Abstract
Sandalwood and agarwood essential oils are rare natural oils comprising fragrant terpenoids that have been used in perfumes and incense for millennia. Increasing demand for these terpenoids, coupled with difficulties in isolating them from natural sources, have led to an interest in finding alternative production platforms. Here, we engineered the budding yeast Saccharomyces cerevisiae to produce fragrant terpenoids from sandalwood and agarwood. Specifically, we constructed strain FPPY005_39850, which overexpresses all eight genes in the mevalonate pathway. Using this engineered strain as the background strain, we screened seven distinct terpene synthases from agarwood, sandalwood, and related plant species for their activities in the context of yeast. Five terpene synthases led to the production of fragrant terpenoids, including α-santalene, α-humulene, δ-guaiene, α-guaiene, and β-eudesmol. To our knowledge, this is the first demonstration of β-eudesmol production in yeast. We further improved the production titers by downregulating ERG9, a key enzyme from a competing pathway, as well as employing enzyme fusions. Our final engineered strains produced fragrant terpenoids at up to 101.7 ± 6.9 mg/L. We envision our work will pave the way for a scalable route to these fragrant terpenoids and further establish S. cerevisiae as a versatile production platform for high-value chemicals.
1. Introduction
Terpenoids are a large family of natural products with diverse biological functions and an incredible array of industrial applications, including being used as vitamins, pigments, medicine, and fragrances [1,2]. Notable examples of terpenoids currently used for human consumption include the anticancer agent taxol, the antimalarial drug artemisinin, the powerful antioxidant astaxanthin, and the essential oils from sandalwood and agarwood. Production of these compounds by extraction from natural sources largely fails to scale up to meet industrial scale and is dependent on seasonal conditions. These factors, coupled with the increasing demands for terpenoids, have led to an interest in finding an alternative production platform [3,4,5]. Notably, the production of terpenoids in the yeast Saccharomyces cerevisiae has emerged as an attractive alternative, with several terpenoids (such as farnesene [6] and artemisinin [7]) already making their way into the market [8].
Despite their vast structural and functional diversity, all terpenoids are synthesized from the isomeric five-carbon (C5) precursors isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP), which, in yeast and mammals, are produced by the mevalonate (MEV) pathway (Figure 1) [2]. The MEV pathway involves the condensation of three molecules of acetyl-CoA to 3-hydroxy-3- methylglutaryl CoA (HMG-CoA) and subsequent reduction to MEV by HMG-CoA reductase. From MEV, three phosphorylations and one decarboxylation produce IPP, and isomerization to DMAPP completes the pathway. The IPP and DMAPP precursors are then condensed to lengthen the carbon chain in C5 increments to form geranyl pyrophosphate (GPP, C10) and farnesyl pyrophosphate (FPP, C15). In S. cerevisiae, this condensation step is catalyzed by the enzyme FPP synthase (FPPS) [9]. Finally, dedicated terpene synthases convert these prenyl pyrophosphate intermediates to monoterpenes (C10) and sesquiterpenes (C15), and, in the process, generate structural diversity as well as biological activities of the resulting terpenoids (Figure 1). In sandalwood (Santalum album Linn), santalene synthase converts FPP to α-santalene as a major product and small amounts of β-santalene and α-exo-bergamotene [10,11]. In Agarwood (Aquilaria spp.), several dedicated terpene synthases exist to convert FPP into a variety of fragrant terpenoids including α-humulene, δ-guaiene and β-eudesmol [12,13,14].
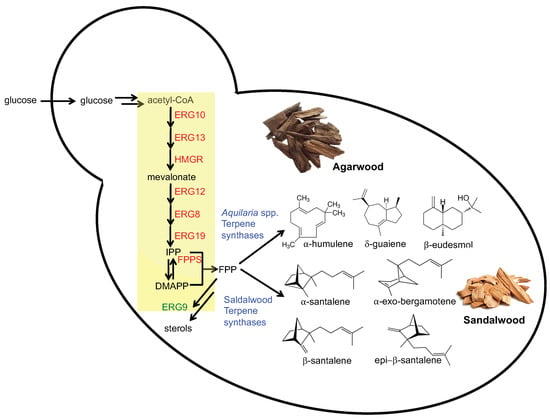
Figure 1.
Metabolic engineering of S. cerevisiae for production of fragrant terpenoids from agarwood and sandalwood. Mevalonate pathway enzymes (yellow box, highlighted in red) were overexpressed to increase the pool of the key intermediate farnesyl pyrophosphate (FPP). Terpene synthase enzymes (highlighted in blue) were heterologously expressed in the engineered yeast to produce the fragrant terpenoids. Expression of the squalene synthase enzyme ERG9 was repressed to minimize the diversion of FPP towards sterol biosynthesis. ERG10, Acetyl-CoA C-acetyltransferase (acetoacetyl-CoA thiolase); ERG13, 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) synthase; HMGR, HMG-CoA reductase; ERG12, Mevalonate kinase; ERG8, Phosphomevalonate kinase; ERG19, Mevalonate pyrophosphate decarboxylase; FPPS, farnesyl pyrophosphate synthetase (synthase); ERG9, squalene synthase.
In this work, we engineered the budding yeast S. cerevisiae to produce fragrant terpenoids found naturally in sandalwood and agarwood. We chose S. cerevisiae as the production host for several reasons. First, S. cerevisiae is genetically tractable; thus, genetic tools for metabolic pathway manipulation are abundant. Second, the construction, isolation, and analysis of S. cerevisiae mutant strains can be performed with relative ease. Third, S. cerevisiae has a long and proven track record in various industrial applications, and the fermentation of S. cerevisiae has previously been manipulated to produce numerous heterologous metabolites including other terpenoids. Finally, S. cerevisiae can be cultivated in a chemically defined medium and exhibits fast growth rates, thus facilitating scaling-up processes. To enable S. cerevisiae to produce fragrant terpenoids, we first overexpressed the entire mevalonate pathway which comprises eight genes that leads to the production of farnesyl pyrophosphate (FPP). Using the resulting engineered strain as the background strain, we then screened seven distinct terpene synthase enzymes from agarwood, sandalwood, and related plant species using a plasmid-based expression system. We further improved the production titer of the fragrant terpenoids by minimizing the production of sterol which also uses FPP as an intermediate. Furthermore, we employed enzyme fusion to bring FPP synthase and the terpene synthase into proximity. We envision our work will pave the way for a scalable route to these fragrant terpenoids that are hard to obtain from their natural sources.
2. Materials and Methods
2.1. Yeast Strain, Media, and Transformation
The yeast strains used in this study were constructed from the haploid strain of S. cerevisiae BCC39850 and strain WWY005 [15] (Table 1). The CRISPR-Cas9 plasmids used in this study for genome editing were generated from pRPR1-gRNA-handle and p414-TEF1p-Cas9-CYC1t [16]. The yeast expression plasmids used in this study were generated from pRSII426 and pRSII416 [17]. The primers used for plasmid construction, strain construction, strain verification, and qPCR are listed in Table S1. A detailed description of plasmid and strain construction is also provided in the Supplementary Materials (Supplementary File S1). Yeast and bacterial strains were stored in 25% glycerol at −80 °C. E. coli was grown in a Luria-Bertani medium. Ampicillin at 100 μg/mL was added to the medium when required. Yeast strain without plasmid was cultivated in YPD medium (10 g/L yeast extract, 20 g/L Bacto Peptone, and 20 g/L glucose). Yeast cells were transformed using the previously described LiAc/SS Carrier DNA/PEG method [18,19]. Selection of yeast transformants was done on a yeast minimal medium (6.7 g/L of Yeast Nitrogen Base (Difco), 20 g/L glucose, and a mixture of appropriate nucleotide bases and amino acids with dropouts (i.e., CSM-HIS for HIS3 selection and CSM-URA for URA3 selection).

