortunately, some microorganisms become resistant to many traditionally used food preservatives. In addition, the resistance of pathogens to antibiotics is a serious threat to modern health systems worldwide [9,10]. Antimicrobial compounds produced by food-grade microorganisms are a promising, sustainable alternative [9,11,12,13]. Many organic acids are powerful antimicrobial compounds. Propionic acid is one of the most important and commercially valuable volatile fatty acids which is utilized in many sectors such as medical, pharmaceutical, perfumes and detergents. Propionates are also used as preservatives in food, especially for bread protection against Bacillus mesentericus, and in animal feed production [10,14,15]. Propionic acid is the most common commercial grain preservative used as a fungal growth inhibitor and has been utilized for many years as a fungistat by directly adding it to various stored agricultural products [16]. Today, this valuable antimicrobial organic acid is obtained mainly by petrochemical route, as the low productivity of propionic acid and the cost of its recovery from various aqueous solutions and fermentation broths limit its commercialization [15,17,18,19]. However, there is increasing interest in propionic acid synthesis from alternative sources due to concerns over the sustainability of the crude oil supply and the environmental pressure caused by the petrochemical industry [20]. Organic acid production in the microbial pathway from agricultural and industrial wastes is an environmentally friendly and renewable process [21]. Propionic acid bacteria (PAB) are a group of bacteria fermenting cheese and producing propionic acid [17,22]. PAB are robust microorganisms with remarkable adaptability to technological and physiological stress conditions [12]. Traditionally, PAB have been used as one of the starter cultures for Swiss-type cheese to produce the characteristic flavor and eyes. Nowadays PAB have also found applications in agriculture and in the production of other food products as protective cultures and are prospective living organisms because of their probiotic potential [23,24,25,26]. According to genome sequence analyses, the family Propionibacteriaceae is divided into three genera: Propionibacterium spp., Acidipropionibacterium spp. and Cutibacterium spp. [26]. Two of them—Propionibacterium (P.) spp. and Acidipropionibacterium (A.) spp.—are dairy-related microorganisms, mainly inhabiting dairy (milk, cheese) and silage environments [24]. No references related to possible concerns for human or animal safety of P. freudenreichii and A. acidipropionici have been reported. Therefore, these microorganisms have been assigned QPS (Qualified Presumption of Safety) status by the European Food Safety Authority [26,27]. The metabolic activity of PAB enriches the final product with organic acids, vitamins (B2, B12, K and folate) and other nutrients, thus improving the stability and nutritional value of food products [24,25,26,28,29,30]. Due to the presence of specific extracellular metabolites, PAB fermentation products have antimicrobial activity. Among them, propionic and acetic acids are the main compounds. Some of the PAB strains also produce bacteriocins that inhibit growth of certain bacteria [24,31,32,33]. Bacteriocins produced by several Gram-positive microorganisms (including PAB bacteria) produce bacteriocins that differ from the original definition, as many of them have broader inhibitory spectra [26]. Apart from organic acids and bacteriocins, other PAB metabolites are known to have antibacterial or antifungal activity, such as diacetyl, acetoin, 2-pyrrolidine-5-carboxylic acid and 3-phenyl lactic acid [29]. Therefore, fermentation broths (hereinafter referred as ‘fermentates’) containing various compounds with antimicrobial properties can be used as complex biopreservatives against contamination by a wide range of microorganisms [13,17,22]. For example, the MicroGARD® brand biopreservatives are produced from skimmed milk powder, dextrose or wheat starch fermentates to target a broad spectrum of microorganisms [34]. The antimicrobial activity of this biopreservative is related to metabolites produced by P. freudenreichii subsp. shermanii [8]. Another product, Inhibit 3600 Dairy, obtained from whey cultured with P. freudenreichii, inhibits mold, some yeasts and Gram-negative bacteria [26,35]. Danilova et al. [36] found that the broth produced by P. freudenreichii Pr4 is effective against B. subtilis, B. cereus, P. putida and E. coli at neutral pH. Fermentation metabolites of PAB have also been used and tested in agriculture. For example, the treatment of corn seeds with the fermentate (the so-called ‘culture liquid’) of two PAB strains resulted in almost complete inhibition of bacteria, yeasts and molds when used in seed storage [37]. Nevertheless, researchers acknowledge that there is relatively little knowledge of the antimicrobial properties of PAB metabolites, indicating that additional research is needed to assess the potential application of bacteriocin-producing strains of PAB as food biopreservatives and probiotics [26]. In the first part of this study, we aimed to investigate selected PAB strains (A. acidipropionici, P. cyclohexanicum, P. freudenreichii, A. jensenii and A. thoenii) for organic acid and biomass production ability by fermenting lactose and lactate. In the second part of this research, we studied the antimicrobial efficacy of selected fermentates on the inhibition of pathogens: E. coli, K. pneumoniae, P. aeruginosa, B. subtilis and S. aureus, and we also tried to discover whether the inhibition of pathogens is mainly due to the effect of acids or if specific proteins (bacteriocins) are also involved.
2. Materials and Methods
2.1. Microbial Cultures
PAB bacterial strains used in the present study (listed in Table 1) were selected from the major dairy PAB species [26,28]. Cultures were obtained from the Leibniz Institute DSMZ (Braunschweig, Germany) culture collection (German Collection of Microorganisms and Cell Cultures, GmbH).

Table 1.
Microorganisms used in the study.
2.2. Maintenance and Propagation of Microbial Cultures
PAB cultures were propagated according to the Antone et al. [18] study with small modifications. PAB were cultivated on sodium lactate agar plates (containing grams per liter: casein peptone tryptic digest 10, yeast extract 5, sodium lactate 5 and agar 15) as suggested by DSMZ culture collection [38] and incubated anaerobically for 14 days at 30 ± 0.5 °C using an anaerostat and anhydrous anaerobic gas generator (Oxoid AN0035, Hampshire, UK) bags. The PAB cultures were subcultured three times before inoculation. One colony of each strain was transferred from a sodium lactate agar plate to a sterile Falcon tube containing 50 mL of fresh medium and incubated for 2 days at 30 ± 0.5 °C with gentle agitation. Cells were incubated in a semi-synthetic medium with the following composition (per L): sodium DL-lactate 0.18 M, yeast extract 6.06 g, diammonium hydrogen phosphate 2 g, potassium dihydrogen phosphate 1 g, magnesium sulfate (heptahydrate) 0.01 g, calcium chloride (dihydrate) 0.01 g, cobalt(II) chloride (hexahydrate) 0.01 g, iron(II) sulfate (heptahydrate) 0.005 g and manganese(II) sulfate (monohydrate) 0.0025 g. Salts were added according to the method of Coral [22]. Microbial cells from the pre-culture were harvested by centrifugation (10 min at 4629× g), washed twice with sterile water and resuspended in an appropriate amount of water before inoculation so that the biomass optical density (OD) at 600 nm (OD600) was 0.05 absorbance units (AU) at the start of the experiment. OD was determined in a 1 cm cuvette with spectrophotometer Halo XB-10 (Dynamica, Mablethorpe, UK).
Propagation of pathogenic microorganisms for pre-culture was carried out in liquid LB broth (containing grams per L: peptone 10, yeast extract 5 and sodium chloride 0.5) at 30 ± 1 °C with continuous shaking (180 rpm). Overnight culture cell suspensions were washed twice with sterile water and standardized to ensure a suspension density of OD600 0.1 per well at the start of the experiment (T0).
2.3. Fermentation Experiment—Media and Culture Conditions
Fermentation was carried out with PAB strains A, C, E, J and T in the 100 mL glass flasks. For fermentation, 2 types of growth media were used containing lactose or Na lactate with a similar molar concentration (0.18 M), as well as minerals and yeast extract at the same concentrations as in the medium for preparation of inoculums. Flasks were filled up to 100 mL total volume of broth, sealed with sterile cotton plugs and covered with Al foil to reduce media evaporation. Blank samples without added bacteria were studied as a negative control (O). Flasks were placed at 30 ± 1 °C temperature and incubated without agitation for 210 h. At the end of the experiment, pH and OD of the fermentate were measured. Supernatant samples were harvested by centrifugation for 20 min at 4 °C and 21,420× g and kept at −18 °C until further analysis of metabolites.
