The Dynamics of Single-Cell Nanomotion Behaviour of Saccharomyces cerevisiae in a Microfluidic Chip for Rapid Antifungal Susceptibility Testing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Cultivation
2.2. Microfluidic Chip Construction
2.3. Computational Simulation of Fluid Flow in the Microfluidic Chip
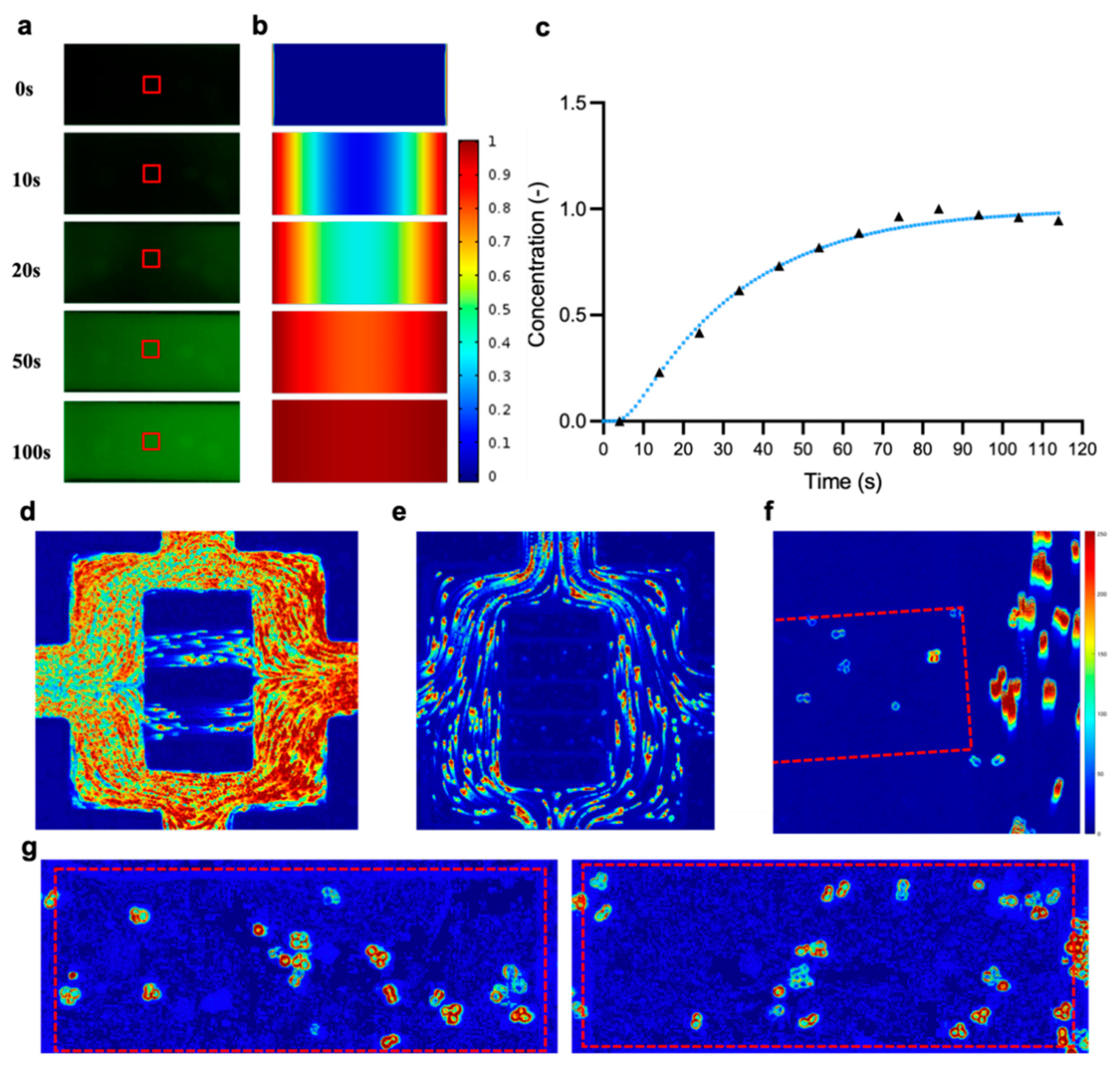
2.4. Determination of the Diffusion Coefficient in the Imaging Chamber
2.5. Microfluidic Chip Setup
2.6. Nanomotion Measurement and Analysis
3. Results
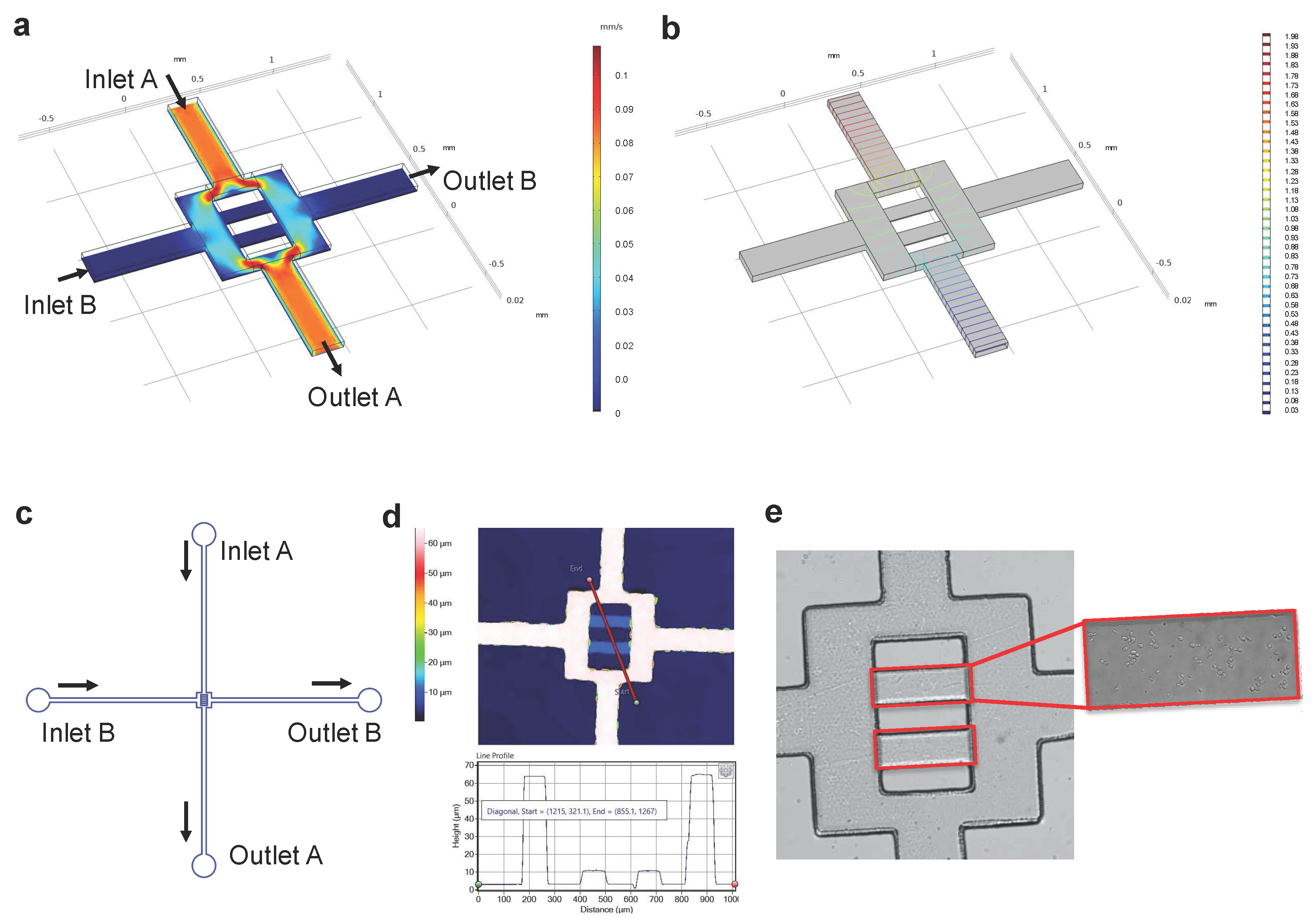
3.1. Design and Construction of the Microfluidic Chip
3.1.1. Modelling Fluid Flow and Mass Transport in the Microfluidic Chip
3.1.2. Microfluidic Chip Construction
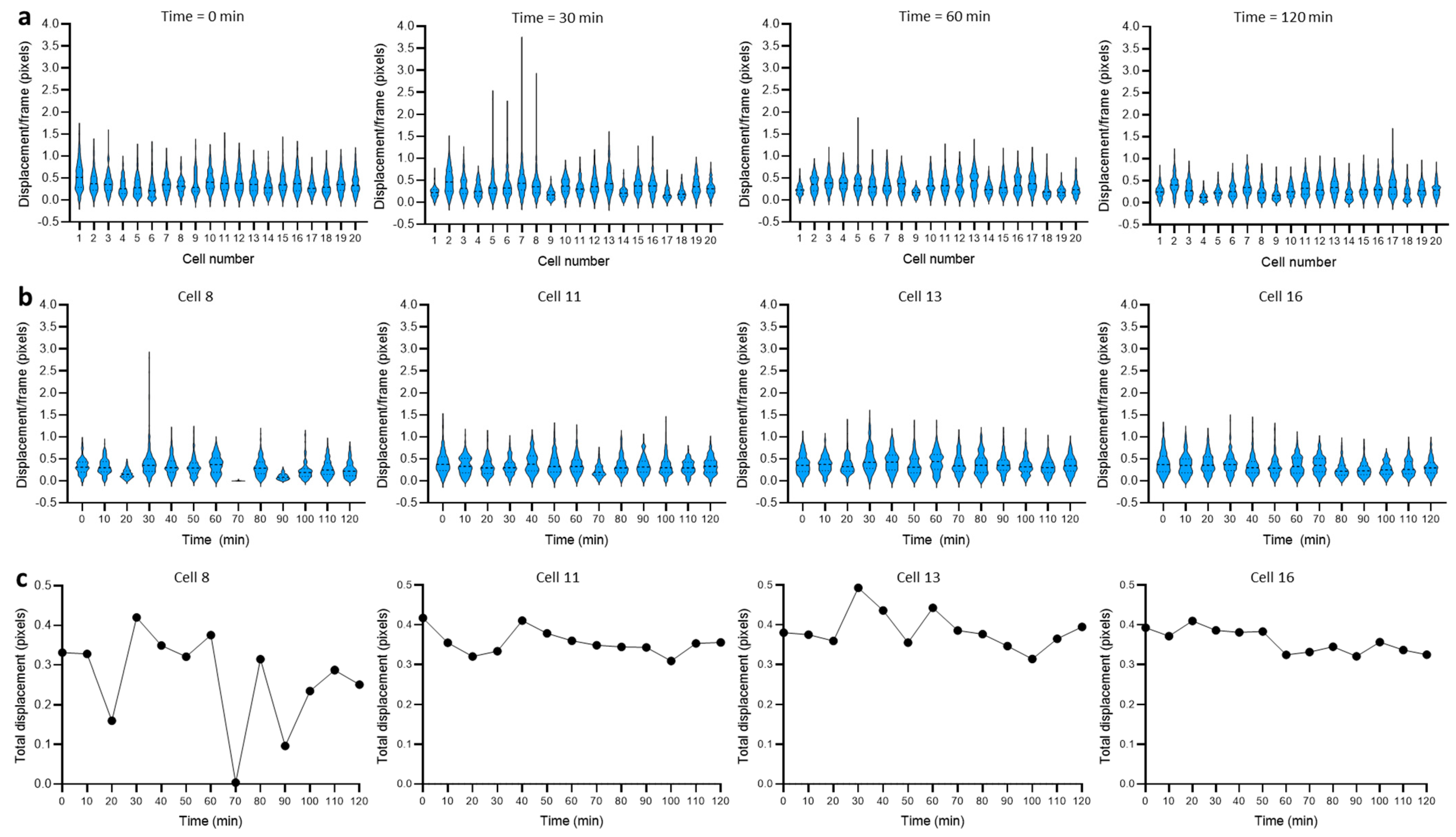
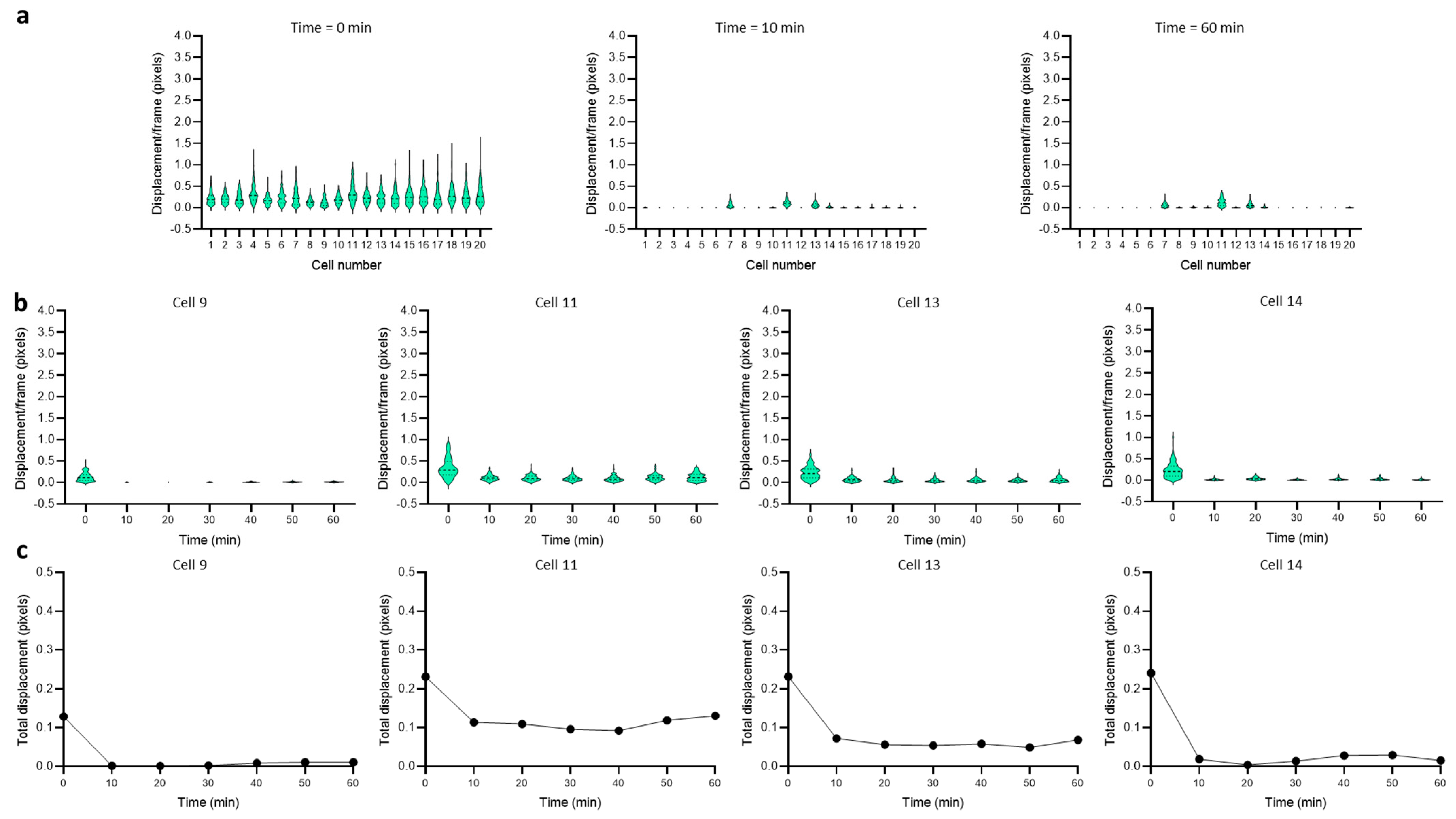
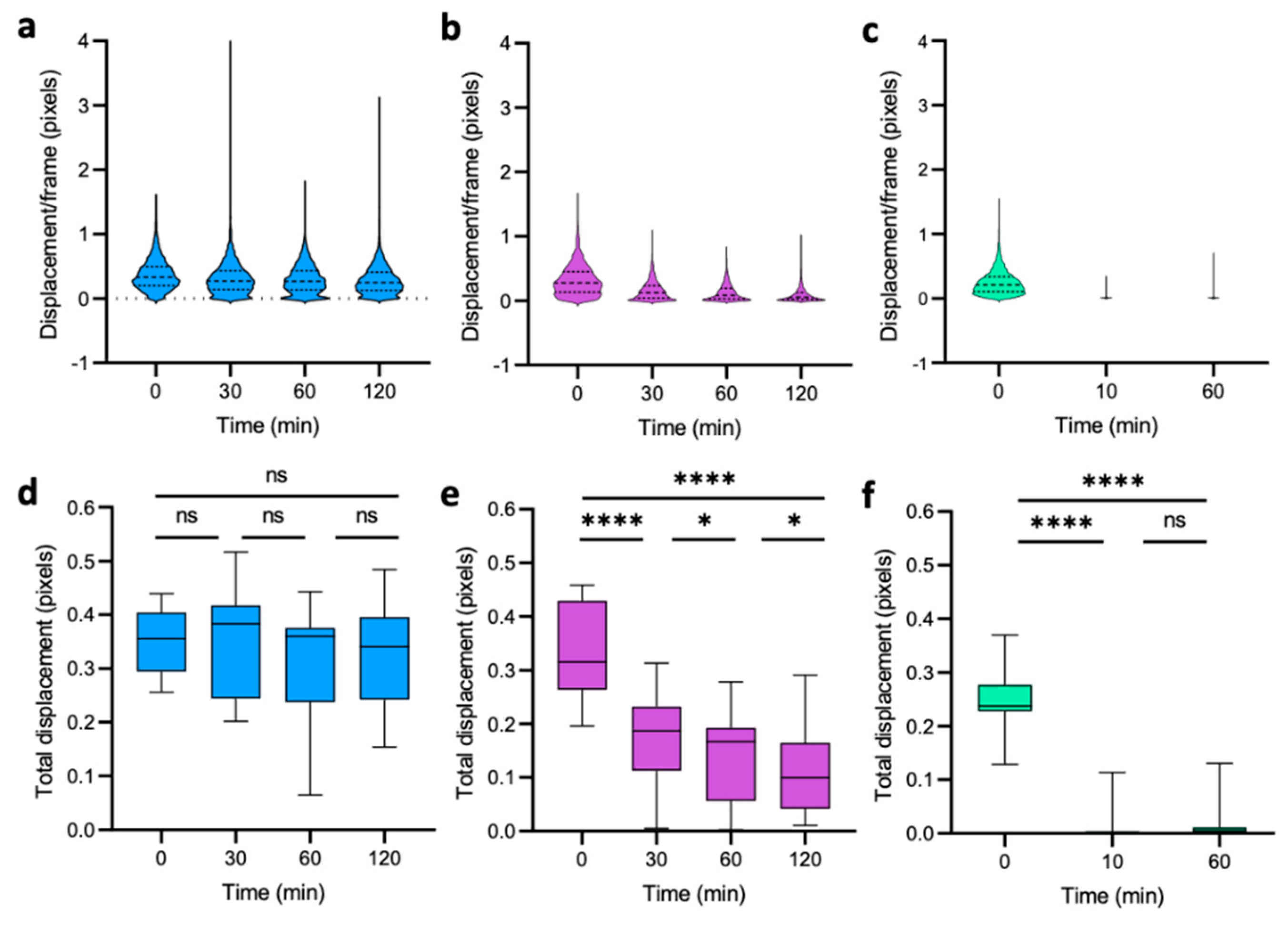
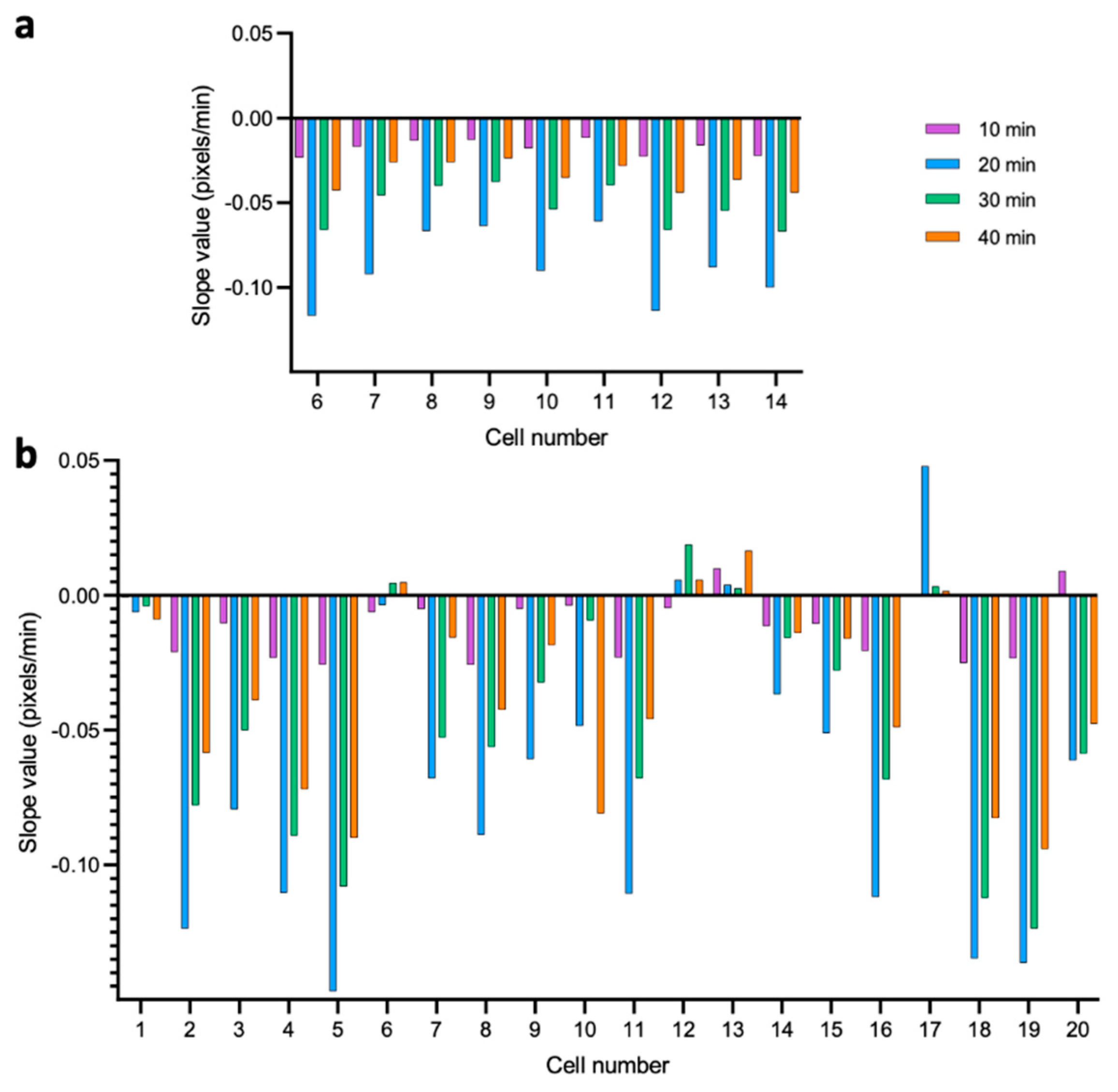
3.2. Dynamics of Single-Cell Nanomotion upon Treatment by a Killing Compound
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and multi-national prevalence of fungal diseases—Estimate precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef] [PubMed]
- Denning, D.W. Calling upon all public health mycologists: To accompany the country burden papers from 14 countries. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 923–924. [Google Scholar] [CrossRef] [PubMed]
