Design of 5′-UTR to Enhance Keratinase Activity in Bacillus subtilis
Abstract
1. Introduction
2. Materials and Methods
2.1. Gene, Plasmids and Strains
2.2. Medium and Culture Conditions
2.3. Keratinase Activity
2.4. Analysis of 5′-UTR Secondary Structure
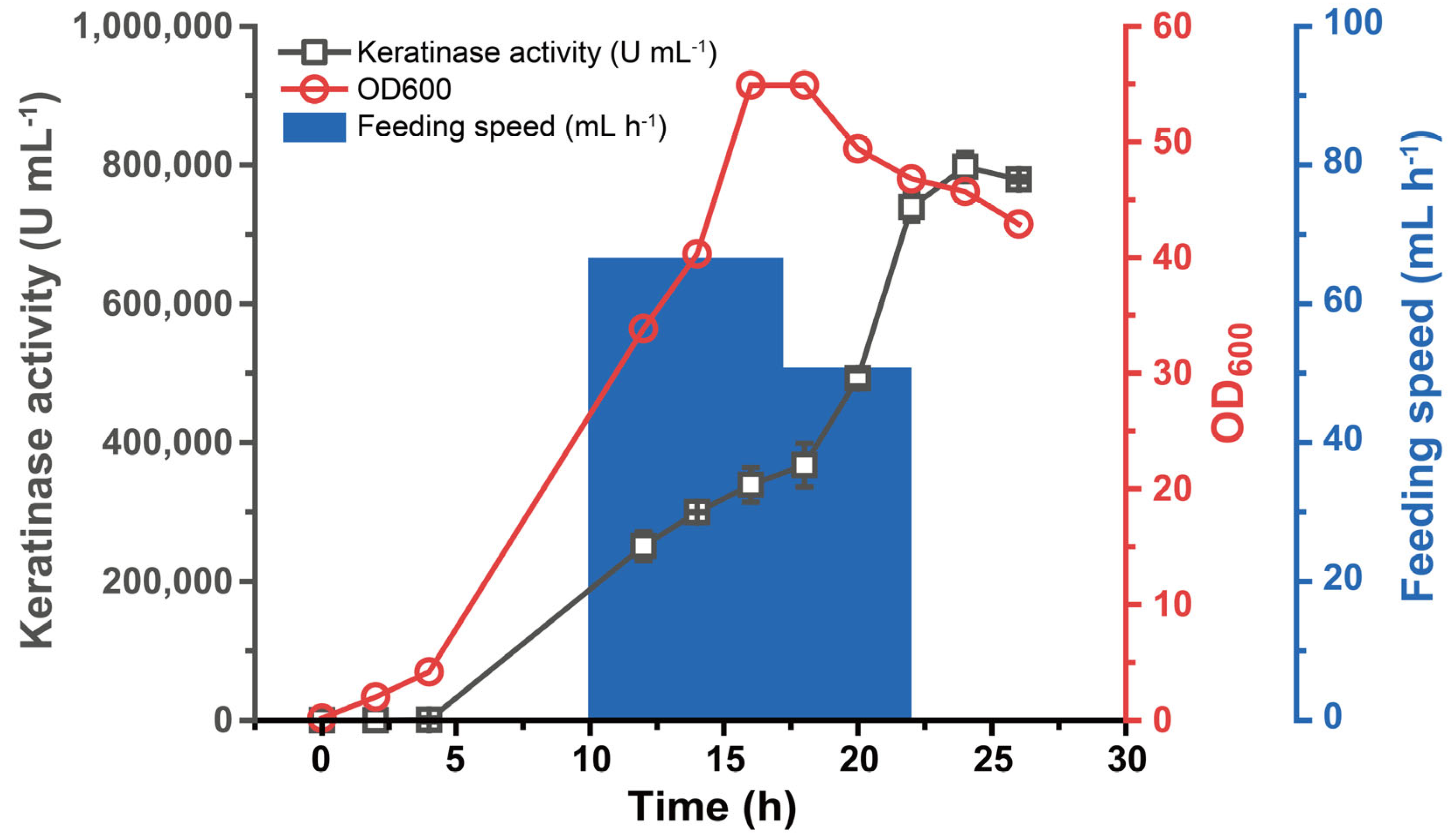
2.5. Fermentation Performance Validation in Fermenter
3. Results and Discussion
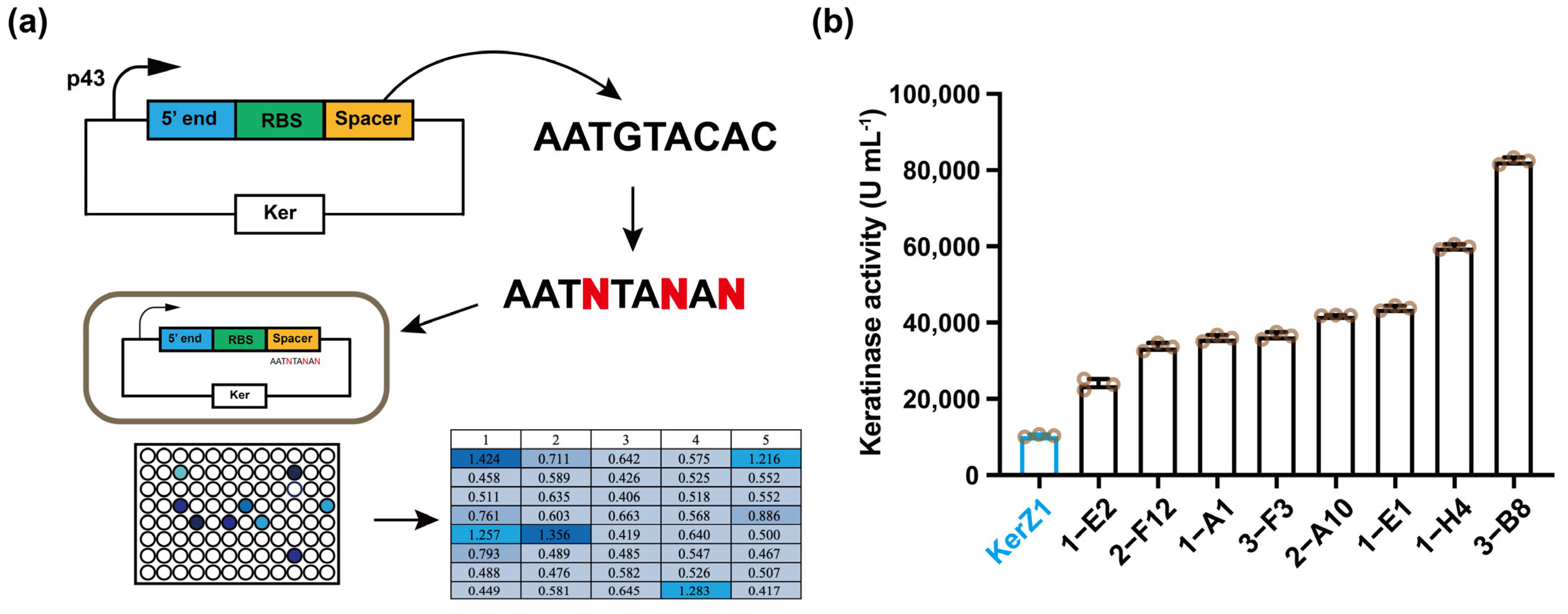
3.1. Replacement of Spacer Sequence C/G to A/T
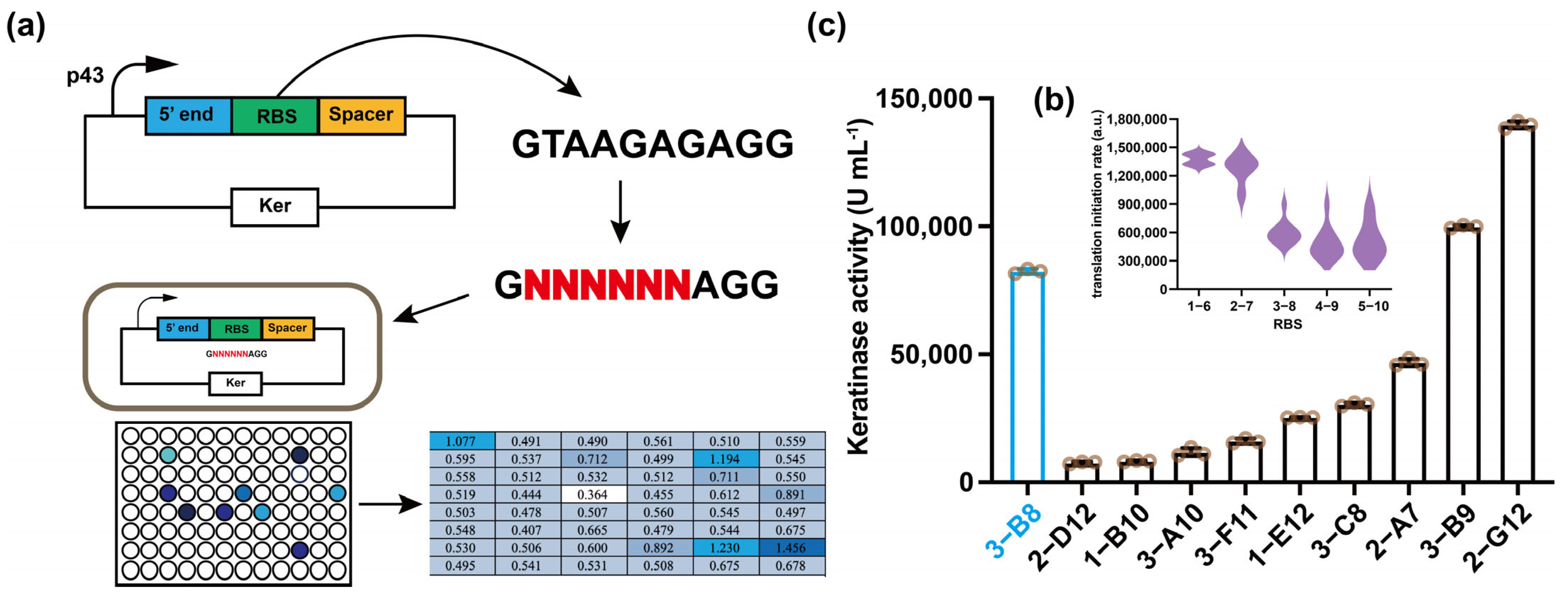
3.2. Screening of Ribosome Binding Sites (RBS)
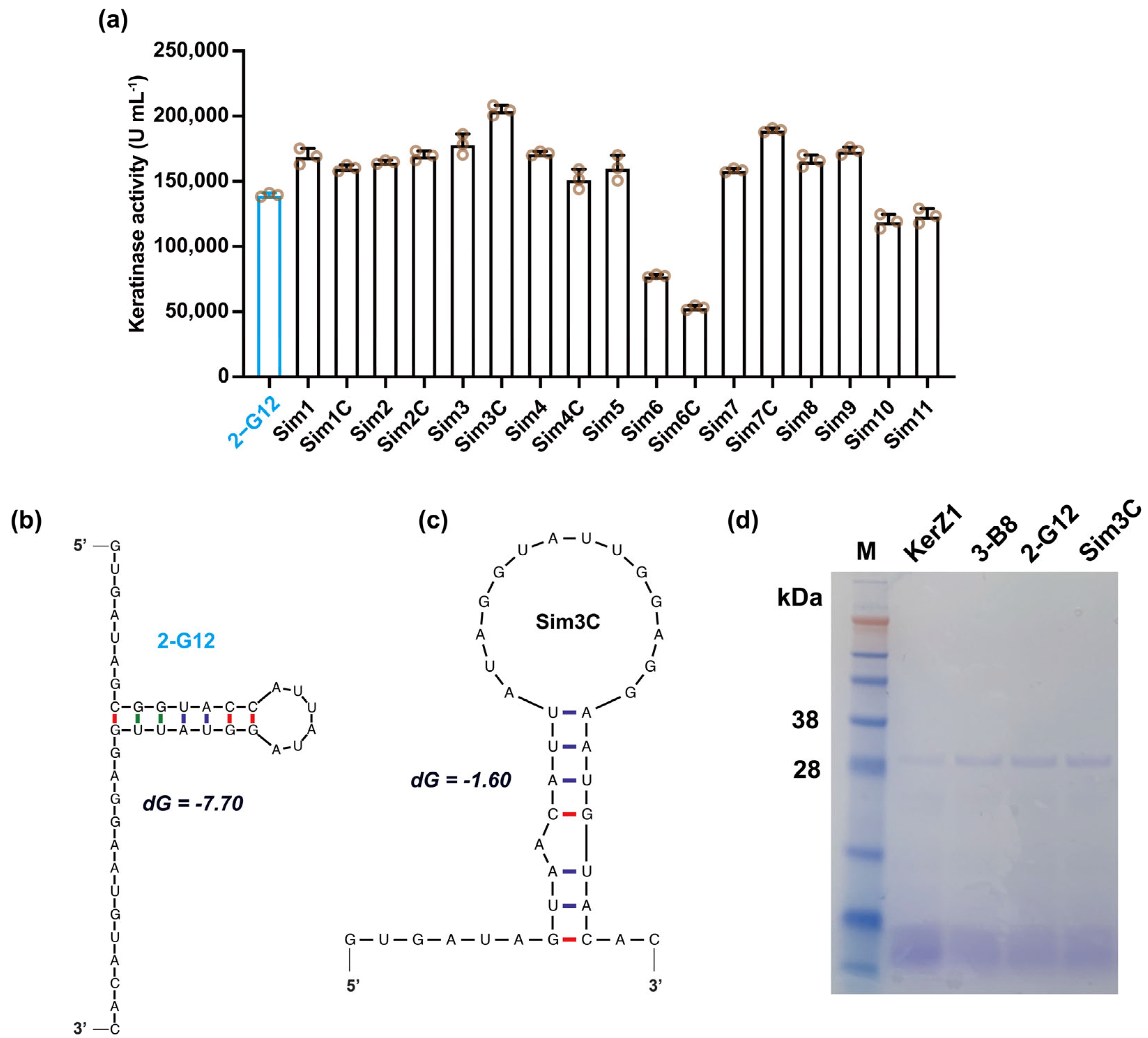
3.3. Simplification of the 5′ End Sequence
3.4. Fermentation Performance of B. subtilis WB600-SP-R-D
4. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, B.; Yang, W.; McKittrick, J.; Meyers, M.A. Keratin: Structure, mechanical properties, occurrence in biological organisms, and efforts at bioinspiration. Prog. Mater. Sci. 2016, 76, 229–318. [Google Scholar] [CrossRef]
- Meyers, M.A.; Chen, P.-Y.; Lin, A.Y.-M.; Seki, Y. Biological materials: Structure and mechanical properties. Prog. Mater. Sci. 2008, 53, 1–206. [Google Scholar] [CrossRef]
- Donato, R.K.; Mija, A. Keratin Associations with Synthetic, Biosynthetic and Natural Polymers: An Extensive Review. Polymers 2019, 12, 32. [Google Scholar] [CrossRef] [PubMed]
- Callegaro, K.; Brandelli, A.; Daroit, D.J. Beyond plucking: Feathers bioprocessing into valuable protein hydrolysates. Waste Manag. 2019, 95, 399–415. [Google Scholar] [CrossRef]
- Brandelli, A.; Daroit, D.J.; Riffel, A. Biochemical features of microbial keratinases and their production and applications. Appl. Microbiol. Biotechnol. 2010, 85, 1735–1750. [Google Scholar] [CrossRef]
