Abstract
Low-molecular-mass iron-reducing compounds (IRCs) were produced by entomopathogenic endophytic fungi Lecanicillium sp. ATA01 in liquid cultures. The extracellular hydrophilic extract contained three IRCs formed by peptides, iron and phenolate structures with molecular masses of 1207, 567 and 550 Da. These compounds were able to chelate and mediate the reduction of Fe+3 to Fe+2 and oxidized recalcitrant lignin-model substrates such as veratryl alcohol (VA), 2,6-dimethoxyphenol (DMP), and 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid (ABTS) with or without hydrogen peroxide. Besides, IRCs can promote the degradation of chlorophenols. The maximal degradation of p-chlorophenol, 2,4-dichlorophenol, 2,4,6-trichlorophenol, and pentachlorophenol was conducted at optimal degradation conditions for IRCs (pH 3.5, iron 100 mM, and H2O2 10 mM). Furthermore, Fenton-like reactions using the synthetic iron chelates DTPA and EDTA and free Fe+2 and Fe+3 were also carried out in order to compare with the reaction mediated by IRCs. The ferric IRCs displayed the ability to enhance the hydroxylation of chlorophenols as a part of a degradation mechanism of the IRC-assisted Fenton reaction. The complexed iron was more efficient than free iron in the Fenton-like reaction, and between them, the fungal chelates were more efficient than the synthetic mill chelates.
1. Introduction
The highly toxic and persistent environmental pollutants known as chlorophenol (CP) compounds (and derivatives) are commonly used in the manufacture of industrial products such as dyes, drugs and pesticides. The excessive use of these industrial products and their discharge in effluents have led to the contamination of soil and water [1]. The CPs are aromatic ring structures of benzene rings containing a hydroxyl (-OH) and a chlorine atom (-Cl), which include monochlorophenols (o-Chlorophenol, C6H5ClO), polychlorophenols (-ClnOH), chloronitrophenols (-NO2), chloraminophenols (-NH2) and chloromethylphenols (-CH3). The acute toxicity of these compounds lies in their resistance to biodegradation, tendency to bioaccumulate, and carcinogenic, mutagenic, and cytotoxic properties [2]. Several physicochemical and biological methods have been used for the removal of CPs from the environment [3]. However, microbial degradation techniques are considered an interesting cost-effective and eco-friendly approach [1]. Despite advances in this topic, there are still gaps of knowledge about the mechanisms of degradation of CPs, the toxicity of their derivatives as well as their degradation by-products [4].
Endophytic fungi of healthy trees have been recognized as priority colonizers that initiate wood decay [5]. Moreover, the ability to produce lignocellulolytic enzymes and wood biodegradation under controlled conditions has been previously reported [6]. However, little is known about the enzymatic and non-enzymatic mechanisms present in the endophytic fungi isolated from extreme environments, the ability to degrade complex natural polymers (e.i., lignocellulose, chitin, keratin) and its emerging potential and versatility for the biodegradation of recalcitrant pollutants [7]. In this sense, the wood-decay mechanisms displayed by white- and brown-rot fungi, besides enzymes, involve the activity of low-molecular-mass oxidants such as lignin-peroxidase (LiP) mediators, organic-acid Mn+3 chelates, laccase mediators, peroxyl radicals produced from lipid peroxidation, and iron-reducing compounds (IRC) (the hydroxyl-radical production via Fenton chemistry, eq.1). A two-step process is carried out: (i) an oxidative-radical-based system via the IRCs produced by extracellular Fenton chemistry Equation (1), and (ii) the non-enzymatic disruption of the lignocellulose matrix by hydroxyl-radical attack (further detailed reaction in [8]).
Fe2+ + H2O2 + H+→ Fe3+ + OH• + H2O
The induction of extracellular hydroxyl-radical production by while-rot fungi through quinone redox cycling was previously reported by [9]. The non-specific mechanisms used by these fungi to degrade substrates allow them to degrade even complex mixtures of pollutants as well as to produce highly reactive oxygen species [8,10]. The generation of these species depends on the chelating agents and reaction conditions. Additionally, the chelate-modified Fenton process has been reported as very efficient in the degradation of contaminants in water [11]. However, little is known about the influence of some IRCs or their degradation products on the reactivity of iron [12].
It has been reported that not only do IRCs have a high affinity for Fe+3, but importantly, they mediate the reduction of this metal in redox-cycling processes under acidic conditions [13]. Depending on the nature of the chelate, iron concentration and other environmental parameters, Fe+3 may remain bound to the chelate (Equation (2)—reduction process) or can be released in the reduced form of Fe+2 (Equation (3)—oxidation process). The reduced state of iron can react with hydrogen peroxide, resulting in the generation of oxygen radicals through Fenton-type reaction mechanisms [14].
R + Fe2+ → R− + Fe3+
R• + Fe3+ → R+ + Fe2+
Fe2+ + H2O2 → Fe3+ + OH• + HO−
The advanced oxidation processes (AOPs) such as Fenton promote the decomposition of harmful organic compounds into less-dangerous inorganic compounds and/or biodegradable compounds through the addition of a hydroxyl radical to the aromatic or heterocyclic ring or the addition of a hydroxyl group to the double bonds of unsaturated compounds.
Rodriguez et al. [15] analyzed different examples of the Fenton reaction driven by hydroxy benzenes and its application in the oxidation of lignin- and pulp-bleaching effluents. The catechol compounds 2,3-dihydroxybenzoic and 3,4-dihydroxyphenylacetic acid were used to mediate the reduction of Fe+3 to Fe+2. Moreover, these compounds, as catechol–iron complexes, were able to promote the subsequent oxidation of o-dianisidine.
Paszczynski et al. [16] isolated 4,5-dimethoxycatechol (DMC) and 2,5-dimethoxyhydroquinone (DMH) from the brown-rot fungus Gloeophyllum trabeum. DMC and DMH may serve as ferric chelates, oxygen-reducing agents, and redox-cycling molecules, which would include working as electron-transport carriers in Fenton-type reactions. The evidence for an extracellular, hydroquinone-driven Fenton-type reaction involving DMH in the biodegradative mechanism of G. trabeum was reported [13]. On the other hand, extracellular oxidation may occur due to ascomycetes laccases or hydroxyl-radical attacks [17].
The Lecanicillium species belong to a group of entomopathogenic fungi and are among the most studied and virulent fungi related to the order Hypocreales (Fungi: Ascomycota). Previous reports have recognized their ability to secrete cell-wall-degrading enzymes such as chitinases and lipases [18] as well as extracellular Mn-peroxidase [19], which is useful during the decomposition of complex biopolymers. The Lecanicillium species are ubiquitous and are distributed in a wide range of habitats including temperate, alpine and antarctic ecosystems [20]. Currently, there is great interest in exploring the potential use of Lecanicillium species not only as an efficient mycoinsecticide [21] but also in the hydrocarbon biodegradation of hazardous pollutants, including polycyclic aromatic hydrocarbons, anthraquinone-type dyes, and oil [19,22]. As a result, the endophytic fungi Lecanicillium ATA01 (Ascomycota, Hypocreales), which was isolated from the extreme environment of the Atacama Puna Plateau, was selected as the target species for chlorophenol degradation.