- Geddes-McAlister, J.; Shapiro, R.S. New pathogens, new tricks: Emerging, drug-resistant fungal pathogens and future prospects for antifungal therapeutics. Ann. N. Y. Acad. Sci. 2019, 1435, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Beardsley, J.; Halliday, C.L.; Chen, S.C.-A.; Sorrell, T.C. Responding to the emergence of antifungal drug resistance: Perspectives from the bench and the bedside. Future Microbiol. 2018, 13, 1175–1191. [Google Scholar] [CrossRef] [Green Version]
- Willaert, R. Micro- and Nanoscale Approaches in Antifungal Drug Discovery. Fermentation 2018, 4, 43. [Google Scholar] [CrossRef] [Green Version]
- Lemke, A.; Kiderlen, A.F.; Kayser, O. Amphotericin B. Appl. Microbiol. Biotechnol. 2005, 68, 151–162. [Google Scholar] [CrossRef]
- Bellmann, R. Pharmacodynamics and Pharmacokinetics of Antifungals for Treatment of Invasive Aspergillosis. Curr. Pharm. Des. 2013, 19, 3629–3647. [Google Scholar] [CrossRef]
- Pfaller, M.A. Antifungal Drug Resistance: Mechanisms, Epidemiology, and Consequences for Treatment. Am. J. Med. 2012, 125, S3–S13. [Google Scholar] [CrossRef]
- Pianalto, K.M.; Alspaugh, J.A. New horizons in antifungal therapy. J. Fungi 2016, 2, 26. [Google Scholar] [CrossRef]
- Nivoix, Y.; Ubeaud-Sequier, G.; Engel, P.; Leveque, D.; Herbrecht, R. Drug-Drug Interactions of Triazole Antifungal Agents in Multimorbid Patients and Implications for Patient Care. Curr. Drug Metab. 2009, 10, 395–409. [Google Scholar] [CrossRef]
- Lewis, R.E. Current concepts in antifungal pharmacology. In Mayo Clinic Proceedings; Elsevier Ltd.: Amsterdam, The Netherlands, 2011; Volume 86, pp. 805–817. [Google Scholar]