Table 1.
Engineered strains generated in this study.
2.2. RNA Isolation and Transcript Quantification
Engineered strains were pre-cultured in 5 mL aliquots in an SC yeast minimal medium (1X yeast nitrogen base, 2% glucose, complete supplement mixture (CSM)) overnight and used to inoculate 5 mL fresh SC-mixed carbon source medium (1× yeast nitrogen base, 1.8% galactose, 0.2% glucose, complete supplement mixture (CSM)) in 50 mL Falcon tubes to achieve an initial OD600 of 0.05. The cultures were grown at 30 °C and 250 rpm in an orbital shaking incubator. After 24 h, a 1.2–1.5 mL aliquot of each culture was collected and centrifuged for 5 min at 3000× g. The pellets were washed with 5 mL of distilled water. Total RNA was extracted using the QIAgen RNeasy Kit under the manufacturer’s protocol. Contaminating genomic DNA was removed from the RNA samples by DNaseI (Thermo Fisher Scientific, Waltham, MA, USA) digestion using the manufacturer’s protocol. The RNA quantity was analyzed using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, NC, USA), and samples were stored at −80 °C until RT-PCR analysis. cDNA was obtained using RevertAid Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA) using the manufacturer′s protocol. Relative expression levels of ERG10, ERG20, tHMG1, ERG8, ERG13, IDI1, ERG19, and ERG12 were quantified using iQ SYBR Green Kit (Bio-Rad, Hercules, CA, USA) on CFX96 Touch Real-time PCR Detection System (Bio-Rad, Hercules, CA, USA). Real-time PCR was performed in triplicate, and TAF10, a gene that encodes a subunit of transcription factor IID (TFIID), was used to normalize the amount of the total mRNA in all samples. Primers for real-time PCR are listed in Table S1.
2.3. Quantification of Terpenoid Production in Engineered Strains
Engineered strains were pre-cultured in 5 mL aliquots in a yeast minimal medium SC-Ura (1X yeast nitrogen base, 2% glucose, complete supplement mixture (CSM) without uracil) overnight and used to inoculate 10 mL fresh SC-Ura-mixed carbon source medium (1X yeast nitrogen base, 1.8% galactose, 0.2% glucose, complete supplement mixture (CSM) without uracil) in 50-mL Falcon tubes to achieve an initial OD600 of 0.05. Yeast cultures were overlaid with 2 mL dodecane to reduce evaporation of the terpenoid compounds. The cultures were grown at 30 °C and 250 rpm in an orbital shaking incubator. Samples were taken at 72 h to determine the production of terpenoid compounds. The identification of terpenoid compounds dissolved in the dodecane layer was determined using Gas Chromatography-Mass Spectrometry (GC-MS, Agilent 7000 GC/MS Triple Quad, Agilent, Santa Clara, CA, USA). Briefly, 500 μL of dodecane from the cultures was analyzed on GC-MS using an HP-5MS 5% Phenyl Methyl Silox column. Samples were run using a previously described method [20] with some differences. The GC program was as follows: an initial temperature of 140 °C, followed by ramping to 180 °C at a rate of 2 °C/min, then ramping to 300 °C at a rate of 20 °C/min, where the temperature was held for 5 min. The quantification of terpenoid compounds dissolved in the dodecane layer was determined using GC with flame-ionization detection (GC-FID, Shimadzu 2014 AFSC, Shimadzu, Kyoto, Japan) using a Restek Rxi-5sil MS column with the same GC program as above. The external calibration curves of the standards α-humulene and β-eudesmol were plotted to quantify the terpenoid components. One mg/mL stock solution of each standard was accurately prepared. Each stock solution was serially diluted by factors of 10, 50, 250, and 1250. The resulting solutions were detected by GC-FID to construct external calibration curves.
3. Results and Discussion
3.1. Overexpression of the Entire Mevalonate Pathway Genes in S. cerevisiae
We began by creating strain FPPY005, which overexpresses all eight mevalonate pathway genes ERG10, ERG13, tHMG1 (encodes truncated HMGR), ERG12, ERG8, ERG19, ERG20, and IDI1 (Table 2, Figure 2). To do this, we constructed yeast expression cassettes for the mevalonate pathway genes with each construct containing two unique genes driven by the galactose-inducible bidirectional PGAL1/10 promoter. Three copies of the gene encoding truncated HMGR (tHMG1) are integrated because the enzyme was previously shown to be rate-limiting [21]. We opted to use the native terminator for each gene (i.e., ERG10 terminator was used for ERG10 ORF) to minimize the number of fragments required for homologous recombination. URA3 selectable marker was chosen as it allows for simple recycling of the marker via loxP-Cre recombination and counter-selection with 5′-fluoroorotic acid (5′-FOA) [22]. The gene cassettes were placed in the shuttle vector (pRSII416) for ease of cloning.

Table 2.
Plasmids generated in this study.
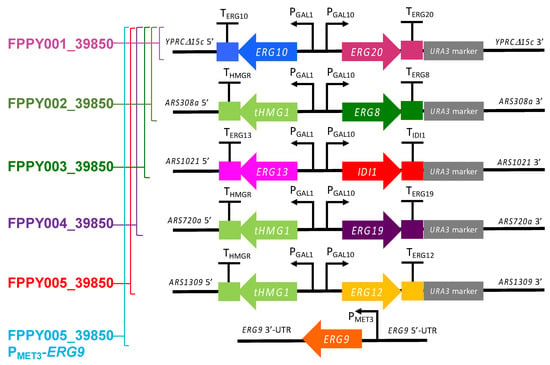
Figure 2.
Pathway design for the construction of FPP-overproducing S. cerevisiae strain. Block arrows designate mevalonate pathway genes; block squares are endogenous terminators; thin arrows are endogenous promoters; grey rectangles are native auxotrophic selectable markers (the URA3 selectable marker is flanked by loxP sites). The integration sites (YPRCΔ15c, ARS308a, ARS1021, ARS720a, and ARS1309) for the gene constructs are marked.
The gene expression cassettes for the upregulation of the mevalonate pathway were sequentially integrated into the genome of BCC39850hlu (S. cerevisiae BCC39850 with triple deletions) at designated sites, as shown in Figure 2 (YPRCΔ15c, ARS308a, ARS1021, etc.). Integration of gene expression cassettes at these sites has previously been shown to lead to high gene expression levels [23].