2.4. Fermentation Metabolites, pH and Biomass Analyses
The content of organic acids and lactose in culture supernatants was determined by HPLC using the chromatograph Agilent 1100 (Agilent Technologies, Santa Clara, CA, USA). Samples were pre-treated according to the method of [39] with modifications: 0.5 mL of the supernatant was mixed with 1 mL of 0.02 N H2SO4, and then samples were vortexed for 20 s and frozen for at least 2 h for protein precipitation at −18 ± 1 °C temperature. Before HPLC analysis, samples were melted, and 0.5 mL of deionized water was added; then samples were centrifuged at 4 °C 21,420× g for 10 min, and the supernatants were filtered through 0.22 µm pore-sized nylon filters. HPLC analyses were performed as follows: acetic, propionic, pyruvic, formic and fumaric acids were determined using a Supelcogel C-610H (Sigma Aldrich, Burlington, MA, USA) column (30 cm × 7.8 mm ID) using a diode array detector with UV 210 nm. 10 µL of the sample was injected into the column whose temperature was 30 °C; mobile phase 0.1% H3PO4 (m/v); flow rate 0.5 mL·min−1. Lactic and succinic acids were determined on a Shodex SH1011 (Yokohama, Japan) column (300 mm × ID 8.0 mm), preceded by a corresponding guard column SH-G (50 mm × 6 mm ID), column temperature 50 °C; an Agilent diode array UV detector at a wavelength of 210 nm was used. The mobile phase was 0.005 M H2SO4 with a flow rate of 0.6 mL·min−1; the amount of sample injected into the column was 5 µL. Lactose was determined by Shodex column SP 0810 (ID 8 mm, length 300 mm), preceded by a corresponding guard column SP-G (50 mm × 6 mm ID), column temperature 80 °C. The amount of sample injected into the column was 5 µL. The mobile phase was deionized water with a flow rate of 0.5 mL·min−1; a refractive index detector was used. The peaks of analytes were identified by the retention times of the standards of each analyte. Peak area was used to quantify metabolites against standards. pH was measured by a pH meter Jenway 3520 (Barloworld Scientific Ltd., Staffordshire, UK) on the day of inoculation and at the end of fermentation. Biomass was determined from a standard curve of absorbance (OD) versus dry weight (1 unit of absorbance (OD600) = 0.19 g·L−1 dry weight). OD 600 was determined using a spectrophotometer Halo XB-10 (Dynamica, UK). Dry weight was determined by centrifugation of biomass, washing twice with distilled water, and drying 10 mL of suspension in an oven at 104°C for 16 h according to the previously published method [40]. All chemicals were of analytical or HPLC purity. Reagents, salts, acids and media components were purchased from Sigma Aldrich (USA), except for yeast extract and agar (Biolife, Monza, Italy). Sodium lactate was purchased as 60% (by mass) synthetic syrup suitable for cell culture. Proteinase K enzyme (activity > 600 U·mL−1) was obtained from Thermo Fisher Scientific (Waltham, MA, USA) and used in the reaction mixture where the enzyme concentration was 0.2 mg·mL−1.
2.5. Acid and Biomass Outcomes Calculations
Theoretical titers of produced acids from lactate were calculated using the following equation:
where:
Tacid theor.—the theoretical titer of acid produced (g·L−1);
W—amount of lactate consumed (g);
MW—molar mass (g·mol−1); lactic acid was used in the calculations;
C—coefficient = 2/3 = 0.667 for propionic acid and 1/3 = 0.333 for acetic acid according to literature (Equation (7)).
Theoretical titers of produced acids from lactose were calculated using the following equation:
where:
Tacid theor.—the theoretical titer of acid (g·L−1);
W—amount of lactose consumed (g);
MW—molar mass (g·mol−1);
C—coefficient = 4/3 = 1.333 for propionic acid and 2/3 = 0.667 for acetic acid according to literature (see Equation (8)).
The substrate conversion efficiency was calculated according to the formula:
where:
SCE—substrate conversion efficiency (%);
Tacid observ.—acid titer observed (g·L−1);
Tacid theor.—the theoretical titer of acid (g·L−1).
The acid yield was calculated according to the formula:
where:
Yacid—acid yield (g·g−1);
Tacid observ.—acid titer observed (g·L−1);
Sconsumed—consumed substrate (lactose or lactate) (g·L−1).
The biomass yield was calculated according to the formula:
where:
Ybiomass—biomass yield (g·g−1);
Bobserv.—biomass produced (g·L−1);
Sconsumed—consumed substrate (lactose or lactate) (g·L−1).
2.6. Preparation of Fermentates for the in Vitro Antimicrobial Effects Studies
Fermentates were prepared as follows: supernatants of fermentation broths (thawed 0.5 h at room temperature) were mixed from 3 biological replicates in equal proportions, centrifuged at 21,420× g for 10 min (20 ± 1 °C) and sterilized through 0.22 µm nylon filters. Further, the following filter-sterilized types of fermentates were used in studies of antimicrobial effects: (a) not treated with proteinase—samples labeled as ‘FS’ and (b) treated with proteinase at 50 °C for 20 min, followed by enzyme inactivation at 70 °C for 5 min—the corresponding samples were labeled as ‘FSP’.
2.7. Determination of the Antimicrobial Effects of Fermentates
The antimicrobial effects of the obtained PAB fermentates were tested utilizing a growth inhibition assay in 96-well microplates. The total working volume of each well was 200 µL. The following components were pipetted into each well: 60 µL of 3.3-fold concentrated LB medium, 120 µL of particular fermentate (or sterile water for wells without inhibitors) and 20 µL of a specific foodborne pathogen microbial cell suspension. The microplates were aseptically incubated at 30 ± 0.5 °C for at least 12 h. Biomass growth kinetics were monitored as OD measurements at 600 nm (OD600): (i) every 1 h using a 96-well plate reader (DynaRead, Dynex Technologies, Buštěhrad, Czech Republic) for E. coli, K. pneumoniae and P. aeruginosa or (ii) every 10 min using a 96-well multimode reader (Infinite M200 PRO, TECAN, Männedorf, Switzerland) for B. subtilis and S. aureus (see Table 1).
Pathogen survival rate was calculated based on OD measurements after 10 h of incubation, as a percentage of the optical density of the suspension with added fermentate relative to the optical density of the suspension without added fermentate. The antimicrobial (inhibitory) effect was evaluated according to the authors’ set scale as follows: 0–30% of surviving cells—strong, 31–60% of surviving cells—moderate and 61–100% of surviving cells—weak inhibition.
The ratio of the undissociated acid concentration at a given pH was calculated as in [40] according to the Henderson–Hasselbalch equation:
where:
[A−]—concentration of the dissociated acid;
[HA]—concentration of the undissociated acid;
pKa—pKa value of the organic acid (negative base-10 logarithm of the acid dissociation constant);
pH—pH value of the solution (media).
2.8. Determination of Minimum Inhibitory Concentrations (MICs) of Organic Acids
The MIC is defined as the lowest concentration of an antimicrobial agent that inhibits the visible growth of a microorganism after overnight incubation [41]. Propagation of pathogens was performed as described previously, except that cell suspensions from overnight cultures were standardized to give an OD600 of 0.05 per well at T0. The range of acid concentrations for the MIC assays were chosen based on previous experiments and included the following (in the mixture of test medium):
- Lactic and formic acids: 4, 6, 8, 10, 12, 20 mM;
- Propionic and acetic acids: 3, 4, 6, 8, 10, 12 mM;
- Blank samples without added acids (with added sterile water) were studied as a negative control to determine the normal growth kinetics of microorganisms without acid inhibition.
The effect of acids on microbial cultures was tested in a 96-well format. The total working volume of each well was 200 µL consisting of: 160 µL of 1.25-fold concentrated LB medium, 20 µL of 10-fold concentrated acid or water if the medium was prepared without inhibitors and 20 µL of the pathogen cell suspension. The microplate was covered and aseptically incubated at 30 ± 0.5 °C. Growth kinetics were monitored by optical absorbance at 600 nm (OD600) using a 96-well multimode reader (Infinite M200 PRO, TECAN, Switzerland) taking measurements every 10 min for a total of 18 h.
The MIC determination was performed in the following steps: (1) The percentage of survivors was calculated from the results of OD600 measurements at the 10 h incubation time point, comparing suspensions with added acids to the suspensions without acid (i.e., positive controls). (2) An approximation plot of survival as a function of acid concentration was created. The MIC was defined as the intersection coordinate with the X-axis (0% survival) of the approximation graph.
2.9. Statistical Analyzes
The statistical analyses were performed using SPSS 26 (IBM Corporation, Chicago, IL, USA) and Microsoft Office Excel (Redmond, WA, USA) (2007). A two-tailed, paired Student’s t-test was used to test the difference between the two independent groups. To test for a difference between more than two independent groups, one-way ANOVA with Tukey’s post hoc was performed. All assays were performed in at least three trials, and the results presented are mean ± standard deviation (S.D.) unless otherwise stated. Differences were considered statistically significant at p < 0.05.
3. Results
3.1. Results of Propionic Acid Fermentation Study—Substrates and Bacterial Strains
To compare the potential of selected strains of PAB to metabolize milk by-products, we tested their fermentation performance in a ‘model medium’ with lactose or lactate as the main carbon sources thus mimicking different types of whey (sweet or acid, respectively). The selected PAB strains are known for their significant propionic acid production potential [24,26]. We measured organic acid and biomass production and assessed the medium pH. Results of the produced acids, pH, as well as propionic and acetic acid ratios (P/A ratios) are summarised in Table 2, but results of the biomass production are shown in Figure 1.

Table 2.
The concentration of organic acids and pH of propionic acid bacterial fermentates.