The aim of this study pursued the potential application of low-molecular-mass iron-reducing compounds (IRCs), which are produced by the endophytic fungi Lecanicillium ATA01, as promoters of the bioremediation of chlorophenol contamination through the Fenton reaction. To achieve this, in the present work the degradation of the chlorinated phenolic compounds p-chlorophenol (p-CP), 2,4-dichlorophenol (2,4-DCP), 2,4,6-trichlorophenol (2,4,6-TCP) and pentachlorophenol (PCP) by IRCs from Lecanicillium ATA01 were investigated. Under the same conditions, free Fe+2 and Fe+3 and the iron complexes DTPA-Fe+2 (diethylenetriaminepentaacetic acid) and EDTA-Fe+2 (ethylenediaminetetraacetic acid) were also carried out as different scenarios for comparison.
2. Materials and Methods
2.1. Culture, DNA Extraction, Amplification, Sequencing, and Phylogenetic Analysis
The ATA01 strain culture was grown at 22 °C in solid PDA (potato dextrose agar) medium for 15 days. Genomic DNA was isolated from a single monosporic colony using a DNeasy blood and tissue kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer’s procedure. PCR amplification of the ITS1-5.8S-ITS2 region was carried out using primers ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) [23]. The PCR product was analyzed on 1.5% agarose gel and subsequently purified with PCR Cleanup kit (Axygen, Corning, NY, USA). The ITS1-5.8S-ITS2 region was sequenced at Macrogen (Inc., Seoul, Korea). The GenBank/EMBL/DDBJ accession number for the ITS1-5.8S-ITS2 region sequence of the strain is MT422210.
2.2. Morphological and Molecular Characterization of Fungal Isolate
Macro- and micro-morphological features of fungal colonies and molecular tools were used in the identification of the fungal isolate. To study the morphological features, agar plugs (6 mm diameter) obtained from single monosporic, two-week-old cultures were inoculated in the center of ten Petri dishes (100 × 17 mm) containing PDA medium and incubated at 25 °C in darkness. Macroscopic characteristics such as colony growth and appearance of mycelia in potato-dextrose-broth (PDB) medium were recorded. For micro-morphological features, the size and shape of macro-and microconidia, phialide size and their arrangement were recorded (n = 15). For the microscopic characterization, micro-cultures were prepared on slides covered by a thin layer of water agar and incubated in a wet chamber at 22 °C for a week. Then, a cover slide was placed on the fungal growth top and the dimensions of 50 conidia were recorded. The measurements were made using a Leica DMRB research microscope fitted with a Moticam 2300 digital camera.
The phylogenetic analysis was performed using the PhyML 3.3 available on the ATGC bioinformatics platform (http://www.atgc-montpellier.fr/ access date: 26 January 2022). The best substitution model for the maximum-likelihood analysis was found by the “Smart Model Selection” (SMS) software based on the Bayesian information criterion [24]. The best-fit model of nucleotide substitution according to the Bayesian information criteria (BIC) was GTR, which was used for the maximum-likelihood-tree reconstructions using IQ_TREE v1.6.10 [25]. Branch-support values were provided with 1000 bootstrap replicates by the SH-like approximate-likelihood-ratio test (SH-aLRT); the ultrafast bootstrap [26] and approximate-Bayes test [27] were implemented in IQ-TREE. Trees were visualized using iTOL v4 (https://itol.embl.de/ access date: 28 January 2022; [28]).
2.3. Production, Purification, and Characterization of IRCs
Liquid cultures of Lecanicillium ATA01 were grown in 20 g of malt extract (Difco, NJ, USA) and 5 g of peptone (Difco, Difco, NJ, USA) per liter, in 5 L Erlenmeyer flasks at 22 °C in stationary conditions. The cultures were incubated for 20 days, which is the time required to yield maximum extracellular IRCs. Cultures were collected and filtered through Whatman No. 2 filter paper. The crude extract was concentrated and lyophilized from 1 L to 50 mL, followed by ultrafiltration in an Amicon system (Amicon Corp., Lexington, MA, USA)with a nominal-molecular-weight cut-off of 3000 Da. After that, the crude extract of low-molecular-mass compounds was acidified to pH 2.5 with concentrated HCl, followed by solvent extraction with an equal volume of ethyl acetate. Ethyl-acetate fractions were combined and evaporated to dryness, and the residue was resuspended in deionized and distilled water. The Amberlite XAD-4 (Organo, Japan) ion-chromatography resin was used to eliminate ions and other impurities. Subsequent rinses were performed with deionized water (Fraction A) and methanol (Fraction B) as mobile phases. Individual fractions were collected, combined, and lyophilized to 5 mL. Figure 1 shows the schematic procedure for low-molecular-weight iron-reducing-compound (IRC) isolation from Lecanicillium ATA01. Purification of fraction A was performed by HPLC-RP using a Jasco PU 980 detector (Japan Spectroscopic, Tokyo, Japan) provided with a UV lamp (Jasco UV-970) and an integrator (Jasco 807-IT). A reverse-phase column, Cosmosil C18-AR (5 μm, 4.6 mm i.d. × 25 cm, Nakarai tesque, Kyoto, Japan), and the mobile phase of methanol and 0.08% phosphate buffer (10:90, flow rate 1.0 mL/min) were used. The chromatograms were carried out in isocratic conditions.
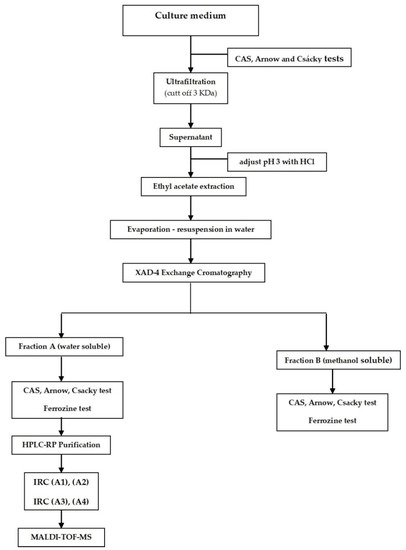
Figure 1.
Isolation procedure for low-molecular-weight iron-reducing compounds (IRCs) produced by Lecanicillium isolate ATA01.
The IRCs isolated from Lecanicillium sp. ATA01 were termed desferri-IRCs and ferri-IRCs to discriminate between the desferri and ferri iron-complex forms, respectively. To detect the production and the occurrence of a functional group of IRCs, the Chrome Azurol-S (CAS) universal assay [29], Csáky assay [30] and Arnow assay [31] were used for the detection of the total, hydroxamate type and catechol type of iron chelates, respectively. The reduction of Fe+3 to Fe+2 as well as the Fe+2 concentration in low-molecular-weight IRCs was measured using a modified ferrozine method [32]. The detection of reducing sugar in the IRC was determined by using a modification of the Somogyi–Nelson assay [33] using a glucose standard. Protein concentration was determined using the protein-microassay commercial kit BioRad reagent following a Bradford assay [34]. Bovine serum albumin (BSA) was used as a protein standard.