- Fang, Z.; Yong, Y.-C.; Zhang, J.; Du, G.; Chen, J. Keratinolytic protease: A green biocatalyst for leather industry. Appl. Microbiol. Biotechnol. 2017, 101, 7771–7779. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, D.; Zhang, X. Enzymatic decolorization of melanoidins from molasses wastewater by immobilized keratinase. Bioresour. Technol. 2019, 280, 165–172. [Google Scholar] [CrossRef]
- Peng, Z.; Mao, X.; Mu, W.; Du, G.; Chen, J.; Zhang, J. Modifying the Substrate Specificity of Keratinase for Industrial Dehairing to Replace Lime-Sulfide. ACS Sustain. Chem. Eng. 2022, 10, 6863–6870. [Google Scholar] [CrossRef]
- Gupta, R.; Sharma, R.; Beg, Q.K. Revisiting microbial keratinases: Next generation proteases for sustainable biotechnology. Crit. Rev. Biotechnol. 2013, 33, 216–228. [Google Scholar] [CrossRef]
- Verma, A.; Singh, H.; Anwar, S.; Chattopadhyay, A.; Tiwari, K.K.; Kaur, S.; Dhilon, G.S. Microbial keratinases: Industrial enzymes with waste management potential. Crit. Rev. Biotechnol. 2017, 37, 476–491. [Google Scholar] [CrossRef]
- Qiu, J.; Wilkens, C.; Barrett, K.; Meyer, A.S. Microbial enzymes catalyzing keratin degradation: Classification, structure, function. Biotechnol. Adv. 2020, 44, 107607. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Gong, J.-S.; Qin, J.; Li, H.; Li, H.; Xu, Z.-H.; Shi, J.-S. The tale of a versatile enzyme: Molecular insights into keratinase for its industrial dissemination. Biotechnol. Adv. 2020, 45, 107655. [Google Scholar] [CrossRef] [PubMed]
- Daroit, D.J.; Brandelli, A. A current assessment on the production of bacterial keratinases. Crit. Rev. Biotechnol. 2014, 34, 372–384. [Google Scholar] [CrossRef]