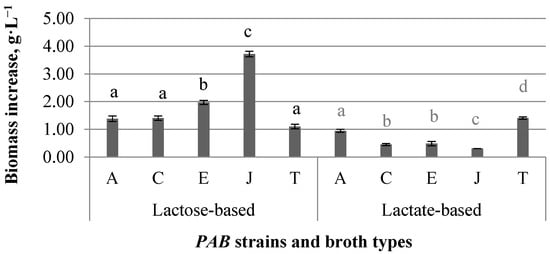
Figure 1.
Biomass produced by different PAB strains in broths with added lactose or lactate as carbon sources, data are averaged from at least three independent, parallel 210 h-long cultivations; error bars represent the standard deviation of biomass increase of at least three independent fermentations (n = 3); a, b, c, d—different small letters on bars show that biomass produced by PAB strains grown on the same substrate are different (p < 0.05).
As expected, propionic acid was the main metabolite of PAB regardless of the broth carbon source. Acetic and succinic acids were the main by-products, which is in agreement with previous research elsewhere [12,19]. Lactic acid was produced only by strains C and T in lactose broth. The amount of fumaric and formic acid detected in both-type fermentates was less than 0.01 g·L−1 and 0.04 g·L−1 (results not shown). The type of substrate significantly influenced the concentrations of acids, as well as pH and biomass production. Significantly more propionic (p < 0.05) and acetic (p < 0.05) acids were produced in lactate broths. The final pH of lactate fermentates compared to lactose-based was significantly higher (6.49 ± 0.04 vs. 4.16 ± 0.21, resp., p < 0.05).
When PAB were cultivated in Na lactate broth, the lactate was practically depleted. The concentration of propionic and acetic acids was around 7.2–8.6 g·L−1 and 3.3–3.9 g·L−1, accordingly. Amounts of acids produced in lactate broths differed across the strains (Table 2). Regarding the main metabolite—propionic acid—concentration in fermentates, the highest outcome was reached by PAB strains A, C, J and T being similar (p ≥ 0.05); at the same time, for strain E, it was significantly lower (p < 0.05) compared to strains A and T. The highest acetic acid outcome was reached by strains A, C and T. The highest succinic acid concentration of 1.34 g·L−1 was determined in the fermentate of strain T—it was significantly higher (p < 0.05) than all other strains that were producing succinic acid at quantities less than 0.5 g·L−1. The P/A ratio in most lactate broth fermentates was slightly above 2 regardless of the PAB strain used (i.e., around 2.2). The pH of the broths at the end of fermentation was around 6.4–6.6, being close to the neutral. As the initial pH of the broths was ~6.2, no acidification had been taking place there.
When PAB were grown in broth with added lactose, the amount of propionic acid produced was around 3.5–4.3 g L−1; the acetic acid concentration was 1.4–2.2 g·L−1. Amounts of acids produced in lactose broths differed across the strains. The P/A ratio in lactose-derived fermentates had a larger amplitude compared to lactate-derived. It was significantly higher (p < 0.05) for PAB strains A, C and T (i.e., 2.3–2.6) than strains E and J (~1.95). The pH of the broths at the end of fermentation was around 3.8–4.4. The significantly lower compared to other strains’ pH (p < 0.05) was reached by strains C and T (pH 3.84 ± 0.02 and 4.06 ± 0.01, respectively); the pH differences were significant also between both strains C and T (p < 0.05). Both mentioned strains (C and T) also produced lactic acid (2.40 ± 0.12 and 0.17 ± 0.02 g·L−1, resp.) in lactose-based broth.
Biomass growth was observed in both carbon source substrate mediums (Figure 1). For all PAB strains, except strain T, it was significantly higher (p < 0.05) when the carbon source was lactose. For the strain T, it was similar in both mediums (p ≥ 0.05). Average biomass increase comparing biomass concentration at inoculation and the end of the experiment was around 200-fold in lactose, while 75-fold in the lactate medium; therefore, lactose is better as a carbon source for biomass production. Also biomass yield was higher in lactose-based broths and was strain-dependent (Table 3). In the lactate medium, strain T had the highest (p < 0.001) biomass yield, while strain J had the lowest (p < 0.05). In lactose-supplemented broth, the highest outcome of biomass was produced by strain J (p < 0.05) but the lowest was produced by strains A, C and T (p < 0.05). We calculated the quantity of consumed substrates, outcomes of propionic and acetic acids, as well as substrate conversion efficiencies. Results are shown in Table 3.

Table 3.
Substrate consumption, conversion efficiency and outcomes of main acids and biomass by different PAB strains cultivated in lactose- or lactate-based broths.
Results show that the amount of lactose consumed was 8.6–12.4 g·L−1 or 12.6-20.0% of the initial lactose amount; significant differences were observed between strains C and E (p < 0.05). Lactate conversion efficiency to propionic acid was 82–99%; it was strain-dependent (p < 0.05). Lactose conversion efficiency to propionic acid was lower (64–84%), being similar for all strains (p ≥ 0.05) (Table 3).
3.2. Results of Propionic Acid Bacteria Antimicrobial Effects Study
The PAB fermentates used in the inhibition test (lactate fermentates of PAB species E, T and A) were selected based on our previous experiments, considering the lactate fermentates with the most promising results (results not published). To investigate whether bacteriocins may be involved in the inhibition of pathogens, the antimicrobial activity of proteinase K-treated fermentates was tested. In addition, minimum inhibitory concentrations of pure organic acids—lactic, propionic, acetic and formic acid—against the aforementioned foodborne pathogens were determined. This experiment aimed to find out the effect of PAB fermentates on the inhibition of pathogens and also to find out whether the inhibition is mainly due to the effect of acids or specific proteins (bacteriocins) are involved. According to the information available in the literature, many bacteriocins are thermally stable [11,23]; therefore, proteinase K digestion was performed to eliminate their possible activity. We hypothesized that if proteinase treatment reduces the inhibitory effect of the tested product, we can assume that proteins (most likely bacteriocins) contribute to the inhibition. The antimicrobial effects of the fermentates produced by PAB were tested.
3.2.1. Pathogen Growth Dynamics throughout the Total Incubation Period (12 h)
Pathogen growth dynamics reflecting the antimicrobial effects of PAB fermentates are shown in Figure 2.
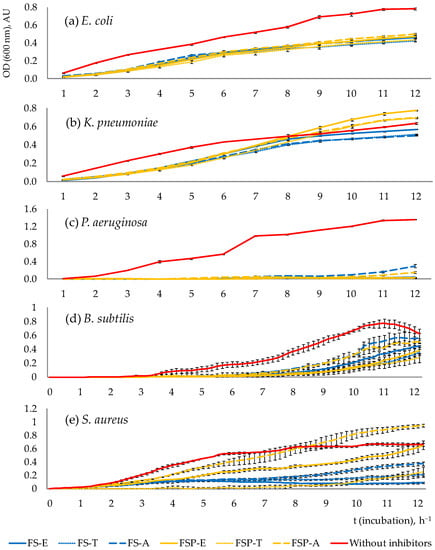
Figure 2.
Growth dynamics of E. coli (a), K. pneumoniae (b), P. aeruginosa (c), B. subtilis (d) and S. aureus (e) in a 12 h inhibition test. Error bars represent the standard deviation of optical density at 600 nm of at least three independent wells (n = 3); AU- absorbance units; FS-E, FS-T, FS-A—filter-sterilised fermentates of PAB strains Propionibacterium freudenreichii (E), Acidipropionibacterium thoenii (T) and Acidipropionibacterium acidipropionici (A); FSP-E, FSP-T, FSP-A—filter-sterilised and treated with proteinase K fermentates obtained from the respective PAB strains.
The results show that the growth of pathogens in the presence of fermentates was weakened in all cases. Inhibition, which was observed with samples with added proteinase-treated fermentates, indicates that the inhibition was assured mostly by acids. E. coli, P. aeruginosa and B. subtilis were inhibited by both types of fermentates—untreated (FS-E,T,A) and treated with proteinase (FSP-E,T,A) throughout the total incubation period (12 h). K. pneumoniae was inhibited by both types of fermentates during the first 7–8 h and then only by proteinase-untreated fermentates (FS-E,T,A). S. aureus was inhibited by all fermentates practically throughout the 12 h period, except for the fermentate FSP-A, which inhibited its growth only in the first 7 h.
Interestingly, in some samples with added FSP-type fermentates, a pathogen growth-promoting effect was observed (compared to samples without added fermentates and to samples with FS-fermentates)—after the first hours of inhibition, some FSP fermentates treated with proteinase stimulated the growth of pathogens (see Figure 2b,e for K. pneumoniae and S. aureus). The more intensive growth of pathogens in these samples compared to other samples could be explained by the consumption of released amino acids from proteinase-cleaved proteins (bacteriocins).
3.2.2. The Survival Rate of Pathogens after 10 h of Incubation
The inhibitory effects were evaluated based on the calculated survival rate of pathogenic microorganisms after 10 h of incubation while the exponential phase of growth was still observed. The results are summarized in Table 4.

Table 4.
Antimicrobial effects of PAB fermentates against B. subtilis, E. coli, K. pneumoniae, P. aeruginosa and S. aureus after 10 h incubation.