Phenoloxidase-type activities (PO×) were determined by monitoring the oxidation of recalcitrant lignin-model substrates as follows: (hereby as acronyms) DMP (2,6-dimethoxyphenol, Fluka, Honeywell Specialty Chemicals Seelze GmbH, Seelze, Germany); ABTS (2,2′-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid), Sigma-Aldrich, St. Louis, MO, USA), and VA (veratryl alcohol: 3,4-Dimethoxyphenyl, Sigma Aldrich, St. Louis, MO, USA). The oxidation of ABTS (ε420 = 3.6 × 104 M−1 cm−1) was monitored by determining the increase in the absorbance (λ420 nm), using 0.5 mM of the substrate in 0.1 M Na acetate and pH 5. The oxidation of 2,6-dimethoxyphenol (10 mM) was measured by monitoring the increase of the absorbance at 270 nm due to malonate–Mn+3 complex formation (ε270 = 11.59 mM−1 cm−1) in 50 mM sodium-malonate buffer containing 0.2 mM MnSO4 and 0.1 mM H2O2 at pH 4.0. The oxidation of veratryl alcohol was measured by the appearance of veratraldehyde (ε310 = 9.30 mM−1 cm−1). The assay was performed in 100 mM sodium-tartrate buffer at pH 3, containing 2.0 mM in the presence of 0.4 mM H2O2. Oxidation reactions were started by adding H2O2. Control assays were carried out in the absence of the IRC-Fe solution of H2O2. All oxidation rates were determined at 30 °C in a double-beam spectrophotometer and were reported as the average of four replicates. Phenoloxidase-like activities were expressed as µkat mL−1, and this is defined as the amount of enzyme or substance that converts 1 mol of substrate per second. The low-molecular-weight compound with the higher Fe+3/Fe+2 reduction capacity as well as POx activity was used in the following chlorophenol-degradation experiments.
The MALDI-TOF-MS mass-spectrometry technique was used to characterize low-molecular-weight iron-reducing compounds. All mass-spectra data were recorded according to the procedure described for the Pepseptive Voyager Elite MALDI-TOF-MS mass spectrometer. The matrix used was α-cyano-4-hydroxycinnamic acid (CHCA). The mass spectra were collected under the following conditions: linear mode, accelerating voltage = 20 kV, 50 ns of pulse delay and 203 averaged laser pulses. Enzymatic digestion by using CPY 1 (carboxypeptidase 1) was performed on IRCs to demonstrate the presence of protein or peptide structures.
2.4. Optimization of Chlorophenol Degradation by IRC
All the chlorinated chemical compounds—p-chlorophenol, 2,4-dichlorophenol, 2,4,6-trichlorophenol and pentachlorophenol—were purchased from Tokyo Kasei Kogyo (Tokyo, Japan). New experiments were conducted to optimize the degradation properties of desferri-IRC. The reaction conditions were as follows: 1 mL of d-IRC extracts, 25 μM p-CP, 1 mM Fe+3, 1 mM H2O2 and 20 mM phosphate buffer at pH 7.
2.5. DTPA-Fe+2 and EDTA-Fe+2 Iron Complexes
Iron chelates, DTPA-Fe+2 and EDTA-Fe+2 were formed by overnight incubation under N2 atmosphere, according to the previously described methodology [35].
2.6. Degradation of Chlorophenols
After optimization of the reaction conditions of desferri-IRC, all subsequent reactions with the desferri-IRC and synthetic chelates were carried out in the dark in triplicate sets with a 3 mL reaction volume of 20 mM phosphate buffer (pH 3.5) containing desferri-IRC (1 mL), 25 μM chlorinated compounds, 10 mM H2O2 and 500 μM Fe+3. The reaction was incubated under agitation conditions, in darkness, at 25 °C and 120 rpm in a water-bath shaker. After the prescribed time, the samples were analyzed by HPLC.
2.7. Monitoring of Chlorophenol Degradation by High-Performance Liquid Chromatography (HPLC)
Reactions were monitored by HPLC using a Jasco PU 980 (Spectroscopic, Tokyo, Japan) equipped with a UV detector (Jasco UV-970) and an integrator (Jasco 807-IT). A reverse-phase column, Cosmosil C18-AR (5 μm, 4.6 mm i.d. × 25 cm, Nakalai tesque, Kyoto Japan), and the mobile phase of methanol and 0.08% phosphate buffer (flow rate 1.0 mL/min) were used. The chromatograms were carried out in isocratic conditions defined for each chlorophenol by using a methanol:phosphoric-acid (0.08%) ratio, p-CP (50:50), 2,4-DCP (60:40), 2,4,6-TCP (70:30) and PCP (85:15). The figures show the average result of triplicate samples.
2.8. Identification of Degradation Products
The degradation products of p-CP after the p-CP/IRC/Fe+3/H2O2 reaction were derived and analyzed by GC/MS. After the reaction time, the mixture was acidified with HCl to pH 2, saturated with NaCl, and extracted with ethyl acetate. The organic fraction was dried over anhydrous sodium sulfate, evaporated under N2, and analyzed by gas chromatography/mass spectrometry (GC/MS) after acetylation by acetic anhydride/pyridine (1:1, v/v). The products formed were analyzed by GC/MS, HP5971/HP5890 series II (Hewlett Packard, Palo Alto, CA, USA). An HP-1 fused-silica capillary column (0.25 mm × 30 cm) coated with crosslinked methyl siloxane underwent a gradient temperature change: 65 °C for 15 min, 65–120 °C at 35 deg/min, 120–300 °C a 7 deg/min and then 300 °C for 5 min.
2.9. Rate Constant and Yield of Chlorophenol Degradation by Fenton-like Reaction
The exponential phase for each experimental treatment was determined in order to calculate the rate constant of the logarithmic phase of the reaction and the yield of chlorophenol degradation at the beginning of the stationary phase as well as at the end of the total reaction time. The linearization method was applied, and the reaction rate constant was calculated at the logarithmic slope using the following equation: k = logn ((C0 − C1) + 1)/logn (t), where C0 corresponds to the initial percentage of chlorophenol at the beginning reaction in time 0; C1 corresponds to the percentage of chlorophenol in the reaction mixture at the end the logarithmic slope (t = end time logn in h). The rate constants and yield of degradation were determined by assuming a pseudo-first-order model for Fenton-like oxidations.
2.10. Statistical Analyses
Statistics were performed using R version 3.6.3. The effects of the reaction parameters (pH, H2O2, and iron concentrations) on p-PC degradation were analyzed using a univariate ANOVA function [36] in the R package “car”. Normality and homoscedasticity tests were performed using the Shapiro–Wilk and Levene tests. To test the temporal degradation of phenolic compounds, univariate repeated-measure ANOVA [36] was performed using the R package “ez”. Statistical differences among rates of chlorophenol degradation treatments were evaluated using a one-way ANOVA, followed by the Tukey HSP multiple comparisons and a posteriori test using the package dplyr 37. The sphericity assumption was tested using the Mauchly test. If the sphericity assumption was not fulfilled, Greenhouse–Geisser and Huynh–Feldt corrections were applied [37]. Post hoc multiple comparisons were implemented using lsmeans (least-square means) and adjustment of p-values by the Tukey test in the R package “lsmeans”. All statistical analyses were carried out at 95% confidence levels with R statistical software.
3. Results
3.1. Fungal Identification by Morphological and Molecular Characterization
Cultural, morphological, and molecular characterization of the isolate ATA01 confirmed the identification as the entomopathogenic fungus belonging to genus Lecanicillium. Isolate ATA01 presented, after ten days at 20 °C on PDA plates, white cottony colonies reaching 3.5–3.8 cm diameter with a bulky growth in the center up to an average height of 3.6 mm and oval to ellipsoidal microconidia (2.77 ± 0.47 µm × 1.14 ± 0.13 µm). Phialides formed an oblique angle with the conidiophore stipe, 15.74 ± 2.96 µm long, gradually tapering from 1.57 ± 0.27 µm to 0.72 ± 0.11 µm at the tips (Figure 2a). Phialides were singly produced on vegetative hyphae. Conidia were transversely positioned on phialide, formed in a small slimy head. The analysis of these cultural and morphometric characteristics allowed the identification that was performed in the preliminary examination of the isolates to be confirmed, although further studies on the identification and characterization at the molecular level are recommended. The morphological identification was supported by a phylogenetic analysis that showed that this strain had a close relationship with other species of genus Lecanicillium (Figure 2b).