- McCarthy, M.W.; Walsh, T.J. Drugs currently under investigation for the treatment of invasive candidiasis. Expert Opin. Investig. Drugs 2017, 26, 825–831. [Google Scholar] [CrossRef] [PubMed]
- Perfect, J.R. The antifungal pipeline: A reality check. Nat. Rev. Drug Discov. 2017, 16, 603–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuentefria, A.M.; Pippi, B.; Dalla Lana, D.F.; Donato, K.K.; de Andrade, S.F. Antifungals discovery: An insight into new strategies to combat antifungal resistance. Lett. Appl. Microbiol. 2018, 66, 2–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, L.N.; de Mello, T.P.; de Souza Ramos, L.; Branquinha, M.H.; dos Santos, A.L.S. New and Promising Chemotherapeutics for Emerging Infections Involving Drug-resistant Non-albicans Candida Species. Curr. Top. Med. Chem. 2019, 19, 2527–2553. [Google Scholar] [CrossRef] [PubMed]
- Sousa, F.; Ferreira, D.; Reis, S.; Costa, P. Current insights on antifungal therapy: Novel nanotechnology approaches for drug delivery systems and new drugs from natural sources. Pharmaceuticals 2020, 13, 248. [Google Scholar] [CrossRef] [PubMed]
- Mota Fernandes, C.; Dasilva, D.; Haranahalli, K.; McCarthy, J.B.; Mallamo, J.; Ojima, I.; Del Poeta, M. The Future of Antifungal Drug Therapy: Novel Compounds and Targets. Antimicrob. Agents Chemother. 2021, 65, 65. [Google Scholar] [CrossRef] [PubMed]
- Scorzoni, L.; Fuchs, B.B.; Junqueira, J.C.; Mylonakis, E. Current and promising pharmacotherapeutic options for candidiasis. Expert Opin. Pharmacother. 2021, 22, 867–887. [Google Scholar] [CrossRef]