- Prakash, P.; Jayalakshmi, S.K.; Sreeramulu, K. Purification and characterization of extreme alkaline, thermostable keratinase, and keratin disulfide reductase produced by Bacillus halodurans PPKS-2. Appl. Microbiol. Biotechnol. 2010, 87, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Zhang, J.; Song, Y.; Guo, R.; Du, G.; Chen, J. Engineered pro-peptide enhances the catalytic activity of keratinase to improve the conversion ability of feather waste. Biotechnol. Bioeng. 2021, 118, 2559–2571. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Gao, J.; He, J.; Yu, B.; Zheng, P.; Huang, Z.; Mao, X.; Yu, J.; Han, G.; Chen, D. Codon Optimization Significantly Improves the Expression Level of a Keratinase Gene in Pichia pastoris. PLoS ONE 2013, 8, e58393. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Gong, J.-S.; Sun, Y.-X.; Qin, J.; Zhai, S.; Li, H.; Li, H.; Lu, Z.-M.; Xu, Z.-H.; Shi, J.-S. Combining Pro-peptide Engineering and Multisite Saturation Mutagenesis To Improve the Catalytic Potential of Keratinase. ACS Synth. Biol. 2019, 8, 425–433. [Google Scholar] [CrossRef]
- Peng, Z.; Mao, X.; Zhang, J.; Du, G.; Chen, J. Biotransformation of keratin waste to amino acids and active peptides based on cell-free catalysis. Biotechnol. Biofuels 2020, 13, 61. [Google Scholar] [CrossRef]
- Fang, Z.; Zhang, J.; Liu, B.; Jiang, L.; Du, G.; Chen, J. Cloning, heterologous expression and characterization of two keratinases from Stenotrophomonas maltophilia BBE11-1. Process Biochem. 2014, 49, 647–654. [Google Scholar] [CrossRef]
- Cui, W.; Han, L.; Suo, F.; Liu, Z.; Zhou, L.; Zhou, Z. Exploitation of Bacillus subtilis as a robust workhorse for production of heterologous proteins and beyond. World J. Microbiol. Biotechnol. 2018, 34, 145. [Google Scholar] [CrossRef]
- Gingold, H.; Pilpel, Y. Determinants of translation efficiency and accuracy. Mol. Syst. Biol. 2011, 7, 481. [Google Scholar] [CrossRef] [PubMed]