The integration of the five gene expression cassettes should lead to the overexpression of all eight genes in the mevalonate pathways, namely ERG10, ERG13, tHMG1 (encodes truncated HMGR), ERG12, ERG8, ERG19, ERG20 and IDI1. To verify this, we performed real-time PCR (RT-PCR) in the engineered strain FPPY005_39850 and the parental strain BCC39850hlu under inducing (i.e., galactose-containing medium) and non-inducing (i.e., glucose-only medium) conditions (Figure 3). Real-time PCR analysis indicates that all mevalonate pathway genes are up-regulated under inducing conditions (Figure 3A). As expected (given the two additional copies of the tHMG1 gene expression construct), the strongest up-regulation was observed in the expression of the truncated HMGR in strain FPPY005_39850, where we saw 11.5-fold overexpression compared to levels observed in non-inducing conditions (i.e., medium containing glucose as the sole carbon source; (relative expression levels in galactose medium)/(relative expression levels in glucose medium) (Figure 3A). The lowest up-regulation was observed in the expression of IDI1, at 2.5-fold overexpression compared to levels observed in non-inducing conditions. We did not observe any increase in the expression levels of mevalonate pathway genes in the parental strain BCC39850hlu under inducing conditions (Figure 3B). Accumulatively, these results indicate that the integration of the mevalonate pathway gene expression cassettes led to the upregulation of the mevalonate pathway in strain FPPY005_39850.
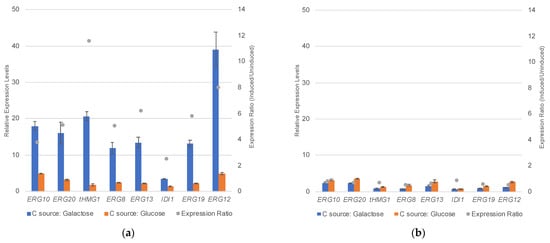
Figure 3.
Real-time PCR (RT-PCR) analysis of mevalonate pathway genes in engineered strains FPPY005_39850 (a) and BCC39850hlu (b).
3.2. Screening of Terpene Synthases in FPPY005_39850
Having verified that the mevalonate pathway is up-regulated in strain FPPY005_39850, we next turned our attention to identifying efficient terpene synthase enzymes for the production of fragrant agarwood and sandalwood terpenoids in the context of yeast. Specifically, we screened seven distinct terpene synthases from agarwood, sandalwood, and related species with previously reported enzymatic activities [5]. These include three sesquiterpene synthase genes (AsSesTPS, AsSesTPS1 and AsASS1) from Aquilaria sinensis, α-humulene synthase gene (AcHS1) from Aquilaria crassna, δ-guaiene synthase gene (AmdGS1) from Aquilaria microcarpa, β-eudesmol synthase gene (ZzBES2) from Zingiber zerumbet and santalene synthase gene (ClTPS2 or ClSanSyn) from Clausena lansium (Table 2). We constructed the gene cassettes for the expression of seven distinct terpene synthase genes. For these gene expression cassettes, we used codon-optimized and synthesized genes to ensure robust expression in yeast. As with the mevalonate gene expression cassettes, we used the galactose-inducible bidirectional PGAL1/10 promoter to drive the expression of the terpene synthase genes. However, we used the THIS5 terminator as the universal terminator. The gene expression cassettes were placed in the shuttle vector (pRSII426), which contains a 2 micron origin of replication, to ensure that the plasmid is maintained in the cell at a high-copy number.
We transformed strain FPPY005_39850 with plasmids harboring the individual terpene synthase expression cassette and measured the production of fragrant terpenoids using GC-MS (for product identification) and GC-FID (for product quantification). To minimize the evaporation of the fragrant terpenoids, the cultures were overlaid with 20% dodecane. Gratifyingly, we observed the production of several fragrant terpenoids including α-humulene, δ-guaiene, and a small amount of α-guaiene from agarwood and α-santalene from sandalwood in our engineered strains (Figure 4, Figure 5 and Figures S1–S3). For the production of agarwood terpenoids, FPPY005_39850 overexpressing AsSesTPS1 produced α-humulene at a titer of 14.6 ± 0.4 mg/L (Figure 4), while the strain overexpressing AcHS1 produced α-humulene at a titer of 22.3 ± 0.7 mg/L (Figure 5). FPPY005_39850 overexpressing AmdGS1 produced δ-guaiene at a titer of 14.5 ± 0.7 mg/L and a trace amount of α-guaiene (Figure S1). FPPY005_39850 overexpressing ZzBES2 produced β-eudesmol at a titer of 16.5 ± 0.5 mg/L (Figures S3–S4). Importantly, to our knowledge, this is the first demonstration of β-eudesmol production in the yeast system. For the production of sandalwood terpenoids, the engineered strain overexpressing ClTPS2 produced α-santalene at a titer of 6.3 ± 0.2 mg/L.
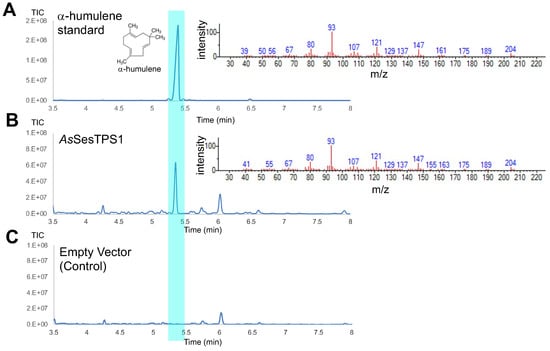
Figure 4.
Total ion chromatograms (left panel) and mass spectra (right panel) obtained from GC-MS analysis of an authentic standard for α-humulene (A) and dodecane samples from the cultivation of strains FPPY005_39850 harboring pRSII426-Gal1/10-AsSesTPS1 (B) and FPPY005_39850 harboring the empty vector pRSII426-Gal1/10 (C).
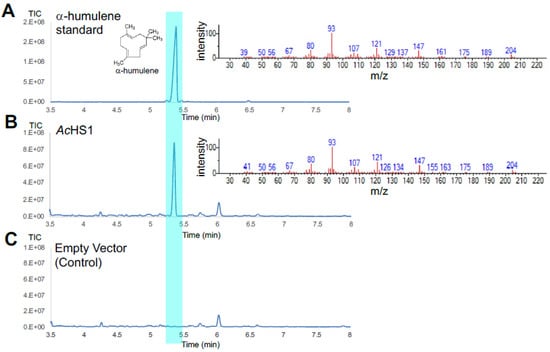
Figure 5.
Total ion chromatograms (left panel) and mass spectra (right panel) obtained from GC-MS analysis of an authentic standard for α-humulene (A) and dodecane samples from the cultivation of strains FPPY005_39850 harboring pRSII426-Gal1/10-AcHS1 (B) and FPPY005_39850 harboring the empty vector pRSII426-Gal1/10 (C).