- Posteraro, B.; Torelli, R.; De Carolis, E.; Posteraro, P.; Sanguinetti, M. Antifungal susceptibility testing: Current role from the clinical laboratory perspective. Mediterr. J. Hematol. Infect. Dis. 2014, 6, e2014030. [Google Scholar] [CrossRef] [Green Version]
- Campbell, J.; McBeth, C.; Kalashnikov, M.; Boardman, A.K.; Sharon, A.; Sauer-Budge, A.F. Microfluidic advances in phenotypic antibiotic susceptibility testing. Biomed. Microdevices 2016, 18, 103. [Google Scholar] [CrossRef] [Green Version]
- Kohler, A.C.; Venturelli, L.; Longo, G.; Dietler, G.; Kasas, S. Nanomotion detection based on atomic force microscopy cantilevers. Cell Surf. 2019, 5, 100021. [Google Scholar] [CrossRef]
- Longo, G.; Alonso-Sarduy, L.; Rio, L.M.; Bizzini, A.; Trampuz, A.; Notz, J.; Dietler, G.; Kasas, S. Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nat. Nanotechnol. 2013, 8, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Kasas, S.; Ruggeri, F.S.; Benadiba, C.; Maillard, C.; Stupar, P.; Tournu, H.; Dietler, G.; Longo, G. Detecting nanoscale vibrations as signature of life. Proc. Natl. Acad. Sci. USA 2015, 112, 378–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villalba, M.I.; Stupar, P.; Chomicki, W.; Bertacchi, M.; Dietler, G.; Arnal, L.; Vela, M.E.; Yantorno, O.; Kasas, S. Nanomotion Detection Method for Testing Antibiotic Resistance and Susceptibility of Slow-Growing Bacteria. Small 2018, 14, 1702671. [Google Scholar] [CrossRef] [PubMed]