- Espah Borujeni, A.; Salis, H.M. Translation Initiation is Controlled by RNA Folding Kinetics via a Ribosome Drafting Mechanism. J. Am. Chem. Soc. 2016, 138, 7016–7023. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, I.; Belardinelli, R.; Rodnina, M.V. Translation initiation in bacterial polysomes through ribosome loading on a standby site on a highly translated mRNA. Proc. Natl. Acad. Sci. USA 2018, 115, 4411–4416. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Li, Y.; Ma, A.; Liu, W.; Wang, H.; Zhuang, G. An efficient design strategy for a whole-cell biosensor based on engineered ribosome binding sequences. Anal. Bioanal. Chem. 2011, 401, 2891–2898. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, S.; Ding, R.; Chen, J.; Du, G.; Li, H.; Zhou, J. Obtaining a Panel of Cascade Promoter-5′-UTR Complexes in Escherichia coli. ACS Synth. Biol. 2017, 6, 1065–1075. [Google Scholar] [CrossRef]
- Zhang, D.; Zhou, C.Y.; Busby, K.N.; Alexander, S.C.; Devaraj, N.K. Light-Activated Control of Translation by Enzymatic Covalent mRNA Labeling. Angew. Chem. Int. Ed. Engl. 2018, 57, 2822–2826. [Google Scholar] [CrossRef]
- Viegas, S.C.; Apura, P.; Martinez-García, E.; de Lorenzo, V.; Arraiano, C.M. Modulating Heterologous Gene Expression with Portable mRNA-Stabilizing 5′-UTR Sequences. ACS Synth. Biol. 2018, 7, 2177–2188. [Google Scholar] [CrossRef]
- Salis, H.M.; Mirsky, E.A.; Voigt, C.A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009, 27, 946–950. [Google Scholar] [CrossRef]
- Cuperus, J.T.; Groves, B.; Kuchina, A.; Rosenberg, A.B.; Jojic, N.; Fields, S.; Seelig, G. Deep learning of the regulatory grammar of yeast 5′ untranslated regions from 500,000 random sequences. Genome Res. 2017, 27, 2015–2024. [Google Scholar] [CrossRef]