3.3. Improving Terpenoid Production via Downregulation of ERG9 Gene Expression
In order to improve the production titer of the fragrant terpenoids, we explored two complementary strategies: (1) down-regulation of ERG9 expression and (2) fusing the FPP synthase and the terpene synthase enzymes. In the first strategy, ERG9 encodes squalene synthase, which catalyzes the first committed step of ergosterol biosynthesis (Figure 1). Since the enzyme uses FPP as a substrate, downregulation of its expression was hypothesized to divert the metabolic flux away from the production of ergosterol to the product of interest. In previous works, ERG9 downregulation has led to improvement in the production of several high-value terpenoids such as artemisinin, casbene, and lycopene [21,24,25]. In order to downregulate ERG9 expression, we used the CRISPR/Cas9 system to replace the endogenous ERG9 promoter with an L-methionine repressible promoter from MET3 (PMET3) in strain FPPY005_39850, resulting in strain FPPY005_39850 PMET3-ERG9. In parallel, we also created an analogous strain named WWY005 PMET3-ERG9, which has the same genetic modifications as strain FPPY005_39850 PMET3-ERG9 [15]. The only difference between the two strains is that WWY005 PMET3-ERG9 is derived from the laboratory strain CEN.PK2-1C while strain FPPY005_39850 PMET3-ERG9 is derived from strain BCC39850.
We transformed strains WWY005 PMET3-ERG9 and WWY005 with the plasmid overexpressing ClTPS2 and measured the production of α-santalene by the resulting strains in media either with or without L-methionine supplementation (Figure 6). Without L-methionine supplementation, strain WWY005 PMET3-ERG9 harboring the ClTPS2 overexpression plasmid produced α-santalene at a titer of 12.1 ± 0.1 mg/L, a 94% improvement in titer compared to the level observed in strain WWY005 harboring the same ClTPS2 overexpression plasmid (6.3 ± 0.2 mg/L). The titer of α-santalene was further improved to 17.3 ± 0.1 mg/L when 1 mM L-methionine was supplemented to the fermentation medium. Our results suggest that ERG9 downregulation is a viable strategy for improving fragrant terpenoid production.
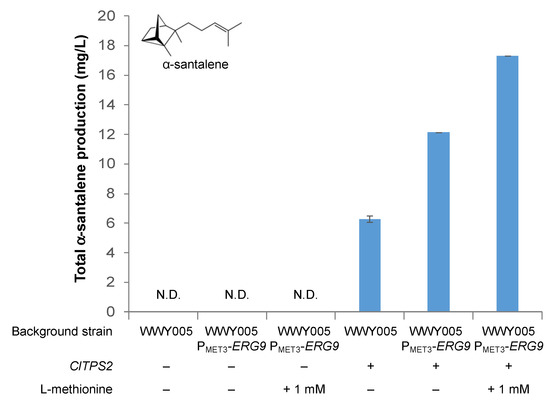
Figure 6.
Effects of ERG9 repression on α-santalene production in engineered strains. + indicates that the gene was overexpressed. – indicates that the gene was not overexpressed. N.D. not detected.
We obtained an even more dramatic improvement in α-humulene production when ERG9 was repressed (Figure 7). Specifically, without L-methionine supplementation, strain FPPY005 PMET3-ERG9 harboring the AcHS1 overexpression plasmid produced α-humulene at a titer of 94.1 ± 3.3 mg/L, a 5.0-fold improvement in titer compared to the level observed in strain FPPY005 harboring the same AcHS1 overexpression plasmid (18.7 ± 0.8 mg/L). Interestingly, the addition of 1 mM L-methionine to the production media did not further increase α-humulene titer (as was the case for α-santalene production) but rather decreased the titer to 78.1 ± 5.3 mg/L, a 17% decrease compared to no additional L-methionine supplementation. Accumulatively, these results underscore the importance of fine-tuning the level of ERG9 repression. Moreover, similar results in both strains (FPPY005_39850 PMET3-ERG9 and WWY005 PMET3-ERG9) suggest that the strategy of ERG9 repression to improve terpenoid production is general and may not be specific to the background strain.
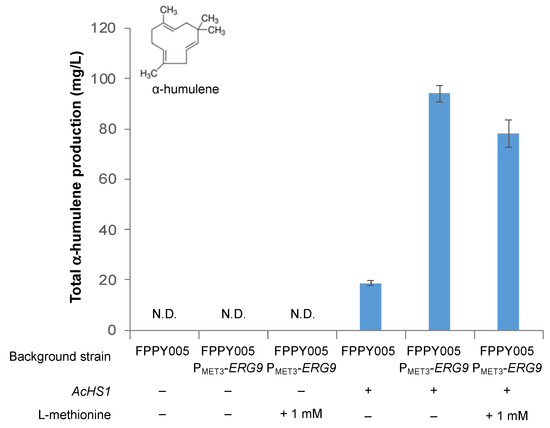
Figure 7.
Effects of ERG9 repression on α-humulene production in engineered strains. + indicates that the gene was overexpressed. – indicates that the gene was not overexpressed. N.D. not detected.
3.4. Improving Fragrant Terpenoid Production via Enzyme Fusion
Having demonstrated that ERG9 repression can improve the production titer of fragrant terpenoid, we next turned to the second strategy of fusing the FPP synthase and the terpene synthase enzymes. Previous works have shown that creating a spatial constraint between the enzyme and the substrate can increase the efficiency of the metabolic pathway resulting in a higher product titer [26]. This can be achieved by one of the following approaches: engineering fusion proteins, using protein scaffolds, and compartmentalizing metabolic pathways. In this work, we focused on fusion proteins as the two latter approaches risk either overburdening the cells with expressing additional scaffold proteins (which could deplete cellular resources that could otherwise go to making the compound of interest) or obtaining poor product titer due to the lack of suitable organelle(s) and its corresponding transporters for transporting the FPP substrate in and the terpenoid product out. Enzyme fusion improves the flux toward FPP and further to the terpenoid of interest by enabling substrate channeling, which is the direct migration of the product of one enzyme to the active site of the second enzyme [27,28]. Substrate channeling is faster than random diffusion of the intermediate through the bulk solution.
For this strategy, we constructed enzyme fusions of FPPS and each of the selected terpene synthases (AcHS1, AmdGS1, ZzBES2, and ClTPS2). A large body of work (see review [28]) demonstrates that the linker regions between the functional domains can influence the expression levels [29,30], folding and stability [31,32,33,34], as well as biological activities of the fusion enzymes [35,36,37]. In this study, we used two of the most commonly employed flexible linkers, the Gly-Ser-Gly (GSG) and the Gly-Gly-Gly-Gly-Ser (GGGGS) linkers [34,38,39], to fuse FPPS and the terpene synthases. As enzyme fusion can affect enzyme folding and stability, we also took structure prediction of the resulting enzyme fusions into consideration. Recent development in machine learning has led to significant advances in structural biology [40,41]. In particular, AlphaFold (along with the subsequent release AlphaFold v2) has solved a long-standing challenge of predicting structures of a wide variety of proteins with remarkable accuracy using only the amino acid sequences as input [42,43]. Using AlphaFold v2, we obtained predicted structures for the four selected terpene synthases and FPPS—as both standalone enzymes as well as fusion enzymes—and verified that the fused enzymes maintained the overall structures of the individual enzymes (Figures S5–S16).