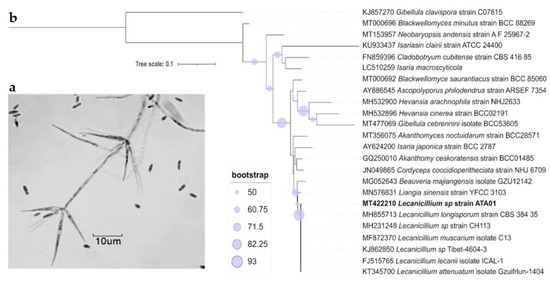
Figure 2.
Fungal identification by morphological and molecular characterization. (a) Phialides forming an oblique angle with the conidiophore stipe, and (b) maximum-likelihood phylogenetic tree, of 24 sequences from nearest genus and species of Lecanicillium, based on the partial internal transcribed spacer (ITS) inferred using IQ-TREE with GTR nucleotide-substitution model and 1000 bootstrap replicates.
3.2. Production and Characterization of IRCs
Following the isolation and purification steps described in Figure 1, two fractions were collected and termed fraction A (water soluble) and fraction B (methanol soluble). Then CAS, Arnow, Csáky, Somogyi–Nelson, Bradford and ferrozine assays, as well as phenoloxidase-type activities were performed with both fractions. The ability to reduce Fe+3 to Fe+2 as well as the oxidation of substrates were mainly found in fraction A while fraction B did not significantly react. Fraction A obtained marked positive reaction in the CAS, Arnow, Bradford and Ferrozine assays, while the Csáky and Somogyi–Nelson assays were negative. The reaction between the CAS reagent and iron-reducing compounds showed that almost 80% of the CAS-Fe+3 blue-color complex (λmax 630 nm) disappeared while at the same time the CAS reaction solution became a purple-reddish color after 10 min. This result demonstrated the ability to remove and the high affinity for iron present in CAS-reagent complex by IRCs. The positive reactions in the CAS and Arnow assays show that IRCs are iron chelators that contain phenolate-type moieties with iron-reducing ability.
Figure 3 shows the changes in absorbance at 562 nm (Ferrozine assay) during the reaction mixtures containing the desferri-IRC before and after free Fe+3 addition. The absorbance showed a small increment when ferrozine was added to the reaction mixture in the presence only of desferri-IRC. A rapid increment in the absorbance was observed after Fe+3 addition. The absorption spectra did not change over 40 and 80 s before and after iron addition, respectively. The rate of ferri-IRC complexing was improved when air and O2 were supplied to the reaction mixture.
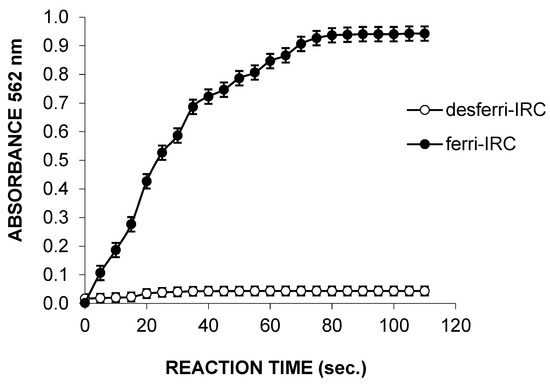
Figure 3.
Reduction of Fe+3 to Fe+2 by desferri-IRCs using ferrozine reagent.
Figure S1 (Supplementary material) shows an HPLC chromatogram of fraction A containing four compounds defined as A1, A2, A3 and A4, which are hydrophilic. Each peak was separated, recovered and phenoloxidase-type activities were assayed. The higher POx-type activities were found in peaks A1, A2, and A4, while A3 did not show activity. Phenoloxidase-type activities were not detected in the crude culture broth, although the purified extract showed a positive reaction. The phenoloxidase-type activities of ferri-IRCs were higher with H2O2 than without H2O2. In all cases, phenoloxidase-type activities for VA, 2,6-DMP and ABTS substrates were 7, 3.6, and 1.8 times higher, respectively, in the presence of hydrogen peroxide than in its absence (Figure 4).
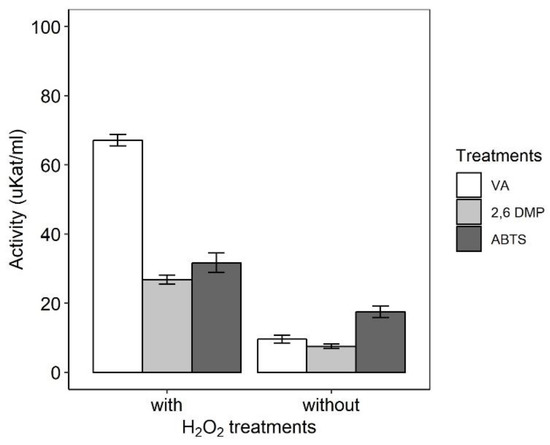
Figure 4.
Phenoloxidase-type activities mediated by ferri-IRCs in the presence (with) or absence (without) of H2O2. Symbols: (VA) veratryl alcohol, (2,6 DMP) 2,6-dimethoxyphenol and (ABTS) 2,2′-azino-bis-[3-ethyl-benzothiazoline-(6)-sulphonic acid].
MALDI-TOF-MS was used to characterize and determine the molecular masses of IRCs. The molecular weights of IRCs were A1 = 1207 Da, A2 = 567 Da, A3 = 550 Da and A4 = 522 Da (Supplementary material, Figure S2). The CPY1 digestion and MALDI-TOF-MS analysis confirmed the presence of peptide structures in the IRCs (data not shown).
3.3. Optimization of Chlorophenol Degradation
To optimize the degradation properties of desferri-IRCs (displayed in a previous experiment), the reaction parameters of pH, H2O2, and iron concentration were investigated (Figure 5). For all the degradation experiments, p-CP was used as a model chlorine compound, the parameters were optimized one by one and consequently incorporated into the following experiments. The degradation of p-CP was significantly affected by the main effects of the reaction parameter (Supplementary material, Table S1). A significant increase in the degradation rate was achieved between pH 2–3.5, whereas at higher pH values, the degradation rate was significantly lower (Figure 5a). The high rate and efficiency of p-CP degradation were detected after 76 h at H2O2 concentrations between 10–50 mM. The rate and efficiency of p-CP degradation significantly decreased when increasing the H2O2 concentration above 100 mM, and at 150 mM of H2O2 p-CP degradation did not occur. Nevertheless, a degradation of 15% was found in the absence of H2O2 in the reaction mixture (Figure 5b). Solutions of p-CP were incubated at various concentrations of Fe+3 (0–125 μM). Significantly higher degradation values (almost 100%) were obtained at 100 and 125 μM Fe+3 after 12 h of incubation. On the contrary, this rate significantly decreased at lower Fe+3, even after 76 h of incubation (Figure 5c). A degradation of 15% was found when iron was not added to the reaction mixture.
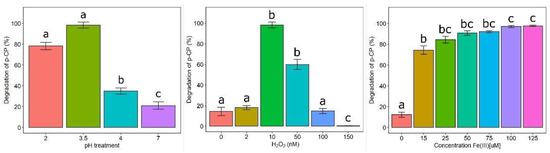
Figure 5.