The antimicrobial efficacy of proteinase K-untreated (‘natural’) fermentates varied depending on the pathogen. The strongest antimicrobial effect of PAB fermentates after 10 h incubation was against P. aeruginosa (whose survival rate was only 2–8%). Less pronounced, still on a rating scale a strong or moderate inhibitory efficiency of PAB fermentates was observed against E. coli, B. subtilis and S. aureus—pathogen survival rate was 52–58%, 21–48% and 11–45%, respectively. The weakest inhibitory efficacy of PAB fermentates was observed against K. pneumoniae (survival rate 84–94%). When different PAB strains were compared, none of them was a distinct leader in all tested pathogens’ inhibition. All PAB strains showed a strong inhibitory effect against P. aeruginosa, a moderate effect against E. coli and weak inhibition of K. pneumonia. Compared to other pathogen inhibition, the inhibition of B. subtilis and S. aureus was more strain-dependent: against B. subtilis the strongest efficacy (less than 30% survivors after 10 h of incubation) was shown by PAB strain T, and against S. aureus by PAB strains E and T. Overall, the weakest (p < 0.05) inhibitory effect was observed for PAB strain A (which can be seen in the inhibition of such microorganisms as P. aeruginosa, B. subtilis and S. aureus). Differences in the inhibitory effect of the tested PAB strains are shown in the supplementary material (Figure S1).
3.2.3. Antimicrobial Compounds Involved in the Inhibition of Pathogenic Microorganisms
Results show that for P. aeruginosa and B. subtilis inhibition is likely due to acids but not due to protein antimicrobials; the inhibition of E. coli, in general, is also likely due to acids; however, in the case of PAB strain A fermentate, a protein cleaved by the proteinase K could also be involved in the inhibition, but its role could be rather insignificant because calculated difference in survival rates of pathogens (FSP minus FS) is only 4.89% (Table 4). The protein that is cleaved by the proteinase (bacteriocin/-s) is likely to play a significant role in the inhibition of K. pneumoniae and also a major role in the inhibition of S. aureus, whose survival rate differences (FSP minus FS) were 24–27% and 59–83%, accordingly.
3.2.4. Antimicrobial Effects of Pure Organic Acid Exposure
In the next phase of the study, the minimum inhibitory concentrations (MICs) of the pure acids (lactic, propionic, acetic and formic) against the same pathogen species were determined to compare the antimicrobial effects of pure acids and fermentates. Propionic acid and acetic acid were selected as the main PAB metabolic products, while lactic and formic acids were chosen as one of the most popular antimicrobial acids used in the food industry and agriculture. The MICs of organic acids inhibiting E. coli, K. pneumoniae, P. aeruginosa, B. subtilis and S. aureus are summarized in Table 5.

Table 5.
The minimum inhibitory concentrations (MIC) of organic acids against the tested pathogens.
MIC concentrations of the tested acids ranged from 3.6–14.0 mM·L−1. B. subtilis was the most sensitive, but S. aureus was the most resistant microorganism to acid exposure. When the inhibitory effects of individual acids were compared, the lactic acid in most cases was significantly weaker. The results show that against E. coli the strongest (p < 0.05) was formic acid, followed by propionic and acetic acids with similar MICs (p > 0.05), but lactic acid was significantly weaker (p < 0.05). Inhibitory capacity (MIC) of propionic, acetic and formic acids against K. pneumoniae was similar, but lactic acid was significantly weaker (p < 0.05). The same was observed regarding P. aeruginosa and S. aureus. Against B. subtilis, propionic acid had the strongest inhibitory effect (p < 0.05), followed by acetic acid, but formic and lactic acids had the weakest effect. Summarizing the above, the antimicrobial efficacy of organic acids against the tested pathogens can be ranked in the following order:
- E. coli: formic > propionic = acetic > lactic;
- K. pneumonia, P. aeruginosa, S. aureus: propionic = acetic = formic > lactic;
- B. subtilis: propionic > acetic > lactic = formic.
Analyzing the antimicrobial effects of pure organic acid exposure, it should be noted that the effectiveness of organic acids in the inhibition of the growth of microorganisms depends on their dissociation properties represented by the acid dissociation constant (pKa). pKa value describes the pH value at which the acid is available 50% in its dissociated and undissociated form, respectively. The theoretical pKa values of the tested acids can be arranged in the following order: propionic acid (4.87), acetic acid (4.76), lactic acid (3.86) and formic acid (3.75) [42]. Antimicrobial effect by weak acids can be reached by rapid diffusion of undissociated molecules through the cell plasma membrane; dissociation of these molecules within cells liberates protons, thus acidifying the cytoplasm and preventing growth [43,44]. As the pKa value of an organic acid is responsible for its inhibiting effects on microorganisms, the inhibitory potential of acids should follow the same order (from the highest to the lowest: propionic, acetic, lactic and formic acid). At the same time, the actual pH of the fermentates determines the concentration of undissociated acid potentially harmful to microorganisms. The results of broth pH (from the experiment of MICs determination) depending on the concentration of the corresponding acid are shown in Figure 3a. The actual concentration of the undissociated acid might differ from the total acid concentration within the solution. We calculated concentrations of undissociated acids (HA) using the Henderson–Hasselbalch equation; the results are presented in Figure 3b.
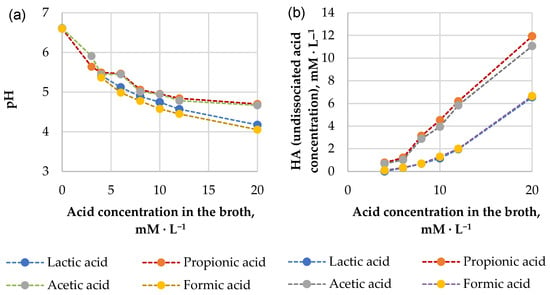
Figure 3.
The relationship between the molar concentration of the acid and the pH of the medium (a) and undissociated acid concentration (b) in broth from the experiment of MICs determination.
It can be seen (Figure 3b) that the concentration of undissociated formic acid and lactic acid in the broths was about two times lower than the concentration of undissociated propionic acid and acetic acid. This aspect might explain why lactic acid at an equivalent pH is less effective when compared with acetic and propionic acids as antimicrobials against tested microorganisms. It can be seen that the HA concentration of formic acid in the pH range from 4 to 6.5 (Figure 3a,b) is also relatively low, but unlike lactic acid, it inhibits the growth of the tested pathogens (except B. subtilis) no less strongly than the active antimicrobial agents (acetic acid and propionic acid). We hypothesize that its antimicrobial effect could be related to its small molecular size, which allows it to diffuse across biological membranes more quickly compared to propionic acid or acetic acid [45]. Therefore, the presence of acids other than lactic acid (e.g., propionic, acetic, formic) increases the preservative effect of the culture that has been described earlier [46].
We compared the inhibitory effects of fermentates and pure acids. Although the acid concentrations in fermentates are many times higher (ca. 104–116 mM propionic acid and 61–64 mM acetic acid) than their corresponding MIC concentrations (4 to 12 mM·L−1), the inhibitory effects of the fermentates can be considered as relatively weak. This discrepancy, as concluded above, seems to be related to the existent concentration of the undissociated acetic or propionic acid within fermentates. It has been known previously that weak-acid preservatives appear to share a common mode of action; despite their various chemical structures, they all become increasingly potent as antimicrobial agents at more acidic pH values [43,44]. The initial pH of the growth media (after mixing the ingredients to start the inhibition test) was 6.8–7.0 for FS-type fermentates and 6.5–6.7 for FSP-type fermentates, and the pH in the growth environment was close to neutral. Using the Henderson–Hasselbalch equation (Equation (6)), we calculated the undissociated acid (HA) concentration of propionic and acetic acid of the given fermentates (e.g., pH 6.5) and the desirable pH to increase antimicrobial properties of fermentate (Table 6).

Table 6.
The concentration of undissociated acids in the fermentates depending on pH.
Obviously, HA concentration was well below MIC concentrations (see MIC concentrations in Table 5 for comparison). Therefore, the reason why the antimicrobial effect of fermentates was weaker than could be expected (knowing their acid concentrations) is related to its relatively high pH (pH = 6.5–6.7), which did not promote protonation of the acids. When we model acidification of the fermentates with the same propionic and acetic acid concentrations to pH 5.5, undissociated form concentrations of acetic and propionic acids increase above their respective MIC concentration, and the antimicrobial properties of the product improve (Table 6).
4. Discussion
4.1. Assessment of the Results of the Fermentation Study
Our results show that both lactose and lactate can be used as the substrate for propionic acid fermentation, confirming literature data [47,48]; however, the fermentative production of propionic acid and acetic acid was considerably higher when lactate was added as the carbon source. As previously described, the fermentation metabolism of PAB differs depending on the C source used. When sugar is available, PAB use the EMP pathway. Fermentation of 3 moles of glucose produces 4 moles of propionic acid, 2 moles of acetic acid, 2 moles of CO2 and 12 moles of ATP. When lactate is the initial substrate, fermentation results in the production of 2 moles of propionic acid, 1 mole of acetic acid, and 1 mole of CO2. This process produces 1 mole of ATP per nine carbons, and therefore propionic acid bacteria usually grow very slowly [47]. Proposed metabolism of pyruvate in P. freudenreichii by Turgay et al. [26] is shown in Figure 4.