In addition to the type of linker used for fusing the enzymes, previous work by Hu and coworkers has shown that the fusion order is also important as it can affect protein folding and the biological activities of these enzymes [44]. Notably, putting FPPS at the N terminus and the terpene synthase (germacrene synthase in this case) at the C terminus led to a germacrene titer up to two-fold higher than the level observed in the other enzyme fusion orientation. Using the work by Hu and coworkers [44] as our guide, we constructed plasmids for expressing these enzyme fusions: FPPS-GSG-AcHS1, FPPS-GGGGS-AcHS1, FPPS-GSG-AmdGS1, FPPS-GGGGS-AmdGS1, FPPS-GSG-ZzBES2, FPPS-GGGGS-ZzBES2, FPPS-GSG-ClTPS2 and FPPS-GGGGS-ClTPS2. Additionally, we constructed plasmids for overexpressing the individual terpene synthases and the FPPS as distinct enzymes as controls to verify that the differences in the terpenoid titers are not due to additional expression of FPPS. We then transformed strain FPPY005 PMET3-ERG9 with these plasmids and measured the resulting strains’ terpenoid levels when grown in a medium containing additional L-methionine supplementation.
Enzyme fusion improved the production titer for only two out of the four terpenoid targets, α-santalene and δ-guaiene (Figure 8). For α-santalene production, additional expression of FPPS led to a 32.6% increase in α-santalene titers compared to the strain overexpressing ClTPS2 alone (37.9 ± 2.8 mg/L vs. 28.5 ± 1.7 mg/L) (Figure 8a). The expression of FPPS and ClTPS2 as fusion enzymes using the GGGGS linker further improved the production titer to 47.6 ± 0.1 mg/L. This is a 66.9% increase over the strain overexpressing ClTPS2 alone and a 25.8% increase over the strain overexpressing FPPS and ClTPS2 as distinct enzymes. Interestingly, expression of the FPPS-GSG-ClTPS2 fusion enzymes lowered α-santalene production when compared to the strain overexpressing FPPS and ClTPS2 as distinct enzymes.
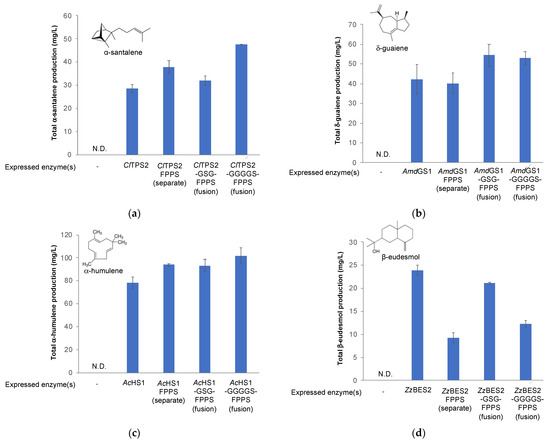
Figure 8.
Improving fragrant terpenoid production via enzyme fusion. (a) Production of α-santalene in engineered strains overexpressing ClTPS2 either alone or as fusion enzymes with FPPS; (b) Production of δ-guaiene in engineered strains overexpressing AmdGS1 either alone or as fusion enzymes with FPPS; (c) Production of α-humulene in engineered strains overexpressing AcHS1 either alone or as fusion enzymes with FPPS; (d) Production of β-eudesmol in engineered strains overexpressing ZzBES2 either alone or as fusion enzymes with FPPS. N.D. not detected.
For δ-guaiene production, additional expression of FPPS did not increase δ-guaiene titer compared to the strain overexpressing AmdGS1 alone (40.2 ± 5.1 mg/L and 42.1 ± 7.5 mg/L, respectively) (Figure 8b). However, the expression of FPPS and AmdGS1 as fusion enzymes improved the production titer to 54.5 ± 5.5 mg/L (for the GSG-linked fusion enzyme) and 52.9 ± 3.3 mg/L (for the GGGGS-linked fusion enzyme), equivalent to approximately 30% increase in titer.
For the remaining two terpenoid targets, α-humulene and β-eudesmol, enzyme fusion did not improve the production titer in a meaningful way. For α-humulene production, the biggest improvement in production titer came from increased expression of FPPS as a standalone enzyme. Specifically, the strain with additional expression of FPPS led to a 20.3% increase in α- humulene titers compared to the strain overexpressing AcHS1 alone (94.0 ± 1.1 mg/L vs. 78.1 ± 5.3 mg/L) (Figure 8c). The expression of FPPS and AcHS1 as fusion enzymes using either the GSG or GGGGS linker did not significantly improve the production titer of α-humulene (93.2 ± 5.1 mg/L and 101.7 ± 6.9 mg/L, respectively). The most surprising results came from the strains producing β-eudesmol (Figure 8d). Additional expression of FPPS led to a 61.4% decrease in β-eudesmol titers compared to the strain overexpressing ZzBES2 alone (9.2 ± 1.2 mg/L vs. 23.9 ± 1.1 mg/L). We were able to revert some of the titer loss by employing the enzyme fusions. Specifically, the expression of FPPS and ZzBES2 as fusion enzymes using the GSG and GGGGS linkers resulted in β-eudesmol titers of 21.1 ± 0.2 mg/L and 12.3 ± 0.7 mg/L, respectively.
Overall, our engineered strains produce fragrant terpenoids at similar levels to previous reports [45,46,47,48]. For example, Zhang and coworkers engineered the yeast S. cerevisiae to produce α-humulene by targeting both the mevalonate pathway enzymes and the α-humulene synthase to the yeast peroxisome, resulting in an engineered strain (M4-Ase) that produced α-humulene at a titer of 76.16 mg/L and 144.91 mg/L in glucose- and galactose-containing medium, respectively [46]. An and coworkers engineered the yeast S. cerevisiae to produce δ-guaiene, α-guaiene, and (−)-rotundone (an oxidized product of α-guaiene) [49]. The authors reported the titer of only (−)-rotundone (~1.2 mg/L) but not the other two compounds. A similar effort to produce δ-guaiene in the bacterium E. coli resulted in a strain with a relatively low titer of 38−42 μg/mL [47]. As indicated above, to our knowledge, this is the first demonstration of β-eudesmol production in the yeast system. Yu and coworkers previously engineered E. coli to produce β-eudesmol by overexpressing ZzBES2 and the mevalonate pathway enzymes [48]. Their engineered E. coli strain produced β-eudesmol at a titer of 100 mg/L in a culture medium supplemented with 500 mg/L of the intermediate mevalonate. While the reported β-eudesmol titer of 100 mg/L is well above the titer achieved in our yeast system (23.9 ± 1.1 mg/L), we contend that the E. coli system’s requirement of exogenous mevalonate supplementation will likely increase the production cost which may make the bioprocess difficult to scale up.
Several strategies exist for improving the product titer of these high-value terpenoids [4,5]. For example, as alluded to earlier, compartmentalization of pathway enzymes can create a spatial constraint between the substrate and pathway intermediates and the enzymes which results in improved efficiency of the metabolic pathway. Dusséaux and coworkers introduced a heterologous mevalonate pathway into the yeast peroxisomes to supplement the endogenous pathway in the cytosol [50]. Their engineered strains produced terpenoids at up to 125-fold yield over cytosolic production. An additional benefit of targeting terpenoid production to the yeast peroxisome is that many terpenoids require further decoration by cytochrome P450 enzymes, which reside at the endoplasmic reticulum (ER). The proximity of the ER to the peroxisomes has been suggested to improve the P450-mediated oxidation of terpenoids [50,51].