(a) Effect of pH on p-CP degradation by IRC/Fe+3/H2O2; (b) Effect of H2O2 concentration on the p-CP degradation by IRC/Fe+3/H2O2; (c) Effect of ferric-iron concentration on p-CP degradation by IRC/Fe+3/H2O2. Finally, different lower-case letters denote significant differences between treatment groups (Tukey post-hoc; p < 0.05; Table S1, supplementary data).
3.4. Degradation of Chlorinated Phenolic Compounds by Ferric-IRCs
At the optimized conditions for p-CP degradation (pH 3.5, H2O2 10 mM and Fe+3, 100 μM), the degradation of other chlorophenol experiments was carried out. The degradation of different chlorinated phenolic compounds was significantly affected by the group treatments, time and the interaction between both factors (Supplementary material, Table S2). Figure 6 shows the degradation of chlorinated phenolic compounds mediated by ferri-IRC. Also, kinetic parameters of Fenton -mediated chlorophenol degradation by low-molecular-weight iron-reducing compounds are showed (Table 1). No significant differences were found between p-CP, 2,4-DCP and 2,4,6-TCP (Supplementary material, Table S3). It can be observed that p-CP was completely degraded by ferri-IRCs in 7 h, while the degradation rates of 2,4-DCP and 2,4,6-TCP were 76% and 75%, respectively (Figure 6). After 16 h, the 100% degradation of 2,4-DCP and 2,4,6-TCP was observed (Figure 6). PCP degradation was significantly lower than the other chlorophenols. Even after 46 h of incubation, the removal of PCP was 63% (Figure 6; Supplementary material, Table S3).
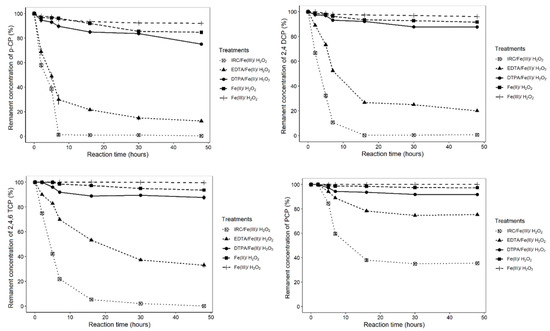
Figure 6.
Remanent concentration of chlorophenols (p-CP, 2,4-DCP, 2,4,6-TCP and PCP) after reaction with IRC-Fe+2, EDTA-Fe+2, DTPA-Fe+2, free Fe+2 and Fe+3.

Table 1.
Kinetic parameters of Fenton-mediated chlorophenol degradation by low-molecular-weight iron-reducing compounds.
3.5. Degradation of Chlorophenols by f-IRC and Synthetic Iron Chelates
To compare the degradation of phenolic chlorinated compounds by the ferri-IRC and synthetic iron chelates, degradation experiments using the complexes EDTA-Fe+2, DTPA-Fe+2, and free Fe+2 and Fe+3 were carried out. Non-chelated iron, which is an aqueous complex, will be named free iron. From previous experiments, it was found that the ferric complexes DTPA-Fe+3 and EDTA-Fe+3 did not significantly react, and they were not included in the subsequent experiments. All the reactions were performed at the same incubation conditions (25 μM chlorophenol, 10 mM H2O2, 100 μM Fe+3, pH 3.5).
The degradation of p-CP was significantly affected by the group treatments, time and the interaction between both factors (Supplementary material, Table S2). Significantly increased degradation was observed for the ferri-IRC and EDTA-Fe+2 (Supplementary material, Table S3). The complete degradation of p-CP by the ferri-IRC was observed after 7 h, whereas the degradation by EDTA-Fe+2 was 87% after 48 h (Figure 6, Table 1). The degradations by DTPA-Fe+2, free Fe+2 and free Fe+3 were significantly lower than the ferri-IRC and EDTA-Fe+2 with values of 25%, 15.3% and 8%, respectively, even after 48 h of incubation (Figure 6; Table 1; Supplementary material, Tables S3 and S4).
The degradation of 2,4-DCP was significantly affected by the group treatments, time and the interaction between both factors (Supplementary material, Table S2). The 2,4-DCP degradation was significantly higher by the ferri-IRC compared to another ferric complex, and complete degradation was observed at 16 h (Figure 6; Supplementary material, Tables S3 and S4). The degradation by EDTA-Fe+2 was significantly higher than DTPA-Fe+2, free Fe+2 and free Fe+3 (Supplementary material, Tables S3 and S4). After 48 h of incubation, chlorophenols were degraded by 80% for EDTA-Fe+2, 12.2% for DTPA-Fe+2 and 8.6% for free Fe+2 (Figure 6, Table 1). Moreover, this degradation percentage decreased to 4.3% when free Fe+3 was used (Figure 6, Table 1).
The degradation of 2,4,6-TCP was significantly affected by the group treatments, time and the interaction between both factors (Supplementary material, Table S2). Similar to 2,4-DCP, the degradation was significantly higher by ferri-IRC compared to another ferric complex (Supplementary material, Tables S3 and S4). The 2,4,6-TCP was completely degraded by the ferri-IRC after 48 h of incubation (Figure 5). The degradation by EDTA-Fe+2 was significantly higher than DTPA-Fe+2, free Fe+2 and free Fe+3 (Supplementary material, Table S3). At the end of the experiment, the degradation by EDTA-Fe+2, DTPA-Fe+2 and free Fe+2 was 65%, 11% and 5.6%, respectively (Figure 6, Table 1). The degradation by Fe+3 was very low (3.5%), but not significantly different to that oif free Fe+2 and free Fe+3 (Supplementary material, Tables S3 and S4).
The degradation of PCP by all the tested systems was low compared to the degradation of the other chlorophenol compounds. The degradation of PCP was significantly affected by the group treatments, time and the interaction between both factors (Supplementary material, Table S2). After 48 h, the degradation by ferri-IRC and EDTA-Fe+2 was significantly higher than DTPA-Fe+2, free Fe+2 and free Fe+3 with 72% and 30%, respectively (Figure 6; Table 1; Supplementary material, Table S3). DTPA-Fe+2 and free Fe+2 were not effective at the degradation of PCP. Moreover, Fe+3 did not degrade PCP to any extent. However, these last three ferric complexes were not significantly different (Supplementary material, Tables S3 and S4).
The products of p-CP degradation were analyzed by HPLC and GC-MS. The ferri-IRC degraded about 90% of p-CP in 10 h and after that reaction, the mixture became dark brown and the color continued to change with time. When sodium dithionite was added to the mixture, all the colors immediately disappeared. Both the HPLC and GC-MS results showed that p-CP was mainly converted to 4-chloro-catechol after reaction with the ferri-IRC (Figure 7). Therefore, ferri-IRCs can mediate the hydroxylation of chlorophenols.
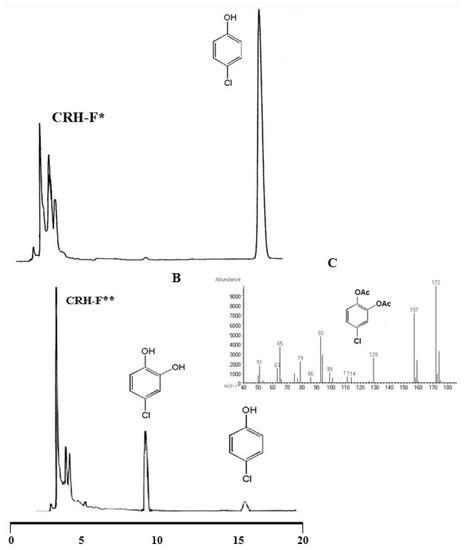
Figure 7.