- Venturelli, L.; Kohler, A.C.; Stupar, P.; Villalba, M.I.; Kalauzi, A.; Radotic, K.; Bertacchi, M.; Dinarelli, S.; Girasole, M.; Pešić, M.; et al. A perspective view on the nanomotion detection of living organisms and its features. J. Mol. Recognit. 2020, 33, e2849. [Google Scholar] [CrossRef]
- Kasas, S.; Malovichko, A.; Villalba, M.I.; Vela, M.E.; Yantorno, O.; Willaert, R.G. Nanomotion Detection-Based Rapid Antibiotic Susceptibility Testing. Antibiotics 2021, 10, 287. [Google Scholar] [CrossRef]
- Syal, K.; Iriya, R.; Yang, Y.; Yu, H.; Wang, S.; Haydel, S.E.; Chen, H.-Y.; Tao, N. Antimicrobial Susceptibility Test with Plasmonic Imaging and Tracking of Single Bacterial Motions on Nanometer Scale. ACS Nano 2016, 10, 845–852. [Google Scholar] [CrossRef]
- Johnson, W.L.; France, D.C.; Rentz, N.S.; Cordell, W.T.; Walls, F.L. Sensing bacterial vibrations and early response to antibiotics with phase noise of a resonant crystal. Sci. Rep. 2017, 7, 12138. [Google Scholar] [CrossRef] [Green Version]
- Syal, K.; Shen, S.; Yang, Y.; Wang, S.; Haydel, S.E.; Tao, N. Rapid Antibiotic Susceptibility Testing of Uropathogenic E. coli by Tracking Submicron Scale Motion of Single Bacterial Cells. ACS Sens. 2017, 2, 1231–1239. [Google Scholar] [CrossRef]
- Bermingham, C.R.; Murillo, I.; Payot, A.D.J.; Balram, K.C.; Kloucek, M.B.; Hanna, S.; Redmond, N.M.; Baxter, H.; Oulton, R.; Avison, M.B.; et al. Imaging of sub-cellular fluctuations provides a rapid way to observe bacterial viability and response to antibiotics. bioRxiv 2018, 460139. [Google Scholar] [CrossRef] [Green Version]