Another strategy that has shown significant promise in improving the terpenoid titer is to increase the availability of the precursor acetyl-CoA [52]. Overexpression of the mevalonate pathway, as employed in our study, may drain the pool of cytosolic acetyl-CoA, which can adversely affect yeast growth, which in turn decreases terpenoid production. Shiba and coworkers have created a pyruvate dehydrogenase bypass to increase cytosolic acetyl-CoA [53]. More recently, Meadows and coworkers devised a synthetic metabolic pathway to increase the cytosolic acetyl-CoA levels for farnesene production [54]. Their pathway has a reduced ATP requirement, reduced carbon loss, and improved pathway redox balance. The engineered strain produced 25% more farnesene than the control strain. We contend that these strategies could be utilized to improve the production titer of fragrant terpenoids in our strains.
4. Conclusions
We engineered the yeast S. cerevisiae to produce fragrant terpenoids in sandalwood and agarwood. Specifically, we constructed strain FPPY005_39850, which overexpresses all eight genes in the mevalonate pathway under the galactose-inducible system. Using this engineered strain as the background strain, we screened seven distinct terpene synthase genes from agarwood, sandalwood, and related plant species using a plasmid-based expression system. Five terpene synthases, namely AsSesTPS1, AcHS1, ClTPS2, AmdGS1, and ZzBES2, led to the production of fragrant terpenoids, including α-humulene, δ-guaiene, α-santalene, and β-eudesmol in yeast. We further improved the production titer of the terpenoids by repressing the expression of ERG9, a key enzyme from a competing pathway, as well as employing enzyme fusion of the FPPS and the corresponding terpene synthase to bring the enzymes into proximity. Our final engineered strains produced fragrant terpenoids at a titer of up to 101.7 ± 6.9 mg/L (in the case of α-humulene). Moreover, to our knowledge, this is the first demonstration of β-eudesmol production in a yeast system. We envision our work will pave the way for a scalable route to these fragrant terpenoids and further establish S. cerevisiae as a versatile production platform for fuels and high-value chemicals.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation8090429/s1, Detailed description of plasmid and strain construction; Table S1: Primers used in this study; Figure S1: Total ion chromatograms and mass spectra obtained from engineered yeast strain producing δ-guaiene; Figure S2: Total ion chromatograms and mass spectra obtained from engineered yeast strain producing α-santalene; Figure S3: GC-FID chromatograms of engineered yeast strain producing β-eudesmol; Figure S4: Mass spectra of the dodecane sample containing β-eudesmol from engineered yeast strain producing β-eudesmol; Figure S5: AlphaFold-predicted structures of the AcHS1 and FPPS as standalone enzymes; Figure S6: AlphaFold-predicted structure of the FPPS-GSG-AcHS1 enzyme fusion; Figure S7: AlphaFold-predicted structure of the FPPS-GGGGS-AcHS1 enzyme fusion; Figure S8: AlphaFold-predicted structures of the AmdGS1 and FPPS as standalone enzymes; Figure S9: AlphaFold-predicted structure of the FPPS-GSG-AmdGS1 enzyme fusion; Figure S10: AlphaFold-predicted structure of the FPPS-GGGGS-AmdGS1 enzyme fusion; Figure S11: AlphaFold-predicted structures of the ClTPS2 and FPPS as standalone enzymes; Figure S12: AlphaFold-predicted structure of the FPPS-GSG-ClTPS2 enzyme fusion; Figure S13: AlphaFold-predicted structure of the FPPS-GGGGS-ClTPS2 enzyme fusion; Figure S14: AlphaFold-predicted structures of the ZzBES2 and FPPS as standalone enzymes; Figure S15: AlphaFold-predicted structure of the FPPS-GSG-ZzBES2 enzyme fusion; Figure S16: AlphaFold-predicted structure of the FPPS-GGGGS-ZzBES2 enzyme fusion. File S1: A detailed description of plasmid and strain construction.
Author Contributions
Conceptualization, W.R.; methodology, P.P., W.S., T.P. and W.R.; validation, P.P. and W.S.; formal analysis, P.P. and W.R.; investigation, P.P. and W.R.; resources, W.R.; data curation, P.P. and W.R.; writing—original draft preparation, W.R. and P.P.; writing—review and editing, W.R.; visualization, W.R.; supervision, W.R.; project administration, W.R. and S.T.; funding acquisition, W.R. All authors have read and agreed to the published version of the manuscript.
Funding
The project was funded by the Biochemicals and Biofuels (2B) project, “Development of an efficient yeast system for the production of high-value terpenoids”, supported by the National Science and Technology Development Agency (grant number P-19-52252).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing is not applicable to this article.
Acknowledgments
We thank Justin W. Henceroth for his critical reading of this manuscript. We also thank Suchat Pongchaiphol for his assistance with terpenoid analysis via GC-FID.
Conflicts of Interest
The authors declare no conflict of interest. The funder had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Zhang, C.; Hong, K. Production of Terpenoids by Synthetic Biology Approaches. Front. Bioeng. Biotechnol. 2020, 8, 347. [Google Scholar] [CrossRef] [PubMed]
- Kung, Y.; Runguphan, W.; Keasling, J.D. From Fields to Fuels: Recent Advances in the Microbial Production of Biofuels. ACS Synth. Biol. 2012, 1, 498–513. [Google Scholar] [CrossRef] [PubMed]
- Belcher, M.S.; Mahinthakumar, J.; Keasling, J.D. New Frontiers: Harnessing Pivotal Advances in Microbial Engineering for the Biosynthesis of Plant-Derived Terpenoids. Curr. Opin. Biotechnol. 2020, 65, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.C.Y.; Keasling, J.D. Production of Isoprenoid Pharmaceuticals by Engineered Microbes. Nat. Chem. Biol. 2006, 2, 674–681. [Google Scholar] [CrossRef]
- Navale, G.R.; Dharne, M.S.; Shinde, S.S. Metabolic Engineering and Synthetic Biology for Isoprenoid Production in Escherichia Coli and Saccharomyces Cerevisiae. Appl. Microbiol. Biotechnol. 2021, 105, 457–475. [Google Scholar] [CrossRef]
- US20080083158A1-Fuel Compositions Comprising Farnesane and Farnesane Derivatives and Method of Making and Using Same-Google Patents. Available online: https://patents.google.com/patent/US20080083158A1/en (accessed on 5 August 2022).