- Xiao, J.; Peng, B.; Su, Z.; Liu, A.; Hu, Y.; Nomura, C.T.; Chen, S.; Wang, Q. Facilitating Protein Expression with Portable 5′-UTR Secondary Structures in Bacillus licheniformis. ACS Synth. Biol. 2020, 9, 1051–1058. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Osterman, I.A.; Evfratov, S.A.; Sergiev, P.V.; Dontsova, O.A. Comparison of mRNA features affecting translation initiation and reinitiation. Nucleic Acids Res. 2013, 41, 474–486. [Google Scholar] [CrossRef]
- Lee, H.-M.; Ren, J.; Kim, W.Y.; Vo, P.N.L.; Eyun, S.-I.; Na, D. Introduction of an AU-rich Element into the 5’ UTR of mRNAs Enhances Protein Expression in Escherichia coli by S1 Protein and Hfq Protein. Biotechnol. Bioprocess Eng. 2021, 26, 749–757. [Google Scholar] [CrossRef]
- Zelcbuch, L.; Antonovsky, N.; Bar-Even, A.; Levin-Karp, A.; Barenholz, U.; Dayagi, M.; Liebermeister, W.; Flamholz, A.; Noor, E.; Amram, S.; et al. Spanning high-dimensional expression space using ribosome-binding site combinatorics. Nucleic Acids Res. 2013, 41, e98. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.R.; Yurovsky, A.; Chen, Y.; Skiena, S.; Futcher, B. Re-annotation of 12,495 prokaryotic 16S rRNA 3’ ends and analysis of Shine-Dalgarno and anti-Shine-Dalgarno sequences. PLoS ONE 2018, 13, e0202767. [Google Scholar] [CrossRef] [PubMed]
- Komarova, E.S.; Chervontseva, Z.S.; Osterman, I.A.; Evfratov, S.A.; Rubtsova, M.P.; Zatsepin, T.S.; Semashko, T.A.; Kostryukova, E.S.; Bogdanov, A.A.; Gelfand, M.S.; et al. Influence of the spacer region between the Shine–Dalgarno box and the start codon for fine-tuning of the translation efficiency in Escherichia coli. Microb. Biotechnol. 2020, 13, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, L.; Shin, H.