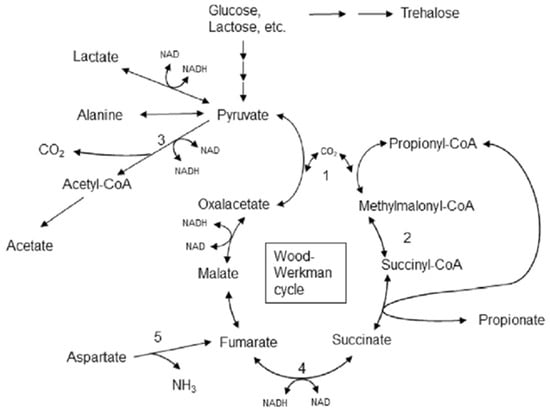
Figure 4.
Lactose and lactate fermentation by propionic acid bacteria ([26], distributed under a Creative Commons Attribution) (1—Methylmalonyl-CoA carboxytransferase, 2—Methylmalonyl-CoA mutase, 3—Pyruvate dehydrogenase, 4—Fumarate reductase, 5—Aspartate ammonia-lyase).
The following equations for the fermentation can be written for the production of the general metabolites via the dicarboxylic acid pathway from lactate (Equation (7)) and lactose (Equation (8)) [47,49]:
3 CH3CHOHCOOH (lactate) → 2CH3CH2COOH (propionate) + 1 CH3COOH (acetate) + 1 CO2 + 1 H2O
3 C12H22O11 (lactose) → 3 C6H12O6 (glucose/galactose) → 4 CH3CH2COOH (propionate) + 2 CH3COOH (acetate) + 2 CO2 + 2 H2O
Equations (7) and (8) show that if the initial lactose and Na lactate molar concentrations are equivalent, amounts of acids produced in the lactose medium theoretically should be higher than in the lactate medium. Initial lactose or lactate molar concentrations were 0.18 mol·L−1; therefore, from this amount of lactose, 0.24 M of propionate should be obtained (separately from both glucose and galactose) but from lactate only 0.12 M of propionate. Nevertheless, results of the present study show that if lactose is utilized as a substrate, concentrations of the resulting acids are significantly lower (p < 0.05)—the concentration of propionate produced in the lactose medium is approximately 2-fold lower than in the Na lactate medium. This low acid production in the lactose-based medium might be explained by the medium acidification, which makes propionic fermentation self-inhibitory [14,50], which therefore inhibited lactose consumption. Indeed, as reviewed by Ranaei et al. [21], several factors influence the propionic acid concentration and productivity (culture conditions, the type of bioreactor scale, temperature), but pH is one of the most important. PAB growth is inhibited both by protons and organic acid anions, as well as by undissociated propionic and acetic acids [37]. Our results show that in lactate-supplemented media acids were produced without the decrease in pH. This could be explained by the buffering properties of Na lactate. This is in line with previous observations that growing PAB in a lactate environment provides a fairly uniform pH [37,48]. Literature also indicates that lactate as a substrate is utilized by PAB much more rapidly than lactose-derived glucose [51]. As reviewed by Ranaei et al. [21], if pH is kept constant, propionic acid yields from lactate are higher than from glucose or lactose; moreover, by-product (succinic acid) production is reduced. Lewis and Yang [48] also observed that compared to fermentations with lactose or glucose at the same pH, lactate gave a higher propionic acid yield, lower biomass yield and lower specific growth rate.
Studies show that the optimal pH for PAB growth is 4.6–5.8; below pH 4.5 growth is stopped [52]. There are various physiological and genetic resistance mechanisms in microorganisms to survive in an acidic environment, including pH homeostasis, alteration of cell membranes, regulation of metabolism and repair of macromolecules [50]. It has been observed that the most sensitive to unfavorable factors (low pH, high concentration of carbon sources and presence of the end products of metabolism) are cells at the beginning of the exponential phase. At the same time, PAB are able to regulate pH by shifting it in the direction favorable for growth, if the medium pH in the chemostat is in the range of 4.4–7.45 [53]. In lactose-based broth, the lowest pH was reached by strains C and T, which also produced lactic acid; therefore, their physiology could be different from other PAB strains. This feature has been observed previously by other researchers reporting that some species of PAB produce lactate from glucose, xylose and arabinose; the quantity of lactate varies as a function of pH, medium composition, conditions of aeration, nature of the strain and other conditions (reviewed by [37]). The Kusano et al. [54] study results indicate that PAB C strain TA-12T, which was isolated from spoiled orange juice, can grow even at pH 3.2 and produces lactic, propionic and acetic acids as a result of glucose fermentation in a molar ratio of 5:4:2, respectively. Results of lactose-based fermentates also show that the strain C also produced significantly more succinic and lactic acid than other strains, but strain T produced more pyruvic acid than other strains (p < 0.05), which can explain significantly lower pH of their fermentates (p < 0.05). As far as organic acid-producing micro-organisms are concerned, a high acid tolerance of industrial strains is particularly necessary being one of the most important criteria for the selection of strains. As concluded previously, the major explanation for the low consumption of lactose is the inhibition of bacteria due to environmental acidification; however, other factors, e.g., strain-dependent properties, could influence the consumption as well. Indeed, the obtained results show that lactose consumption and substrate conversion efficiency are strain-dependent (see Table 3), which is in line with the findings of other studies [37,55]. Improving fermentation yield is the most critical step to achieving economically viable fermentation [15]. The yield of acids can be influenced not only by strain peculiarities or physical factors such as pH but also by temperature and the composition of the growth medium. The lactose metabolism of PAB is influenced by β-galactosidase. Some cations such as Mg2−, Mn2+, Li+, Na+ and K+ acted as stimulators of β-galactosidase activity, whereas, for example, Ca2+ showed an inhibitory effect [56]. Regarding the proportions of produced acids, a higher concentration of propionic acid often is considered an advantage. The ratio of P/A acids demonstrates overall fermentation stoichiometry. By estimating this ratio, we can see if fermentation follows reaction stoichiometry as it is set in Equations (7) and (8). Indeed, fermentation results (Table 2) demonstrate that the P/A ratio was around or slightly above 2 being close to the theoretical (=2) [47,57]. According to the literature data, the P/A ratio tends to vary depending on various circumstances [58,59] including previously mentioned conditions and metabolic characteristics of PAB strains. The increase in CO2 tension can lead to a higher P/A ratio in the fermentation of glucose that can explain the widely varying P/A ratios reported in the literature because the experiments of other investigators have been carried out under varying conditions of pH and with different types of buffers [58]. The presence of the TCA cycle in PAB is another factor that could explain differences in amounts and relative proportions of propionate, acetate and CO2 produced—in the presence of O2 and limiting lactate concentrations, part of the lactate is completely oxidized to CO2 via the TCA cycle [57]. The acidity level of the environment also affects the yields and proportions of acids. Our previous studies [18] and the literature [57] indicate that the formation of propionate is promoted at lower pH, thus increasing the P/A ratio. Differences in P/A ratios may also be due to altered amino acid metabolism of some strains—besides the main propionic acid fermentation pathway, PAB can metabolize aspartate in the presence of lactate with various intermediate stages; as a result, most of the lactate is fermented into acetate and CO2 rather than propionate [55]. Furthermore, production of other acids (succinic, lactic, pyruvic), as well as the growth of biomass, can explain the lower than theoretically possible propionic acid outcome in the present study [26,60,61]. Our results of the biomass yield are in line with literature information regarding ATP generated—when sugar is available, these bacteria use a pathway where 12-fold more ATP is produced [47]. Therefore, if biomass growth is desirable, lactose is better as a carbon source. Neutralization of the environmental acidity during acid production could be a suitable solution to the above-mentioned inhibition problem, as well as to achieve a higher concentration of acids or their salts with respect to the stability of the fermentation product (for example, to prevent the evaporation of undissociated volatile acids). Therefore, by media pH control with the inclusion of buffers or bases for pH adjustment the higher outcome of valuable metabolites and biomass can be reached [3,21].