- Westfall, P.J.; Pitera, D.J.; Lenihan, J.R.; Eng, D.; Woolard, F.X.; Regentin, R.; Horning, T.; Tsuruta, H.; Melis, D.J.; Owens, A.; et al. Production of Amorphadiene in Yeast, and Its Conversion to Dihydroartemisinic Acid, Precursor to the Antimalarial Agent Artemisinin. Proc. Natl. Acad. Sci. USA 2012, 109, E111–E118. [Google Scholar] [CrossRef]
- Paddon, C.J.; Keasling, J.D. Semi-Synthetic Artemisinin: A Model for the Use of Synthetic Biology in Pharmaceutical Development. Nat. Rev. Microbiol. 2014, 12, 355–367. [Google Scholar] [CrossRef]
- Anderson, M.S.; Yarger, J.G.; Burck, C.L.; Poulter, C.D. Farnesyl Diphosphate Synthetase. Molecular Cloning, Sequence, and Expression of an Essential Gene from Saccharomyces Cerevisiae. J. Biol. Chem. 1989, 264, 19176–19184. [Google Scholar] [CrossRef]
- Zhang, X.; Niu, M.; Teixeira Da Silva, J.A.; Zhang, Y.; Yuan, Y.; Jia, Y.; Xiao, Y.; Li, Y.; Fang, L.; Zeng, S.; et al. Identification and Functional Characterization of Three New Terpene Synthase Genes Involved in Chemical Defense and Abiotic Stresses in Santalum Album. BMC Plant. Biol. 2019, 19, 115. [Google Scholar] [CrossRef]
- Jones, C.G.; Moniodis, J.; Zulak, K.G.; Scaffidi, A.; Plummer, J.A.; Ghisalberti, E.L.; Barbour, E.L.; Bohlmann, J. Sandalwood Fragrance Biosynthesis Involves Sesquiterpene Synthases of Both the Terpene Synthase (TPS)-a and TPS-b Subfamilies, Including Santalene Synthases. J. Biol. Chem. 2011, 286, 17445–17454. [Google Scholar] [CrossRef]
- Hashim, Y.Z.H.Y.; Kerr, P.G.; Abbas, P.; Mohd Salleh, H. Aquilaria Spp. (Agarwood) as Source of Health Beneficial Compounds: A Review of Traditional Use, Phytochemistry and Pharmacology. J. Ethnopharmacol. 2016, 189, 331–360. [Google Scholar] [CrossRef] [PubMed]
- Samadi, M.; Zainal Abidin, Z.; Yoshida, H.; Yunus, R.; Awang Biak, D.R. Towards Higher Oil Yield and Quality of Essential Oil Extracted from Aquilaria Malaccensis Wood via the Subcritical Technique. Molecules 2020, 25, 3872. [Google Scholar] [CrossRef] [PubMed]
- Kumeta, Y.; Ito, M. Characterization of Delta-Guaiene Synthases from Cultured Cells of Aquilaria, Responsible for the Formation of the Sesquiterpenes in Agarwood. Plant. Physiol. 2010, 154, 1998–2007. [Google Scholar] [CrossRef]
- Watcharawipas, A.; Sansatchanon, K.; Phithakrotchanakoon, C.; Tanapongpipat, S.; Runguphan, W.; Kocharin, K. Novel Carotenogenic Gene Combinations from Red Yeasts Enhanced Lycopene and Beta-Carotene Production in Saccharomyces Cerevisiae from the Low-Cost Substrate Sucrose. FEMS Yeast Res. 2021, 21, foab062. [Google Scholar] [CrossRef] [PubMed]
- Farzadfard, F.; Perli, S.D.; Lu, T.K. Tunable and Multifunctional Eukaryotic Transcription Factors Based on CRISPR/Cas. ACS Synth. Biol. 2013, 2, 604–613. [Google Scholar] [CrossRef]
- Chee, M.K.; Haase, S.B. New and Redesigned PRS Plasmid Shuttle Vectors for Genetic Manipulation of Saccharomyces Cerevisiae. G3 Genes Genomes Genet. 2012, 2, 515–526. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. Frozen Competent Yeast Cells That Can Be Transformed with High Efficiency Using the LiAc/SS Carrier DNA/PEG Method. Nat. Protoc. 2007, 2, 1–4. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. High-Efficiency Yeast Transformation Using the LiAc/SS Carrier DNA/PEG Method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef]
- Zha, W.; An, T.; Li, T.; Zhu, J.; Gao, K.; Sun, Z.; Xu, W.; Lin, P.; Zi, J. Reconstruction of the Biosynthetic Pathway of Santalols under Control of the GAL Regulatory System in Yeast. ACS Synth. Biol. 2020, 9, 449–456. [Google Scholar] [CrossRef]
- Ro, D.-K.; Paradise, E.M.; Ouellet, M.; Fisher, K.J.; Newman, K.L.; Ndungu, J.M.; Ho, K.A.; Eachus, R.A.; Ham, T.S.; Kirby, J.; et al. Production of the Antimalarial Drug Precursor Artemisinic Acid in Engineered Yeast. Nature 2006, 440, 940–943. [Google Scholar] [CrossRef]
- Gueldener, U.; Heinisch, J.; Koehler, G.J.; Voss, D.; Hegemann, J.H. A Second Set of LoxP Marker Cassettes for Cre-Mediated Multiple Gene Knockouts in Budding Yeast. Nucleic Acids Res. 2002, 30, e23. [Google Scholar] [CrossRef] [PubMed]
- Reider Apel, A.; d’Espaux, L.; Wehrs, M.; Sachs, D.; Li, R.A.; Tong, G.J.; Garber, M.; Nnadi, O.; Zhuang, W.; Hillson, N.J.; et al. A Cas9-Based Toolkit to Program Gene Expression in Saccharomyces Cerevisiae. Nucleic Acids Res. 2016, 45, 496–508. [Google Scholar] [CrossRef]
- Hong, J.; Park, S.-H.; Kim, S.; Kim, S.-W.; Hahn, J.-S. Efficient Production of Lycopene in Saccharomyces Cerevisiae by Enzyme Engineering and Increasing Membrane Flexibility and NAPDH Production. Appl. Microbiol. Biotechnol. 2018, 103, 211–223. [Google Scholar] [CrossRef]
- Callari, R.; Meier, Y.; Ravasio, D.; Heider, H. Dynamic Control of ERG20 and ERG9 Expression for Improved Casbene Production in Saccharomyces Cerevisiae. Front. Bioeng. Biotechnol. 2018, 6, 160. [Google Scholar] [CrossRef] [PubMed]
- Conrado, R.J.; Varner, J.D.; DeLisa, M.P. Engineering the Spatial Organization of Metabolic Enzymes: Mimicking Nature’s Synergy. Curr. Opin. Biotechnol. 2008, 19, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Albertsen, L.; Chen, Y.; Bach, L.S.; Rattleff, S.; Maury, J.; Brix, S.; Nielsen, J.; Mortensen, U.H. Diversion of Flux toward Sesquiterpene Production in Saccharomyces Cerevisiae by Fusion of Host and Heterologous Enzymes. Appl. Environ. Microbiol. 2011, 77, 1033–1040. [Google Scholar] [CrossRef]
- Aalbers, F.S.; Fraaije, M.W. Enzyme Fusions in Biocatalysis: Coupling Reactions by Pairing Enzymes. Chembiochem 2019, 20, 20–28. [Google Scholar] [CrossRef]
- Chen, X.; Bai, Y.; Zaro, J.L.; Shen, W.C. Design of an in Vivo Cleavable Disulfide Linker in Recombinant Fusion Proteins. Biotechniques 2010, 49, 513–518. [Google Scholar] [CrossRef]