Degradation of p-CP mediated by iron reducing compounds (IRC) isolated from Lecanicillium ATA01. (A) HPLC chromatograms of the reaction mixture after 1 h (*), (B) after 10 h (**) and (C) GC/MS spectra of the main degradation product 4-chlorocatechol, formed from p-CP by IRC/Fe+3/H2O2.
4. Discussion
Lecanicillium sp. ATA01 is an entomopathogenic endophytic fungus isolated from the extreme environment of the natural laboratory of the Atacama Puna Plateau and is consequently well-adapted to stressful conditions (i.e., cold temperatures, drought, high radiation, poor soil). Lecanicillium sp. ATA01 has the capacity to degrade recalcitrant compounds, such as chlorophenols, involving low-molecular-mass iron-reducing compounds through a Fenton-like reaction under suitable conditions. As far as we know, this is the first report of a degradative non-enzymatic system displayed by entomopathogenic endophytic fungi inhabiting stressful environments. The degradation of 2,4-DCP by Lecanicillium species was previously reported as a laboratory-viable option [38]. The degradation of anthracene (40%), phenanthrene (63%), and fluorine (81%) within 14 days as well as the production of extracellular peroxidases stimulated by Mn2+ was detected in Lecanicillium aphanocladii [38].
Although the degradation of complex compounds has historically been attributed to enzymatic mechanisms, it has been observed that non-enzymatic processes are led by free hydroxyl as a catalyzer. Due to their size, the enzymes released by microorganisms cannot diffuse easily, but low-molecular-weight compounds such as organic acids in contact with hydrogen peroxide will react, thereby promoting degradation using the OH as a “wedge”. Meanwhile, the extracellularly occurring non-enzymatic mechanisms that are used by the displayed free-radical catalysis could be used as a “low cost” alternative to the chlorophenol compounds and other non-toxic compounds.
The application of ferri-IRC to the degradation of hazardous compounds required adequate equilibrium between Fe+3 and H2O2 concentrations [12]. Thus, it was determined that the optimum conditions for the maximum degradation of chlorinated phenolic compounds by isolated iron chelates were pH 3.5, 100 μM of Fe+3 and 10 mM of H2O2. Similar parameters were determined for the Fenton-mediated indole degradation: 14.47 mmol/L H2O2, pH 3.0 [39].
The optimized concentration of Fe+3 and H2O2 improved the reaction of the ferri-IRC, producing a significant degradation of all the chlorophenols. With higher H2O2 concentrations no degradation was detected, indicating that it might be inhibiting the reactivity of the ferri-IRC reactions. The first important feature is that the efficiency of this reaction is mediated by the ferri-IRC, in terms of the amount and ratio of iron and the hydrogen-peroxide concentration.
The reaction carried out by ferri-IRC was more effective in comparison with the iron complexes such as DTPA-Fe+2, EDTA-Fe+2 and free Fe+2 under the same experimental conditions. Although ferri-IRC without H2O2 promoted the degradation of chlorinated phenolic, it was limited compared to the H2O2-catalyzed reactions. In agreement with the previous report [40], the relationship of Fe+3 and H2O2 has no effect on chlorophenol degradation. Even though ferrous iron and H2O2 produce hydroxyl radicals (Fenton-type reaction), the degradation does not proceed as rapidly as when the IRCs are present in the reaction medium (modulation effect of IRCs).
The effect of the degree of substitution of chlorine in phenols on the oxidation kinetics of the Fenton-type reagent has been demonstrated. In this work, it was shown that the ferri-IRC can degrade chlorophenols and that the degradation rate decreases with increasing substitution of chlorine on the ring. Thus, p-CP was degraded much more rapidly than the other chlorinated compounds. The slow phase of the p-CP decomposition was primarily attributed to the Fe+2 depletion caused by Fe-organic-complex formation, and oxidation was limited by hydrogen peroxide at a pH range of 2–4 [41].
The ferri-IRCs can mediate the hydroxylation of chlorophenols, and catechol was obtained as an intermediate in the degradation pathways of various aromatic compounds [42]. Similar evidence of hydroxylated metabolites derived from the 2,4-dichlorophenol degradation by Gloeophyllum striatum has been provided by Schlosser et al. [43]. The hydroxylation steps have been suggested to be an important reaction in the dechlorination of polychlorophenols in bacteria such as Sphingomonas chlorophenolica [44], as well as in fungi such as P. chrysosporium [44]. Moreover, the same reaction product, 4-chlorocatechol, was found in the mixture without hydrogen peroxide. Methatham et al. [45] reached a degradation of up to 70% with H2O2 and Fe+2 operating at pH 3. Preliminary works have shown that the p-CP degradation in the presence of hydrogen peroxide is approximately 17 times higher than in the absence of hydrogen peroxide.
The p-CP was transformed to 4-chloro-catechol reaction mixtures in the absence of hydrogen peroxide, but at a lower rate. According to previous reports, this result suggests the autoxidation of iron mediated by the deferri-IRC under experimental conditions, with the concomitant production of H2O2 [46] and the promotion of a low rate of chlorophenol degradation.
Previous works suggest that PCP degradation by Lecanicillium ATA01 is possible by combination and step-crosslinking of two pathways of extracellular-oxidative- and intracellular-reductive-dechlorinating reactions [47]. The hydroxylation of p-CP suggests a Fenton-type reaction mediated by the ferri-IRC, which could be involved in part of the extracellular oxidative aerobic degradation based on the Fenton-type reaction displayed by Lecanicillium sp.
There is an increasing interest in the study of the genus Lecanicillium sp. as a biological decomposer of harmful compounds mainly because of the enzymatic mechanisms that are involved in the degradation of oil and some PAHs [19]. The removal of CPs from the environment has been attributed to fungal laccase with an efficiency of 30 to 60% [19], but results from this study have demonstrated that the IRC might mediate the non-enzymatic degradation of CPs between 63% and 100% depending on the molecular-structure complexity, which is a promising scenario for new approaches to bioremediation using extremophilic endophytic fungi.
5. Conclusions
Lecanicillium sp. ATA01 is an entomopathogenic endophytic fungus inhabiting vascular plants (grasses) in the extreme environment of the natural laboratory of the Atacama Puna Plateau, which represents significant potential as a reservoir of microbial genetic resources and biotechnological applications.
Lecanicillium sp. ATA01 produces low-molecular-mass iron-reducing compounds (IRCs) that are characterized as extracellular and hydrophilic compounds that contain peptides, iron and phenolate structures with molecular weights of 1207, 567, and 522 Da as determined by MALDI-TOF-MS. The IRCs of Lecanicillium sp. ATA01 can chelate and mediate the reduction of ferric iron to ferrous iron, as well as oxidize lignin-model substrates such as veratryl alcohol, 2,6-dimethoxyphenol and ABTS both with and without H2O2 (laccase-type activity). The IRCs with higher Fe+3/Fe+2-reduction capacity and remarkable POx activity demonstrated the capacity to degrade chlorophenol compounds in the ferri form under controlled conditions.