- Leonard, H.; Halachmi, S.; Ben-Dov, N.; Nativ, O.; Segal, E. Unraveling Antimicrobial Susceptibility of Bacterial Networks on Micropillar Architectures Using Intrinsic Phase-Shift Spectroscopy. ACS Nano 2017, 11, 6167–6177. [Google Scholar] [CrossRef]
- Willaert, R.G.; Vanden Boer, P.; Malovichko, A.; Alioscha-Perez, M.; Radotić, K.; Bartolić, D.; Kalauzi, A.; Villalba, M.I.; Sanglard, D.; Dietler, G.; et al. Single yeast cell nanomotions correlate with cellular activity. Sci. Adv. 2020, 6, eaba3139. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guizar-Sicairos, M.; Thurman, S.T.; Fienup, J.R. Efficient subpixel image registration algorithms. Opt. Lett. 2008, 33, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radomsky, M.L.; Whaley, K.J.; Cone, R.A.; Saltzman, W.M. Macromolecules released from polymers: Diffusion into unstirred fluids. Biomaterials 1990, 11, 619–624. [Google Scholar] [CrossRef]
- Saltzman, W.M.; Radomsky, M.L.; Whaley, K.J.; Cone, R.A. Antibody diffusion in human cervical mucus. Biophys. J. 1994, 66, 508–515. [Google Scholar] [CrossRef] [Green Version]
- Soeller, C.; Jacobs, M.D.; Donaldson, P.J.; Cannell, M.B.; Jones, K.T.; Ellis-Davies, G.C.R. Application of two-photon flash photolysis to reveal intercellular communication and intracellular Ca2+ movements. J. Biomed. Opt. 2003, 8, 418. [Google Scholar] [CrossRef]
- Casalini, T.; Salvalaglio, M.; Perale, G.; Masi, M.; Cavallotti, C. Diffusion and aggregation of sodium fluorescein in aqueous solutions. J. Phys. Chem. B 2011, 115, 12896–12904. [Google Scholar] [CrossRef] [Green Version]