4.2. Assessment of the Results of Antimicrobial Effects’Study
The tested P. aeruginosa, E. coli, K. pneumonia and S. aureus are representatives of a group of multidrug-resistant bacteria—the so-called ESKAPE list microorganisms [62]. Therefore, the potential threat posed by the tested microorganism species is not only related to food spoilage, but also to the overuse of antibiotics, thus endangering both animal and human life [62]. Results of the present study show that PAB fermentates have the potential to contribute to the protection against the tested pathogens. Even at neutral pH, all tested pathogen species, but especially P. aeruginosa, were inhibited by fermentates. Moreover, with the acidification of fermentates, the antimicrobial effect could be improved as shown through our theoretical calculations or observations elsewhere. The microbial growth dependence on the concentration of undissociated acids is well demonstrated. For example, a pH slightly below the pKa of acetic acid strongly inhibits the growth of Kluyveromyces marxianus, but raising the pH slightly above the pKa of acetic acid greatly reduces its inhibitory potential [40]. Similar effects of pH are observed in other studies—a strong inhibitory effect of PAB fermentates on fungus was seen mainly at pH 3, compared to a weaker effect at pH 5 and no effect at pH 7 [22]. Interestingly, for propionic acid, relative to either pH or acid alone, the combinatorial effects of propionic acid × pH are significantly more growth-inhibitory than any other organic acid and pH combination [63]. In addition, the resulting antimicrobial effect is usually based on a combination of the effects of organic acids and other bacterial metabolites [64]. Our results show that the main principle for inhibition of the pathogenic bacteria is acid formation affirming previous information [7]; however, we observed that other antimicrobial compounds (bacteriocin-like substances) are involved in the inhibition too. Inhibition of S. aureus, especially, depends on the activity of bacteriocins. To a lesser extent bacteriocins are involved in K. pneumonia inhibition too. Warmińska–Radyko and Łaniewska–Moroz [65] also observed the antibacterial activity of bacteriocin-like metabolites synthesized by Propionibacterium acidipropionici towards Gram-negative bacteria belonging to Enterobacteriaceae [64]. Interest in bacteriocins as food biopreservatives is increasing because of their high stability in a wide range of pH values and temperatures [26,64]. Moreover, interest in complex food biopreservatives obtained from natural sources is growing because of the spread of antimicrobial resistance—a global and increasing problem. Moreover, the use of bacteriocin-producing strains is being considered an attractive approach for controlling foodborne pathogens in the gastrointestinal tract; therefore, the knowledge gained by this study may also be useful in evaluating the antimicrobial potential of PAB as prospective probiotic organisms in humans and animals [26,28,29,32,60]. Coliforms (especially the genera Escherichia and Klebsiella) are called opportunistic pathogens and are responsible for a wide variety of infections. At the same time, many species are representatives of normal intestinal flora. Coliforms, S. aureus and P. aeruginosa cause infections not only in humans but also in animals, e.g., mastitis in cows [66]. Some of them have become resistant to the most common antibiotics. Pseudomonas spp. is an aerobic, non-spore-forming, gram-negative, rod-shaped bacteria. Previous studies have shown that these microbes are particularly responsible for the spoilage of dairy products because they produce thermostable lipolytic and proteolytic enzymes that reduce the quality and shelf life of processed milk and dairy products (such as cheese, UHT milk, butter and yogurt) (reviewed by [67,68]). Pseudomonades also spoil a wide range of other food products: meat, fish, water, fruits and vegetables [69]. Regarding health risks, Pseudomonas spp. does not pose a serious risk to public health; however, P. aeruginosa is the most pathogenic species among Pseudomonas spp. Belonging to risk group II, it is an opportunistic pathogen and one of the main bacteria that cause nosocomial infections in hospitals affecting immunocompromised individuals. Therefore, it can also cause serious and even fatal diseases (reviewed by [68,69]). Sorbic and benzoic acids have widespread usage as food preservatives, but at the same time, they have a poor ability to reduce P. aeruginosa growth [63]. Since P. aeruginosa was the microorganism most affected by PAB fermentates in this study, products of PAB fermentation could be of interest in related applications. Moderate or strong inhibition was observed against B. subtilis—a spore-forming bacterium that poses several threats to the food industry because it can tolerate processing and several measures designed to destroy vegetative cells. B. subtilis has been identified as one of the microorganisms capable of forming biofilms that result in food spoilage. It can grow, survive and cause spoilage in meat and potatoes [10]. Spore-forming bacteria also pose the greatest spoilage threat to dairy products, causing serious economic losses, equipment deterioration and reputational damage to food companies [70].
The use of fermentation liquids in the food and agro-industry can have several advantages over the use of concentrated acids. Fermentates can be used in natural or concentrated forms. The combined effect of different antimicrobial compounds on the growth of unwanted microorganisms is likely to result from the interaction of many effects [63]. Knowing the good antifungal activity of PAB [8,30,39], the field of application of such fermented products could be quite wide. For example, PAB food-grade bacterial species are used as bioprotective cultures—an alternative to chemical preservatives, for example, to avoid or delay fungal spoilage of dairy products. As PAB biomass is known to be a source of protein, vitamin B complex, and minerals left in waste bacterium cells, it can be used for increasing the nutritional value of food or animal feed products [1,37,60]. Experimental evaluation should be conducted on a case-by-case basis to determine whether a particular product is an appropriate choice for a particular application. Therefore, further research on the antimicrobial or antifungal effects of PAB fermentates, their forms of use (e.g., direct application on food, in coatings or edible films [9], in the form of microcapsules [71]), as well as their compatibility with different food products or raw materials would be valuable.
5. Conclusions
- The knowledge gained in the present study can help to model the acquirement of acid metabolites and biomass from several PAB strains. The results show that the fermentative production of propionic and acetic acid is considerably influenced by the added carbon source. Lactose broth fermentation resulted in a significant pH reduction, and propionic and acetic acids’ outcomes were significantly lower (p < 0.05) than when using lactate as a carbon source.
- Biomass production in lactose was significantly higher (p < 0.05) than in lactate-based broths.
- When comparing the propionic acid bacteria strains, significant differences were observed for the production of acids and resulting pH in both types of fermentates (p < 0.05). Acid production, lactose consumption, substrate conversion efficiency to acetic acid, as well as biomass production was strain-dependent. At the same time, lactose conversion efficiency to propionic acid and propionic acid yield were similar for all strains (p ≥ 0.05).
- Results show that antimicrobial efficacy is strain-dependent. The strongest inhibitory effect was demonstrated by fermentates of all tested PAB strains against Pseudomonas aeruginosa, by strain Acidipropionibacterium thoenii against Bacillus subtilis and by strains Propionibacterium freudenreichii and Acidipropionibacterium thoenii against Staphylococcus aureus.
- Bacteriocins are likely to play a significant role in the inhibition of Staphylococcus aureus and Klebsiella pneumonia, while Escherichia coli, Pseudomonas aeruginosa and Bacillus subtilis are inhibited by acids. To increase the antimicrobial activity of fermentates, we recommend acidifying the fermentate or fermentate-supplemented medium to at least pH 5.5.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation9010026/s1, Figure S1. The comparison of inhibitory effects of PAB strains towards B. subtilis, E. coli, K. pneumoniae, P. aeruginosa and S. aureus after 10 h incubation.
Author Contributions
Conceptualization, U.A. and J.L.; methodology, J.L.; formal analysis, U.A., J.L., R.S. and M.Z.; investigation, U.A. and J.L.; data curation, U.A.; writing—original draft preparation, U.A., J.L., I.C. and M.Z.; writing—review and editing, U.A., J.L., I.C. and M.Z.; visualization, U.A., J.L. and M.Z.; supervision, I.C. and J.L.; project administration, U.A.; resources and funding acquisition, U.A., J.L., I.C. and M.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the European Regional Development Fund (ERDF) Post-doctoral Research Support Program (project Nr.1.1.1.2/16/I/001) Research application ‘Processing of Whey into Value-Added Products for Food Industry and Agriculture’ (Nr.1.1.1.2./VIAA/2/18/307). The contribution of Janis Liepins was supported by the ERDF, the project ‘Sustainable Microbial Valorisation of Waste Lipids into Biosurfactants’ (project No. 1.1.1.1/19/A/047).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Zandona, E.; Blažić, M.; Režek Jambrak, A. Whey Utilization: Sustainable Uses and Environmental Approach. Food Technol. Biotechnol. 2021, 59, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Chourasia, R.; Phukon, L.C.; Abedin, M.M.; Padhi, S.; Singh, S.P.; Rai, A.K. Whey valorization by microbial and enzymatic bioprocesses for the production of nutraceuticals and value-added products. Bioresour. Technol. Rep. 2022, 19, 101144. [Google Scholar] [CrossRef]