-D.; Chen, R.R.; Li, J.; Du, G.; Chen, J. Pathway engineering of Bacillus subtilis for microbial production of N-acetylglucosamine. Metab. Eng. 2013, 19, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Lv, X.; Liu, Y.; Li, J.; Du, G.; Chen, J.; Rodrigo, L.-A.; Liu, L. Synthetic redesign of central carbon and redox metabolism for high yield production of N-acetylglucosamine in Bacillus subtilis. Metab. Eng. 2019, 51, 59–69. [Google Scholar] [CrossRef]
| Name | Sequence (5′-3′) |
|---|---|
| Strains | |
| JM109 | Escherichia coli |
| WB600 | Bacillus subtilis 168 derivate, missing nprE aprE epr bpr mpr nprB |
| WB600-Ker | Bacillus subtilis WB600 contains plasmid pP43NMK-Ker |
| Plasmids | |
| pP43NMK | Ampr, Kmr, E. coli–B. subtilis shuttle vector |
| pP43NMK-Ker | pP43NMK derivate with B. licheniformis Ker gene under the control of the promoter P43 |
| Primers | |
| Spacer | |
| Spacer-3N-F | TTATAGGTAAGAGAGGAATNTANANATGATGAGGAAAAAGAGTTTTTGGCTTGG |
| Spacer-3N-R | ATTCCTCTCTTACCTATAATGGTACCGCTAT |
| RBS | |
| RBS-6N-F | TAGCGGTACCATTATAGGNNNNNNAGGAATGTATAGATGATGAGGAAAAAGAGTTTTTG |
| RBS-6N-R | CCTATAATGGTACCGCTATCACTTTATATTTTACATAATCG |
| 5′ end | |
| Sim1-F | TAAAATATAAAGTGATAGCGTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim2-F | TAAAATATAAAGTGATAGGTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim3-F | TAAAATATAAAGTGATAGTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim4-F | ATTATGTAAAATATAAAGTATAGTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim5-F | ATTATGTAAAATATAAATATAGTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim6-F | ATGTAAAATATAAAGTGATGCGGTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim7-F | ATGTAAAATATAAAGTGAGCGGTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim1C-F | TAAAATATAAAGTGATAGCGTAACATTATAGGTATTGGAGGAATGTACAC |