- Rosłoń, I.E.; Japaridze, A.; Steeneken, P.G.; Dekker, C.; Alijani, F. Probing nanomotion of single bacteria with graphene drums. bioRxiv 2021, 2021.09.21.461186. [Google Scholar] [CrossRef]
- Howell, A.S.; Lew, D.J. Morphogenesis and the Cell Cycle. Genetics 2012, 190, 51–77. [Google Scholar] [CrossRef] [Green Version]
- Pandey, A.K.; Kumar, M.; Kumari, S.; Kumari, P.; Yusuf, F.; Jakeer, S.; Naz, S.; Chandna, P.; Bhatnagar, I.; Gaur, N.A. Evaluation of divergent yeast genera for fermentation-associated stresses and identification of a robust sugarcane distillery waste isolate Saccharomyces cerevisiae NGY10 for lignocellulosic ethanol production in SHF and SSF. Biotechnol. Biofuels 2019, 12, 40. [Google Scholar] [CrossRef] [Green Version]
- Schiavone, M.; Formosa-Dague, C.; Elsztein, C.; Teste, M.-A.; Martin-Yken, H.; De Morais, M.A.; Dague, E.; François, J.M. Evidence for a Role for the Plasma Membrane in the Nanomechanical Properties of the Cell Wall as Revealed by an Atomic Force Microscopy Study of the Response of Saccharomyces cerevisiae to Ethanol Stress. Appl. Environ. Microbiol. 2016, 82, 4789–4801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanley, D.; Bandara, A.; Fraser, S.; Chambers, P.J.; Stanley, G.A. The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. J. Appl. Microbiol. 2010, 109, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Eigenfeld, M.; Kerpes, R.; Becker, T. Understanding the Impact of Industrial Stress Conditions on Replicative Aging in Saccharomyces cerevisiae. Front. Fungal Biol. 2021, 2, 665490. [Google Scholar] [CrossRef]