- Lakstina, J.; Aboltina, I.; Vanaga, L.; Ciprovica, I.; Jonkus, D.; Zagorska, J.; Cinkmanis, I. The novel solution for acid whey permeate application in animal feeding. Rural Sustain. Res. 2020, 44, 1–7. [Google Scholar] [CrossRef]
- Russo, G.L.; Langellotti, A.L.; Verardo, V.; Martín-García, B.; Di Pierro, P.; Sorrentino, A.; Baselice, M.; Oliviero, M.; Sacchi, R.; Masi, P. Formulation of New Media from Dairy and Brewery Wastes for a Sustainable Production of DHA-Rich Oil by Aurantiochytrium mangrovei. Mar. Drugs 2022, 20, 39. [Google Scholar] [CrossRef] [PubMed]
- Sarenkova, I.; Saez-Orviz, S.; Ciprovica, I.; Rendueles, M.; Diaz, M. Lactobionic acid production by Pseudomonas taetrolens in a fed-batch bioreactor using acid whey as substrate. Int. J. Dairy Technol. 2022, 75, 361–371. [Google Scholar] [CrossRef]
- Caplice, E.; Fitzgerald, G.F. Food fermentations: Role of microorganisms in food production and preservation. Int. J. Food Microbiol. 1999, 50, 131–149. [Google Scholar] [CrossRef]
- Patel, A.; Shah, N. Current scenario of antimicrobial compounds produced by food grade bacteria in relation to enhance food safety and quality. J. Innov. Biol. 2014, 1, 189–194. [Google Scholar]
- Fernandez, B.; Vimont, A.; Desfossés-Foucault, É.; Daga, M.; Arora, G.; Fliss, I. Antifungal activity of lactic and propionic acid bacteria and their potential as protective culture in cottage cheese. Food Control 2017, 78, 350–356. [Google Scholar] [CrossRef]
- Quinto, E.J.; Caro, I.; Villalobos-Delgado, L.H.; Mateo, J.; De-Mateo-Silleras, B.; Redondo-Del-Río, M.P. Food Safety through Natural Antimicrobials. Antibiotics 2019, 8, 208. [Google Scholar] [CrossRef]
- Ajingi, Y.S.; Rodpan, S.; Usman, J.N.; Koga, Y.; Jongruja, N. Synergistic effect of nisin with acetic and propionic acids inactivates Bacillus subtilis on meat and potato. Biocatal. Agric. Biotechnol. 2022, 41, 102317. [Google Scholar] [CrossRef]
- Parada, J.L.; Caron, C.; Medeiros, A.B.; Soccol, C.R. Bacteriocins from lactic acid bacteria: Purification, properties and use as biopreservatives. Braz. Arch. Biol. Technol. 2007, 50, 512–542. [Google Scholar] [CrossRef]
- Poonam, P.S.D.; Tomar, S.K.; De, S.; Singh, R. Multifaceted attributes of dairy propionibacteria: A review. World J. Microbiol. Biotechnol. 2012, 28, 3081–3095. [Google Scholar] [CrossRef]
- Šimat, V.; Čagalj, M.; Skroza, D.; Gardini, F.; Tabanelli, G.; Montanari, C.; Hassoun, A.; Ozogul, F. Chapter Two—Sustainable sources for antioxidant and antimicrobial compounds used in meat and seafood products. In Advances in Food and Nutrition Research; Toldra, F., Ed.; Academic Press: Cambridge, MA, USA, 2021; Volume 97, pp. 55–118. [Google Scholar] [CrossRef]
- Alonso, S.; Rendueles, M.; Díaz, M. Microbial production of specialty organic acids from renewable and waste materials. Crit. Rev. Biotechnol. 2015, 35, 497–513. [Google Scholar] [CrossRef]
- Gonzalez-Garcia, R.A.; McCubbin, T.; Navone, L.; Stowers, C.; Nielsen, L.K.; Marcellin, E. Microbial Propionic Acid Production. Fermentation 2017, 3, 21. [Google Scholar] [CrossRef]
- Rutenberg, R.; Bernstein, S.; Fallik, E.; Paster, N.; Poverenov, E. The improvement of propionic acid safety and use during the preservation of stored grains. Crop Prot. 2018, 110, 191–197. [Google Scholar] [CrossRef]
- Ali, R.; Saravia, F.; Hille-Reichel, A.; Gescher, J.; Horn, H. Propionic acid production from food waste in batch reactors: Effect of pH, types of inoculum, and thermal pre-treatment. Bioresour. Technol. 2020, 319, 124166. [Google Scholar] [CrossRef]
- Antone, U.; Liepins, J.; Zagorska, J.; Cinkmanis, I. Fermentation of milk whey permeate with different dairy propionibacteria strains. In Rural Development: Challenges for Sustainable Bioeconomy and Climate Change, Proceedings of the 10th International Scientific Conference, Kaunas, Lithuania, 21–23 September 2021; Černiauskienė, J., Ed.; Vytautas Magnus University Agriculture Academy: Kaunas, Lithuania, 2021; pp. 1–7. [Google Scholar] [CrossRef]
- Piwowarek, K.; Lipińska, E.; Hać-Szymańczuk, E.; Kieliszek, M.; Ścibisz, I. Propionibacterium spp.—Source of propionic acid, vitamin B12, and other metabolites important for the industry. Appl. Microbiol. Biotechnol. 2018, 102, 515–538. [Google Scholar] [CrossRef]
- Teles, J.C.; Stolle, E.M.; Koloda, S.A.; Barana, A.C. Production of Propionic Acid by Propionibacterium acidipropionici from Agroindustrial Effluents. Braz. Arch. Biol. Technol. 2019, 62, e19180550. [Google Scholar] [CrossRef]
- Ranaei, V.; Pilevar, Z.; Khaneghah, A.M.; Hosseini, H. Propionic Acid: Method of Production, Current State and Perspectives. Food Technol. Biotechnol. 2020, 58, 115–127. [Google Scholar] [CrossRef]
- Coral, J. Propionic acid production by Propionibacterium sp. using low-cost carbon sources in submerged fermentation. Master’s Thesis, The University of Provence, the University of the Mediterranean Sea and Federal University of Parana, Curitiba, Brazil, 2008. [Google Scholar]
- Lyon, W. Characterization and Isolation of a Bacteriocin Produced by a Strain of Propionibacterium thoenii (A Dissertation Submitted to the Graduate Faculty in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy); Iowa State University: Ames, IA, USA, 1992. [Google Scholar] [CrossRef]
- Zarate, G.; Babot, J.D.; Argañaraz-Martínez, E.; Lorenzo-Pisarello, M.J.; Perez Chaia, A. Dairy propionibacteria: Technological importance and probiotic potential for application on human and animal nutrition. In Multidisciplinary Approaches on Food Science and Nutrition for the XXI Century; Filip, R., Ed.; Transworld Research Network: Kerala, India, 2011; pp. 175–213. [Google Scholar]
- Wyk, J.V.; Morkel, R.A.; Dolley, L. Metabolites of Propionibacterium: Techno- and biofunctional ingredients. In Alternative and Replacement Foods; Handbook of Food Bioengineering, Series; Grumezescu, A.M., Holban, A.M., Eds.; Academic Press: London, UK, 2018; Volume 17, pp. 205–260. [Google Scholar]
- Turgay, M.; Bachmann, H.; Irmler, S.; von Ah, U.; Hlich-Wyder, M.F.; Falentin, H.; Deutsch, S.; Jan, G.; Thierry, A. Propionibacterium spp. and Acidipropionibacterium spp.; Reference Module in Food Science; Elsevier: Amsterdam, The Netherlands, 2020; Available online: https://hal.inrae.fr/hal-02640422v1 (accessed on 17 September 2021). [CrossRef]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards); Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Hilbert, F.; et al. Statement on the update of the list of QPS-recommended microbiological agents intentionally added to food or feed as notified to EFSA 16: Suitability of taxonomic units notified to EFSA until March 2022. EFSA J. 2022, 20, 7408. [Google Scholar] [CrossRef]
- Rabah, H.; Rosa do Carmo, F.L.; Jan, G. Dairy propionibacteria: Versatile probiotics. Microorganisms 2017, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.; Deutsch, S.M.; Falentin, H.; Dalmasso, M.; Cousin, F.J.; Jan, G. New insights into physiology and metabolism of Propionibacterium freudenreichii. Int. J. Food Microbiol. 2011, 149, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Hugenholtz, J.; Hunik, J.; Santos, H.; Smid, E. Nutraceutical production by propionibacteria. Le Lait 2002, 82, 103–112. [Google Scholar] [CrossRef]
- Holo, H.; Faye, T.; Brede, D.A.; Nilsen, T.; Ødegård, I.; Langsrud, T.; Brendehaug, J.; Nes, I.F. Bacteriocins of propionic acid bacteria. Le Lait 2002, 82, 59–68. [Google Scholar] [CrossRef]
- Faye, T.; Brede, D.A.; Langsrud, T.; Nes, I.F.; Holo, H. Prevalence of the genes encoding propionicin T1 and protease-activated antimicrobial peptide and their expression in classical propionibacteria. Appl. Environ. Microbiol. 2004, 70, 2240–2244. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thierry, A.; Valence, F.; Deutsch, S.M.; Even, S.; Falentin, H.; Le Loir, Y.; Jan, G.; Gagnaire, V. Strain-to-strain differences within lactic and propionic acid bacteria species strongly impact the properties of cheese—A review. Dairy Sci. Technol. 2015, 95, 895–918. [Google Scholar] [CrossRef]
- MicroGARD® Fermentates. Danisco DuPont. 2022. Available online: https://www.dupontnutritionandbiosciences.com/products/microgard.html (accessed on 10 September 2022).
- Cultured Whey. Inhibit 3600 Dairy. Mezzoni Foods. 2022. Available online: https://www.mezzonifoods.com/shop/cultured-whey/ (accessed on 10 September 2022).
- Danilova, I.V.; Lee, H.; Tourova, T.P.; Ryzhkova, E.P.; Netrusov, A.I. Propionibacterium freudenreichii strains as antibacterial agents at neutral ph and their production on food-grade media fermented by some lactobacilli. J. Food Saf. 2012, 32, 48–58. [Google Scholar] [CrossRef]
- Vorobjeva, I.L. Propionibacteria; Kluver Academic: Dordrecht, The Netherlands, 1999. [Google Scholar]
- Acidipropionibacterium Acidipropionici. Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures GmbH. 2022. Available online: https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium91.pdf (accessed on 15 December 2022).