| Sim2C-F | TAAAATATAAAGTGATAGGTAACATTATAGGTATTGGAGGAATGTACAC |
| Sim3C-F | TAAAATATAAAGTGATAGTAACATTATAGGTATTGGAGGAATGTACAC |
| Sim4C-F | ATTATGTAAAATATAAAGTATAGTAACATTATAGGTATTGGAGGAATGTACAC |
| Sim6C-F | ATGTAAAATATAAAGTGATGCGGTAACATTATAGGTATTGGAGGAATGTACAC |
| Sim7C-F | ATGTAAAATATAAAGTGAGCGGTAACATTATAGGTATTGGAGGAATGTACAC |
| Sim8-F | ATGTAAAATATAAAGTGGCGGTACCATTATAGGTATTGGAGGA |
| Sim9-F | GATTATGTAAAATATAAAGGGCGGTACCATTATAGGTATTGGAGGA |
| Sim10-F | ATTATGTAAAATATAAACGCGGTACCATTATAGGTATTGGAGGA |
| Sim11-F | ATTATGTAAAATATAAAGTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim123-R | TATCACTTTATATTTTACATAATCGCGCGCTTTTTTTC |
| Sim4591011-R | TTTATATTTTACATAATCGCGCGCTTTTTTTCACG |
| Sim678-R | CACTTTATATTTTACATAATCGCGCGCTTTTTTTC |
| Strains | Spacer (5′-3′) | Activity (U mL−1) |
|---|---|---|
| KerZ1 | AATGTACAC | 10,380 |
| 1-A1 1-E1 1-E2 1-H4 2-A10 2-F12 3-B8 3-F3 | AATTTACAT AATTTACAT AATTTACAC AATTTATAA AATTTACAT AATTTACAA AATGTATAG AATTTACAT | 35,935 43,800 23,745 59,845 41,915 33,615 82,435 36,565 |
| Strains | RBS (5′-3′) | Activity (U mL−1) | Translation Initiation Rate (au) |
|---|---|---|---|
| SP | GTAAGAGAGG | 82,435 | 72.08 |
| 1-B10 1-E12 2-A7 2-D12 2-G12 3-A10 3-B8 3-C8 3-F11 | GCTGCACAGG GCTTGCGAGG GGGAAGTAGG GATGGTAAGG GTATTGGAGG GAAAGACAGG GGACGGAAGG GTGTTGCAGG GGGGGCTAGG | 7695 25,350 45,740 7510 139,650 11,060 99,860 30,395 15,710 | 11.63 26.81 362.40 37.82 85.58 23.71 127.08 29.45 121.96 |
| Mutants | 5′-UTR Sequence (5′-3′) |
|---|---|
| 2-G12 | GTGATAGCGGTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim1 | GTGATAGC--GTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim1C | GTGATAGC--GTAACATTATAGGTATTGGAGGAATGTACAC |
| Sim2 | GTGATAG----GTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim2C | GTGATAG----GTAACATTATAGGTATTGGAGGAATGTACAC |