The mechanism of degradation occurs through the generation of hydroxyl radicals and the promotion of hydroxylation by inducing Fenton chemistry. The Lecanicillium sp. ATA01 isolate is very promising for applications in the bioremediation of chlorophenol degradation. Moreover, since the degradative capacity of the generated hydroxyl radicals is not substrate specific, they could also be exploited for bioremediation of other organic pollutants.
The current study proposed the use of low-molecular-weight iron-reducing compounds (IRCs) released by the endophyte fungi Lecanicillium ATA01 for the advanced oxidation process of the recalcitrant compounds, i.e., to take in advance the non-enzymatic processes as a new bioremediation tool for chlorophenol degradation. Our findings provide new evidence of the occurrence of low-molecular-mass iron-reducing compounds in entomopathogenic endophytic fungi that inhabit extreme environments (Atacama Puna Plateau), and the way these IRCs promote Fenton-like reactions under suitable conditions and can degrade hazardous organic compounds opens new possibilities for bioremediation treatments of xenobiotics and contaminants.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation8040147/s1, Figure S1: urification of iron-reducing compounds (IRCs) by high performance liquid chromatography present in fraction A containing four main peaks as follow: A1 = peak #3; A2 = peak #10; A3 = peak #12 and A4 = peak #14, Figure S2: Linear MALDI-TOF-MS analysis for iron reducing compounds (IRCs) A1 = peak #3 (Mw 1207 Da); A2 = peak #10 (Mw 550 Da); A3 = peak #12 (Mw 567 Da) and A4 = peak #14 (Mw 522 Da) obtained from reverse-phase liquid chromatography, Table S1: Summary results of the univariate ANOVA on the effect of the reaction parameter on the p-CP degradation. Statistically significant values (p ≤ 0.05) are indicated in bold, Table S2: Result of the multiple comparison Tukey test among the different of reaction parameter on p-CP degradation by IRC/Fe+3/H2O2. Statistically significant values (p ≤ 0.05) are indicated in bold, Table S3: Results of univariate repeated measures ANOVA for degradation of chlorinated compounds. Since the sphericity assumption was not fulfilled, Greenhouse-Geisser correction (GGe) and Huynh-Feldt correction (HFe) were estimated. The Mauchly test of sphericity is also included, and the results without correction (“sphericity assumed”). Significant p-values are in bold, Table S4: Result of the multiple comparison Tukey test among the different combinations of group treatments on the degradation of chlorinated compounds. Statistically significant values (p ≤ 0.05) are indicated in bold.
Author Contributions
Conceptualization, R.O.-P. and J.R.; Formal analysis, R.O.-P. and P.L.; Funding acquisition, R.O.-P. and J.R. Investigation, R.O.-P., P.L., E.B. and J.R.; Methodology, R.O.-P., P.L. and J.R.; Project administration, R.O.-P.; Supervision, R.O.-P., Validation, R.O.-P., D.I.-R., P.L., E.B. and C.P.; Visualization, E.B. and P.L.; Writing–original draft, R.O.-P., P.L., E.B. and D.I.-R.; Writing–review & editing, R.O.-P., P.L., E.B., C.P. and M.A. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financed by AIST fellow MITI 1997 (Japan), Fondecyt No 1960227, Renewables Resources Laboratory, Center of Biotechnology, master’s degree program, Faculty of Forestry Sciences, University of Concepcion, Chile. This work was partially financed by the Projects High Altitude Laboratory ATA No 1799, ATA No 2095, ATA No 20992 of the Ministry of Education of Chile, Universidad de Atacama, Chile, and Fondecyt Iniciación No 11190754 (National Agency of Research and Development, ANID).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
R. Oses wants thanks to professional staff working in the Hydrospheric Environmental Protection Department–National Institute for Resources and Environment (NIRE) for their advice and technical support. This work is dedicated to the memory of Jaime Rodríguez Gutiérrez († February 2017), past Director of Center of Biotechnology, and Jaime Baeza Hernández († February 2012), past Director of Research, University of Concepcion.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Arora, P.K.; Bae, H. Bacterial degradation of chlorophenols and their derivatives. Microb. Cell Factories 2014, 13, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Igbinosa, E.; Odjadjare, E.; Chigor, V.; Igbinosa, I.H.; Emoghene, A.O.; Ekhaise, F.O.; Igiehon, N.O.; Idemudia, O.G. Toxicological Profile of Chlorophenols and Their Derivatives in the Environment: The Public Health Perspective. Sci. World J. 2013, 2013, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pera-Titus, M.; García-Molina, V.; Baños, M.A.; Giménez, J.; Esplugas, S. Degradation of chlorophenols by means of advanced oxidation processes: A general review. Appl. Catal. B Environ. 2004, 47, 219–256. [Google Scholar] [CrossRef]
- Czaplicka, M. Sources and transformations of chlorophenols in the natural environment. Sci. Total Environ. 2004, 322, 21–39. [Google Scholar] [CrossRef] [PubMed]
- Oses, R.; Valenzuela, S.; Freer, J.; Sanfuentes, E.; Rodriguez, J. Fungal endophytes in xylem of healthy Chilean trees and their possible role in early wood decay. Fungal Divers 2008, 33, 77–86. Available online: https://www.fungaldiversity.org/fdp/sfdp/33-4.pdf (accessed on 11 February 2022).