- Le Lay, C.; Coton, E.; Le Blay, G.; Chobert, J.M.; Haertlé, T.; Choiset, Y.; Van Long, N.N.; Meslet-Cladière, L.; Mounier, J. Identification and quantification of antifungal compounds produced by lactic acid bacteria and propionibacteria. Int. J. Food Microbiol. 2016, 239, 79–85. [Google Scholar] [CrossRef]
- Martynova, J.; Kokina, A.; Kibilds, J.; Liepins, J.; Scerbaka, R.; Vigants, A. Effects of acetate on Kluyveromyces marxianus DSM 5422 growth and metabolism. Appl. Microbiol. Biotechnol. 2016, 100, 4585–4594. [Google Scholar] [CrossRef]
- Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 48, 5–16. [Google Scholar] [CrossRef]
- Latham, K.G.; Ferguson, A.; Donne, S.W. Influence of ammonium salts and temperature on the yield, morphology and chemical structure of hydrothermally carbonized saccharides. SN Appl. Sci. 2019, 1, 1–13. [Google Scholar] [CrossRef]
- Lambert, R.J.; Stratford, M. Weak-acid preservatives: Modelling microbial inhibition and response. J. Appl. Microbiol. 1999, 86, 157–164. [Google Scholar] [CrossRef]
- Moresi, M.; Parente, E. Fermentation (Industrial)|Production of some organic acids (citric, gluconic, lactic, and propionic). In Encyclopedia of Food Microbiology, 2nd ed.; Batt, C.A., Tortorello, M.L., Eds.; Academic Press: Cambridge, MA, USA, 2014; pp. 804–815. [Google Scholar] [CrossRef]
- Frallicciardi, J.; Melcr, J.; Siginou, P.; Marrink, S.J.; Poolman, B. Membrane thickness, lipid phase and sterol type are determining factors in the permeability of membranes to small solutes. Nat. Commun. 2022, 13, 1605. [Google Scholar] [CrossRef]
- Hassan, A.N.; Frank, J.F. Ch.6 Starter cultures and their use. In Applied Dairy Microbiology, 2nd ed.; Marth, E.H., Steele, J., Eds.; Marcel Dekker Inc.: New York, NY, USA; CRC Press: Basel, Switzerland, 2001; p. 185. [Google Scholar] [CrossRef]
- Ciani, M.; Comitini, F.; Mannazzu, I. Ecological Processes. Fermentation. In Encyclopedia of Ecology; Jorgensen, S.E., Fath, B., Eds.; Elsevier Science: Amsterdam, The Netherlands, 2018; pp. 310–321. [Google Scholar] [CrossRef]
- Lewis, V.P.; Yang, S.T. Propionic acid fermentation by Propionibacterium acidipropionici: Effect of growth substrate. Appl. Microbiol. Biotechnol. 1992, 37, 437–442. [Google Scholar] [CrossRef]
- Fitz, A. Über Spaltpilzgärung, IV. Ber. Deut. Chem. Ges. 1878, 11, 1896–1899. [Google Scholar]
- Guan, N.; Liu, L. Microbial response to acid stress: Mechanisms and applications. Appl. Microbiol. Biotechnol. 2020, 104, 51–65. [Google Scholar] [CrossRef]
- Theron, M.M.; Lues, J.F.R. Organic Acids and Food Preservation, 1st ed.; CRC Press: Basel, Switzerland, 2010. [Google Scholar] [CrossRef]
- Blanc, P.; Goma, G. Propionic acid fermentation: Improvement of performances by coupling continuous fermentation and ultrafiltration. Bioprocess. Biosyst. Eng. 1987, 2, 137–139. [Google Scholar] [CrossRef]
- Ibragimova, S.I.; Sakharova, Z.V. Sviaz’ mezhdu udel’noĭ skorost’iu rosta i kolichestvom potreblennogo substrata u Probionibacterium shermanii [Relation between specific growth rate and amount of consumed substrate in Propionibacterium shermanii]. Dokl. Akad. Nauk. SSSR 1974, 219, 988–989. [Google Scholar]
- Kusano, K.; Yamada, H.; Niwa, M.; Yamasato, K. Propionibacterium cyclohexanicum sp. nov., a new acid-tolerant omega-cyclohexyl fatty acid-containing propionibacterium isolated from spoiled orange juice. Int. J. Syst. Bact. 1997, 47, 825–831. [Google Scholar] [CrossRef]
- Thierry, A.; Berthier, F.; Gagnaire, V.; Kerjean, J.R.; Lopez, C.; Noël, Y. 10. Eye Formation and Swiss-Type Cheeses. In Technology of Cheesemaking, 2nd ed.; Law, B.A., Tamime, A.Y., Eds.; Blackwell Publishing Ltd.: Hoboken, NJ, USA, 2010; pp. 360–383. [Google Scholar]
- Zarate, G.; Perez Chaia, A.; Guillermo, O. Some characteristics of practical relevance of the β-galactosidase from potential probiotic strains of Propionibacterium acidipropionici. Anaerobe 2002, 8, 259–267. [Google Scholar] [CrossRef]
- Piveteau, P. Metabolism of lactate and sugars by dairy propionibacteria: A review. Le Lait 1999, 79, 23–41. [Google Scholar] [CrossRef]
- Johns, A.T. The mechanism of propionic acid formation by propionibacteria. J. Gen. Microbiol. 1951, 5, 337–345. [Google Scholar] [CrossRef][Green Version]
- Ahmadi, N.; Khosravi-Darani, K.; Mortazavian, A.M. An overview of biotechnological production of propionic acid: From upstream to downstream processes. Electron. J. Biotechnol. 2017, 28, 67–75. [Google Scholar] [CrossRef]
- Bücher, C.; Burtscher, J.; Domig, K.J. Propionic acid bacteria in the food industry: An update on essential traits and detection methods. Compr. Rev. Food Sci. Food Saf. 2021, 20, 4299–4323. [Google Scholar] [CrossRef] [PubMed]
- Boyaval, P.; Corre, C. Production of propionic acid. Le Lait 1995, 75, 453–461. [Google Scholar] [CrossRef]
- Pulgar, M.J.A. The ESKAPE Bacteria Group and Its Clinical Importance. CLOVER BioSoft. 2019. Available online: https://cloverbiosoft.com/the-eskape-bacteria-group-and-its-clinical-importance/ (accessed on 5 May 2022).
- Bushell, F.M.L.; Tonner, P.D.; Jabbari, S.; Schmid, A.K.; Lund, P.A. Synergistic impacts of organic acids and ph on growth of Pseudomonas aeruginosa: A comparison of parametric and bayesian non-parametric methods to model growth. Front. Microbiol. 2019, 9, 3196. [Google Scholar] [CrossRef] [PubMed]
- Gwiazdowska, D.; Trojanowska, K. Antimicrobial activity and stability of partially purified bacteriocins produced by Propionibacterium freudenreichii ssp. freudenreichii and ssp. shermanii. Le Lait 2006, 86, 141–154. [Google Scholar] [CrossRef]
- Warmińska-Radyko, I.; Łaniewska-Moroz, Ł. Antibacterial activity of different fractions from the culture of Propionibacterium acidipropionici. Pol. J. Food Nutr. Sci. 1999, 49, 23–29. [Google Scholar]
- Cobirka, M.; Tancin, V.; Slama, P. Epidemiology and classification of mastitis. Animals 2020, 10, 2212. [Google Scholar] [CrossRef]
- Rios, V.M. Predictive Food Microbiology—New Models for Safety and Quality Assessment of a Broad Range of Dairy Products. Ph.D. Thesis, Technical University of Denmark, Kongens Lyngby, Denmark, 2019. [Google Scholar]
- Arslan, S.; Eyi, A.; Özdemir, F. Spoilage potentials and antimicrobial resistance of Pseudomonas spp. isolated from cheeses. J. Dairy Sci. 2011, 94, 5851–5856. [Google Scholar] [CrossRef]
- Raposo, A.; Pérez, E.; de Faria, C.T.; Ferrús, M.A.; Carrascosa, C. Food spoilage by Pseudomonas spp.—An overview. In Foodborne Pathogens and Antibiotic Resistance; Singh, O.V., Ed.; John Wiley & Sons: Haboken, NJ, USA, 2016. [Google Scholar] [CrossRef]
- Lopez-Brea, S.G.; Gómez-Torres, N.; Arribas, M.Á. Spore-forming bacteria in dairy products. In Microbiology in Dairy Processing: Challenges and Opportunities; Poltronieri, P., Ed.; John Wiley & Sons: Haboken, NJ, USA, 2017; pp. 11–36. [Google Scholar] [CrossRef]
- Eghbal, N.; Liao, W.; Dumas, E.; Azabou, S.; Dantigny, P.; Gharsallaoui, A. Microencapsulation of natural food antimicrobials: Methods and applications. Appl. Sci. 2022, 12, 3837. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).