| Sim3 | GTGATA------GTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim3C | GTGATA------GTAACATTATAGGTATTGGAGGAATGTACAC |
| Sim4 | GT--ATA------GTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim4C | GT--ATA------GTAACATTATAGGTATTGGAGGAATGTACAC |
| Sim5 | --T--ATA------GTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim6 | GTGAT--GCGGTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim6C | GTGAT--GCGGTAACATTATAGGTATTGGAGGAATGTACAC |
| Sim7 | GTGA----GCGGTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim7C | GTGA----GCGGTAACATTATAGGTATTGGAGGAATGTACAC |
| Sim8 | GTG------GCGGTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim9 | G--G------GCGGTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim10 | ------------GCGGTACCATTATAGGTATTGGAGGAATGTACAC |
| Sim11 | ------------------GTACCATTATAGGTATTGGAGGAATGTACAC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, J.; Zhou, G.; Ji, X.; Zhang, G.; Peng, Z.; Zhang, J. Design of 5′-UTR to Enhance Keratinase Activity in Bacillus subtilis. Fermentation 2022, 8, 426. https://doi.org/10.3390/fermentation8090426
Fang J, Zhou G, Ji X, Zhang G, Peng Z, Zhang J. Design of 5′-UTR to Enhance Keratinase Activity in Bacillus subtilis. Fermentation. 2022; 8(9):426. https://doi.org/10.3390/fermentation8090426
Chicago/Turabian StyleFang, Jun, Guanyu Zhou, Xiaomei Ji, Guoqiang Zhang, Zheng Peng, and Juan Zhang. 2022. "Design of 5′-UTR to Enhance Keratinase Activity in Bacillus subtilis" Fermentation 8, no. 9: 426. https://doi.org/10.3390/fermentation8090426
APA StyleFang, J., Zhou, G., Ji, X., Zhang, G., Peng, Z., & Zhang, J. (2022). Design of 5′-UTR to Enhance Keratinase Activity in Bacillus subtilis. Fermentation, 8(9), 426. https://doi.org/10.3390/fermentation8090426