- Kristanc, L.; Božič, B.; Jokhadar, Š.Z.; Dolenc, M.S.; Gomišček, G. The pore-forming action of polyenes: From model membranes to living organisms. Biochim. Biophys. Acta-Biomembr. 2019, 1861, 418–430. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radonicic, V.; Yvanoff, C.; Villalba, M.I.; Kasas, S.; Willaert, R.G. The Dynamics of Single-Cell Nanomotion Behaviour of Saccharomyces cerevisiae in a Microfluidic Chip for Rapid Antifungal Susceptibility Testing. Fermentation 2022, 8, 195. https://doi.org/10.3390/fermentation8050195
Radonicic V, Yvanoff C, Villalba MI, Kasas S, Willaert RG. The Dynamics of Single-Cell Nanomotion Behaviour of Saccharomyces cerevisiae in a Microfluidic Chip for Rapid Antifungal Susceptibility Testing. Fermentation. 2022; 8(5):195. https://doi.org/10.3390/fermentation8050195
Chicago/Turabian StyleRadonicic, Vjera, Charlotte Yvanoff, Maria Ines Villalba, Sandor Kasas, and Ronnie G. Willaert. 2022. "The Dynamics of Single-Cell Nanomotion Behaviour of Saccharomyces cerevisiae in a Microfluidic Chip for Rapid Antifungal Susceptibility Testing" Fermentation 8, no. 5: 195. https://doi.org/10.3390/fermentation8050195
APA StyleRadonicic, V., Yvanoff, C., Villalba, M. I., Kasas, S., & Willaert, R. G. (2022). The Dynamics of Single-Cell Nanomotion Behaviour of Saccharomyces cerevisiae in a Microfluidic Chip for Rapid Antifungal Susceptibility Testing. Fermentation, 8(5), 195. https://doi.org/10.3390/fermentation8050195