- Oses, R.; Valenzuela, S.; Freer, J.; Baeza, J.; Rodríguez, J. Evaluation of fungal endophytes for lignocellulolytic enzyme production and wood biodegradation. Int. Biodeterior. Biodegrad. 2006, 57, 129–135. [Google Scholar] [CrossRef]
- Parray, J.A.; Shameem, N. Sustainable engineering technologies to promote activities of beneficial microbiome. In Sustainable Agriculture; Academic Pres: San Diego, CA, USA, 2020; pp. 231–275. [Google Scholar] [CrossRef]
- Arantes, V.; Jellison, J.; Goodell, B. Peculiarities of brown-rot fungi and biochemical Fenton reaction with regard to their potential as a model for bioprocessing biomass. Appl. Microbiol. Biotechnol. 2012, 94, 323–338. [Google Scholar] [CrossRef]
- Gόmez-Toribio, V.; García-Martín, A.B.; Martínez, M.J.; Martínez, A.T.; Guillén, F. Induction of Extracellular Hydroxyl Radical Production by White-Rot Fungi through Quinone Redox Cycling. Appl. Environ. Microbiol. 2009, 75, 3944–3953. [Google Scholar] [CrossRef] [Green Version]
- Hammel, K.E.; Kapich, A.N.; Jensen, K.A.; Ryan, Z.C. Reactive oxygen species as agents of wood decay by fungi. Enzym. Microb. Technol. 2002, 30, 445–453. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, M. A critical review of the application of chelating agents to enable Fenton and Fenton-like reactions at high pH values. J. Hazard. Mater. 2018, 362, 436–450. [Google Scholar] [CrossRef]
- Goodell, B.; Jellison, J.; Liu, J.; Daniel, G.; Paszczynski, A.; Fekete, F.; Krishnamurthy, S.; Jun, L.; Xu, G. Low molecular weight chelators and phenolic compounds isolated from wood decay fungi and their role in the fungal biodegradation of wood. J. Biotechnol. 1997, 53, 133–162. [Google Scholar] [CrossRef]
- Kerem, Z.; Jensen, K.A.; Hammel, K. Biodegradative mechanism of the brown rot basidiomyceteGloeophyllum trabeum: Evidence for an extracellular hydroquinone-driven fenton reaction. FEBS Lett. 1999, 446, 49–54. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Mukhopadhyay, M.; Murthy, Z.V.P. Treatment of chlorophenols from wastewaters by advanced oxidation pro-cesses. Sep. Purif. Rev. 2013, 42, 263–295. [Google Scholar] [CrossRef]
- Rodríguez, J.; Parra, C.; Contreras, D.; Freer, J.; Baeza, J. Dihydroxybenzenes: Driven Fenton reactions. Water Sci. Technol. 2001, 44, 251–256. [Google Scholar] [CrossRef]
- Paszczynski, A.; Crawford, R.; Funk, D.; Goodell, B. De novo synthesis of 4, 5-Dimethoxycatechol and 2, 5-Dimethoxyhydroquinone by the brown-rot fungus Gloeophyllum trabeum. Appl. Environ. Microbiol. 1999, 65, 674–679. [Google Scholar] [CrossRef] [Green Version]
- Marco-Urrea, E.; García-Romera, I.; Aranda, E. Potential of non-ligninolytic fungi in bioremediation of chlorinated and polycyclic aromatic hydrocarbons. New Biotechnol. 2015, 32, 620–628. [Google Scholar] [CrossRef]
- Rocha-Pino, Z.; Marín-Cervantes, M.D.C.; Martínez-Archundia, M.; Soriano-Blancas, E.; Revah, S.; Shirai, K. Morphological changes, chitinolytic enzymes and hydrophobin-like proteins as responses of Lecanicillium lecanii during growth with hydrocarbon. Bioprocess Biosyst. Eng. 2012, 36, 531–539. [Google Scholar] [CrossRef]
- Pozdnyakova, N.N.; Varese, G.C.; Prigione, V.; Dubrovskaya, E.V.; Balandina, S.A.; Turkovskaya, O.V. Degradative properties of two newly isolated strains of the ascomycetes Fusarium oxysporum and Lecanicillium aphanocladii. Int. Microbiol. 2018, 22, 103–110. [Google Scholar] [CrossRef] [Green Version]
- Fenice, M. The Psychrotolerant Antarctic Fungus Lecanicillium muscarium CCFEE 5003: A Powerful Producer of Cold-Tolerant Chitinolytic Enzymes. Molecules 2016, 21, 447. [Google Scholar] [CrossRef] [Green Version]
- Jaber, L.R.; Ownley, B. Can we use entomopathogenic fungi as endophytes for dual biological control of insect pests and plant pathogens? Biol. Control 2018, 116, 36–45. [Google Scholar] [CrossRef]
- Radwan, O.; Gunasekera, T.S.; Ruiz, O.N. Draft Genome Sequence of Lecanicillium sp. Isolate LEC01, a Fungus Capable of Hydrocarbon Degradation. Microbiol. Resour. Announc. 2019, 8, e01744-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Anisimova, M.; Gil, M.; Dufayard, J.-F.; Dessimoz, C.; Gascuel, O. Survey of Branch Support Methods Demonstrates Accuracy, Power, and Robustness of Fast Likelihood-based Approximation Schemes. Syst. Biol. 2011, 60, 685–699. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwyn, B.; Neilands, J. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Csáky, T.Z.; Hassel, O.; Rosenberg, T.; Loukamo, S.L.; Turunen, E.; Tuhkanen, A. On the Estimation of Bound Hydroxylamine in Biological Materials. Acta Chem. Scand. 1948, 2, 450–454. [Google Scholar] [CrossRef] [Green Version]
- Arnow, L.E. Colourimetric determination of the components of 3,4-dihydroxyphenylalanine-tyrosine mixtures. J. Biol. Chem 1937, 118, 531–537. Available online: https://www.jbc.org/article/S0021-9258(18)74509-2/pdf (accessed on 11 February 2022). [CrossRef]
- Voelker, B.M.; Sulzberger, B. Effects of Fulvic Acid on Fe(II) Oxidation by Hydrogen Peroxide. Environ. Sci. Technol. 1996, 30, 1106–1114. [Google Scholar] [CrossRef]
- Shao, Y.; Lin, A.H.-M. Improvement in the quantification of reducing sugars by miniaturizing the Somogyi-Nelson assay using a microtiter plate. Food Chem. 2018, 240, 898–903. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Cohen, G.; Sinet, P.M. The fenton reaction between ferrous-diethylenetriaminepentaacetic acid and hydrogen peroxide. FEBS Lett. 1982, 138, 258–260. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org/ (accessed on 11 February 2022).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.A.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Turkovskaya, O.; Dubrovskaya, E.; Grinev, V.; Balandina, S.; Pozdnyakova, N.; Institute of Biochemistry and Physiology of Plants and Microorganisms Ras. Degradative Activity and Production of the Extracellular Peroxidases by Micromycetes with Different Ecological Strategy. Sel’skokhozyaistvennaya Biol. 2019, 54, 65–75. [Google Scholar] [CrossRef]
- Ben Hammouda, S.; Adhoum, N.; Monser, L. Chemical oxidation of a malodorous compound, indole, using iron entrapped in calcium alginate beads. J. Hazard. Mater. 2016, 301, 350–361. [Google Scholar] [CrossRef]
- Ma, J.; Song, W.; Chen, C.; Ma, W.; Zhao, J.; Tang, Y. Fenton Degradation of Organic Compounds Promoted by Dyes under Visible Irradiation. Environ. Sci. Technol. 2005, 39, 5810–5815. [Google Scholar] [CrossRef]
- Barbeni, M.; Minero, C.; Pelizzetti, E. Chemical degradation of chlorophenols with Fenton’s reagent. Chemosphere 1987, 16, 2225–2237. [Google Scholar] [CrossRef]
- Gerginova, M.; Manasiev, J.; Yemendzhiev, H.; Terziyska, A.; Peneva, N.; Alexieva, Z. Biodegradation of Phenol by Antarctic Strains of Aspergillus fumigatus. Z. Für Nat. 2013, 68, 384. [Google Scholar] [CrossRef]
- Schlosser, D.; Fahr, K.; Karl, W.; Wetzstein, H.-G. Hydroxylated Metabolites of 2,4-Dichlorophenol Imply a Fenton-Type Reaction in Gloeophyllum striatum. Appl. Environ. Microbiol. 2000, 66, 3010–3015. [Google Scholar] [CrossRef] [Green Version]
- Copley, S.D. Evolution of a metabolic pathway for degradation of a toxic xenobiotic: The patchwork approach. Trends Biochem. Sci. 2000, 25, 261–265. [Google Scholar] [CrossRef]
- Methatham, T.; Lu, M.-C.; Ratanatamskul, C. Removal of 2,4-dichlorophenol as herbicide’s by-product by Fenton’s reagent combined with an electrochemical system. Desalination Water Treat. 2011, 32, 42–48. [Google Scholar] [CrossRef]
- Cho, N.-S.; Jarosz-Wilkolazka, A.; Leonowicz, A.; Oga, S. Removal of Chlorophenols by Fungal Laccase in the Presence of Aromatic Alcohols. J. Fac. Agric. Kyushu Univ. 2007, 52, 23–27. [Google Scholar] [CrossRef]
- Sutton, H.C.; Vile, G.F.; Winterbourn, C.C. Radical driven fenton reactions—Evidence from paraquat radical studies for production of tetravalent iron in the presence and absence of ethylenediaminetetraacetic acid. Arch. Biochem. Biophys. 1987, 256, 462–471. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).