- Amet, N.; Lee, H.F.; Shen, W.C. Insertion of the Designed Helical Linker Led to Increased Expression of Tf-Based Fusion Proteins. Pharm. Res. 2009, 26, 523–528. [Google Scholar] [CrossRef]
- Reddy, S.T.; Ge, X.; Miklos, A.E.; Hughes, R.A.; Kang, S.H.; Hoi, K.H.; Chrysostomou, C.; Hunicke-Smith, S.P.; Iverson, B.L.; Tucker, P.W.; et al. Monoclonal Antibodies Isolated without Screening by Analyzing the Variable-Gene Repertoire of Plasma Cells. Nat. Biotechnol. 2010, 28, 965–969. [Google Scholar] [CrossRef]
- Bergeron, L.M.; Gomez, L.; Whitehead, T.A.; Clark, D.S. Self-Renaturing Enzymes: Design of an Enzyme-Chaperone Chimera as a New Approach to Enzyme Stabilization. Biotechnol. Bioeng. 2009, 102, 1316–1322. [Google Scholar] [CrossRef] [PubMed]
- Werner, S.; Marillonnet, S.; Hause, G.; Klimyuk, V.; Gleba, Y. Immunoabsorbent Nanoparticles Based on a Tobamovirus Displaying Protein A. Proc. Natl. Acad. Sci. USA 2006, 103, 17678–17683. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Feng, M.G. Bifunctional Enhancement of a Beta-Glucanase-Xylanase Fusion Enzyme by Optimization of Peptide Linkers. Appl. Microbiol. Biotechnol. 2008, 79, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.L.; Xue, C.; Du, J.L.; Ren, M.; Xia, S.; Liu, Z.M. Balancing the Pharmacokinetics and Pharmacodynamics of Interferon-A2b and Human Serum Albumin Fusion Protein by Proteolytic or Reductive Cleavage Increases Its in Vivo Therapeutic Efficacy. Mol. Pharm. 2012, 9, 664–670. [Google Scholar] [CrossRef]
- Guo, H.; Yang, Y.; Xue, F.; Zhang, H.; Huang, T.; Liu, W.; Liu, H.; Zhang, F.; Yang, M.; Liu, C.; et al. Effect of Flexible Linker Length on the Activity of Fusion Protein 4-Coumaroyl-CoA Ligase::Stilbene Synthase. Mol. Biosyst. 2017, 13, 598–606. [Google Scholar] [CrossRef]
- Weimer, T.; Wormsbächer, W.; Kronthaler, U.; Lang, W.; Liebing, U.; Schulte, S. Prolonged In-Vivo Half-Life of Factor VIIa by Fusion to Albumin. Thromb. Haemost. 2008, 99, 659–667. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, S.Z.; Li, J.; Pan, X.; Cahoon, R.E.; Jaworski, J.G.; Wang, X.; Jez, J.M.; Chen, F.; Yu, O. Using Unnatural Protein Fusions to Engineer Resveratrol Biosynthesis in Yeast and Mammalian Cells. J. Am. Chem. Soc. 2006, 128, 13030–13031. [Google Scholar] [CrossRef]
- Klein, J.S.; Jiang, S.; Galimidi, R.P.; Keeffe, J.R.; Bjorkman, P.J.; Regan, L. Design and Characterization of Structured Protein Linkers with Differing Flexibilities. Protein Eng. Des. Sel. 2014, 27, 325–330. [Google Scholar] [CrossRef]
- Subramaniam, S.; Kleywegt, G.J. A Paradigm Shift in Structural Biology. Nat. Methods 2022, 19, 20–23. [Google Scholar] [CrossRef]
- Noé, F.; De Fabritiis, G.; Clementi, C. Machine Learning for Protein Folding and Dynamics. Curr. Opin. Struct. Biol. 2020, 60, 77–84. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhou, Y.J.; Bao, J.; Huang, L.; Nielsen, J.; Krivoruchko, A. Metabolic Engineering of Saccharomyces Cerevisiae for Production of Germacrene A, a Precursor of Beta-Elemene. J. Ind. Microbiol. Biotechnol. 2017, 44, 1065–1072. [Google Scholar] [CrossRef]
- Wang, Y.; Gong, X.; Li, F.; Zuo, S.; Li, M.; Zhao, J.; Han, X.; Wen, M. Optimized Biosynthesis of Santalenes and Santalols in Saccharomyces Cerevisiae. Appl. Microbiol. Biotechnol. 2021, 105, 8795–8804. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, M.; Zhao, G.R.; Lu, W. Harnessing Yeast Peroxisomes and Cytosol Acetyl-CoA for Sesquiterpene α-Humulene Production. J. Agric. Food Chem. 2020, 68, 1382–1389. [Google Scholar] [CrossRef]
- Kurosaki, F.; Kato, T.; Misawa, N.; Taura, F.; Kurosaki, F.; Kato, T.; Misawa, N.; Taura, F. Efficient Production of δ-Guaiene, an Aroma Sesquiterpene Compound Accumulated in Agarwood, by Mevalonate Pathway-Engineered Escherichia Coli Cells. Adv. Biosci. Biotechnol. 2016, 7, 435–445. [Google Scholar] [CrossRef]
- Yu, F.; Harada, H.; Yamasaki, K.; Okamoto, S.; Hirase, S.; Tanaka, Y.; Misawa, N.; Utsumi, R. Isolation and Functional Characterization of a Beta-Eudesmol Synthase, a New Sesquiterpene Synthase from Zingiber Zerumbet Smith. FEBS Lett. 2008, 582, 565–572. [Google Scholar] [CrossRef]
- An, T.; Li, L.; Lin, Y.; Zeng, F.; Lin, P.; Zi, J. Characterization of Guaiene Synthases from Stellera Chamaejasme L. Flowers and Their Application in de Novo Production of (-)-Rotundone in Yeast. J. Agric. Food Chem. 2020, 68, 3214–3219. [Google Scholar] [CrossRef]
- Dusséaux, S.; Wajn, W.T.; Liu, Y.; Ignea, C.; Kampranis, S.C. Transforming Yeast Peroxisomes into Microfactories for the Efficient Production of High-Value Isoprenoids. Proc. Natl. Acad. Sci. USA 2020, 117, 31789–31799. [Google Scholar] [CrossRef]
- Hoepfner, D.; Schildknegt, D.; Braakman, I.; Philippsen, P.; Tabak, H.F. Contribution of the Endoplasmic Reticulum to Peroxisome Formation. Cell 2005, 122, 85–95. [Google Scholar] [CrossRef]
- Zhang, Y.; Nielsen, J.; Liu, Z. Engineering Yeast Metabolism for Production of Terpenoids for Use as Perfume Ingredients, Pharmaceuticals and Biofuels. FEMS Yeast Res. 2017, 17, fox080. [Google Scholar] [CrossRef] [PubMed]
- Shiba, Y.; Paradise, E.M.; Kirby, J.; Ro, D.K.; Keasling, J.D. Engineering of the Pyruvate Dehydrogenase Bypass in Saccharomyces Cerevisiae for High-Level Production of Isoprenoids. Metab. Eng. 2007, 9, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Meadows, A.L.; Hawkins, K.M.; Tsegaye, Y.; Antipov, E.; Kim, Y.; Raetz, L.; Dahl, R.H.; Tai, A.; Mahatdejkul-Meadows, T.; Xu, L.; et al. Rewriting Yeast Central Carbon Metabolism for Industrial Isoprenoid Production. Nature 2016, 537, 694–697. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).