Characterization of Biofilm Microbiome Formation Developed on Novel 3D-Printed Zeolite Biocarriers during Aerobic and Anaerobic Digestion Processes
Abstract
1. Introduction
2. Materials and Methods
2.1. Biocarrier Synthesis
2.2. Brunauer-EMMETT-TELLER (BET) Special Surface Analysis
2.3. Anaerobic Development of Biofilm
2.4. Biochemical Methane Potential (BMP) Assay
2.5. Lab-Scale Wastewater Treatment Set-up and Operation
2.6. Biofilm Extraction
2.7. Tissue Culture Plate (TCP)
2.8. Determination of Physicochemical Parameters
2.9. Determination of Soluble Microbial Products (SMP) and Extracellular Polymeric Substances (EPS)
2.10. DNA Extraction and 16S rRNA Gene Amplicon Sequencing
2.11. Bioinformatics
2.12. Statistics
3. Results
3.1. The Brunauer-EMMETT-TELLER (ΒΕΤ) Special Surface Analysis of the Novel 3D-Printed Biocarriers
3.2. The Effect of the Novel 3D-Printed Biocarriers on Methane Production in Anaerobic Digestion
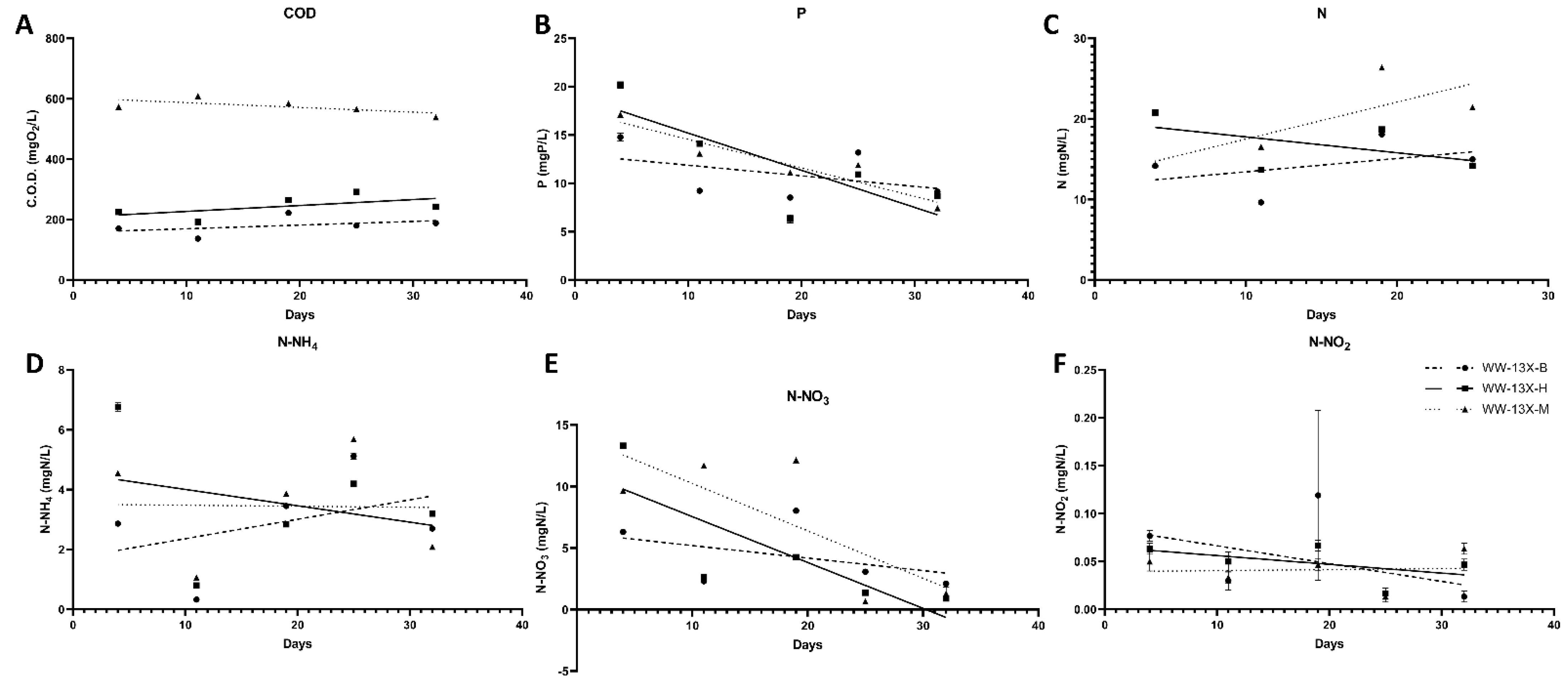
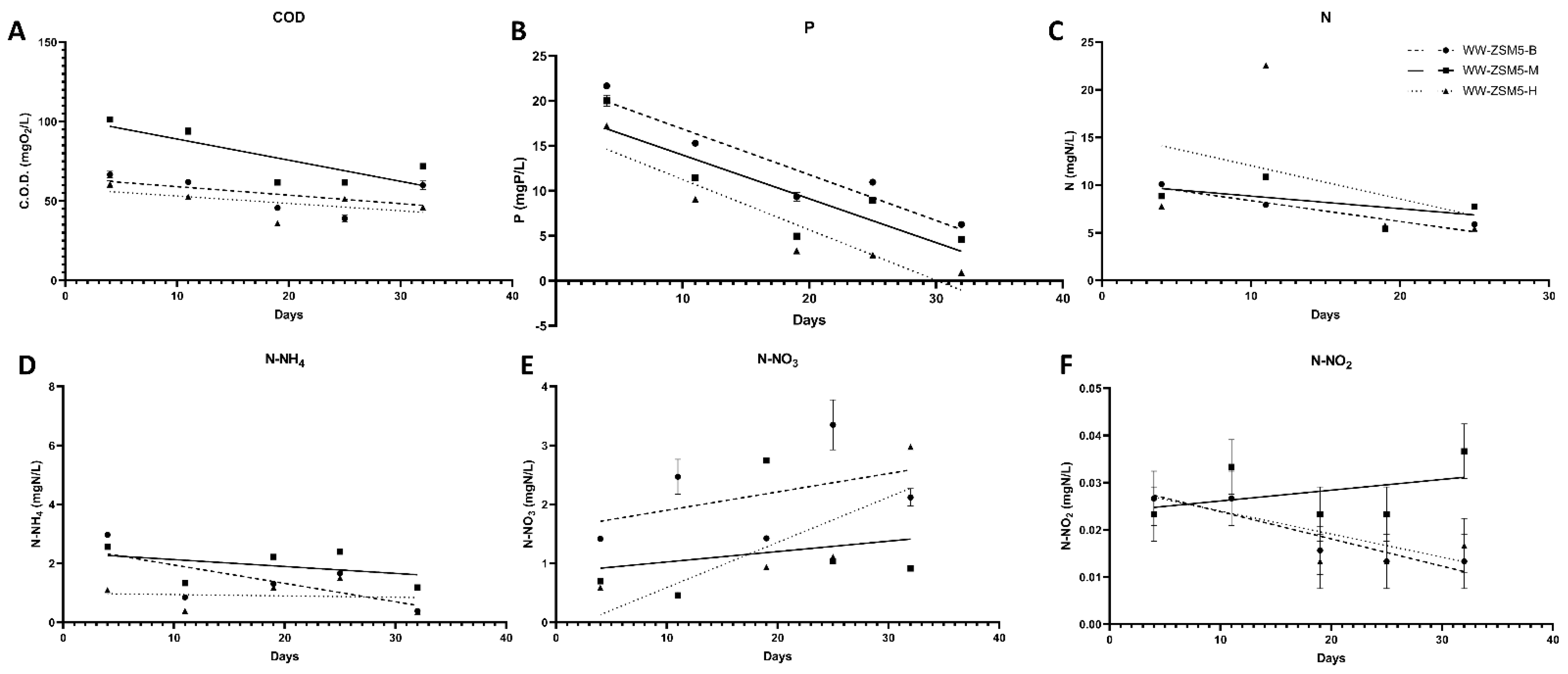
3.3. The Effect of the Novel 3D-printed Biocarriers on Wastewater Treatment in Aerobic Digestion
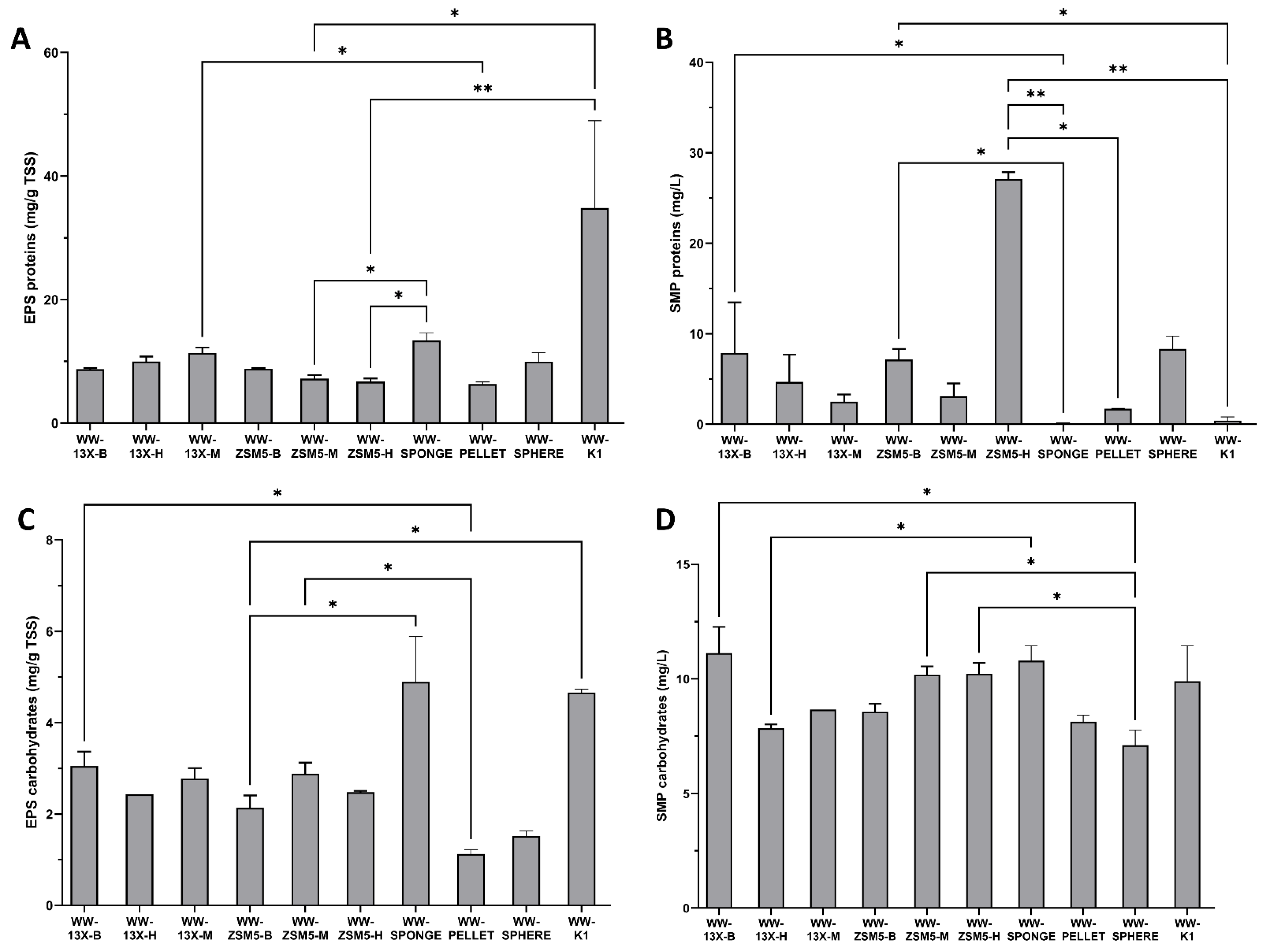
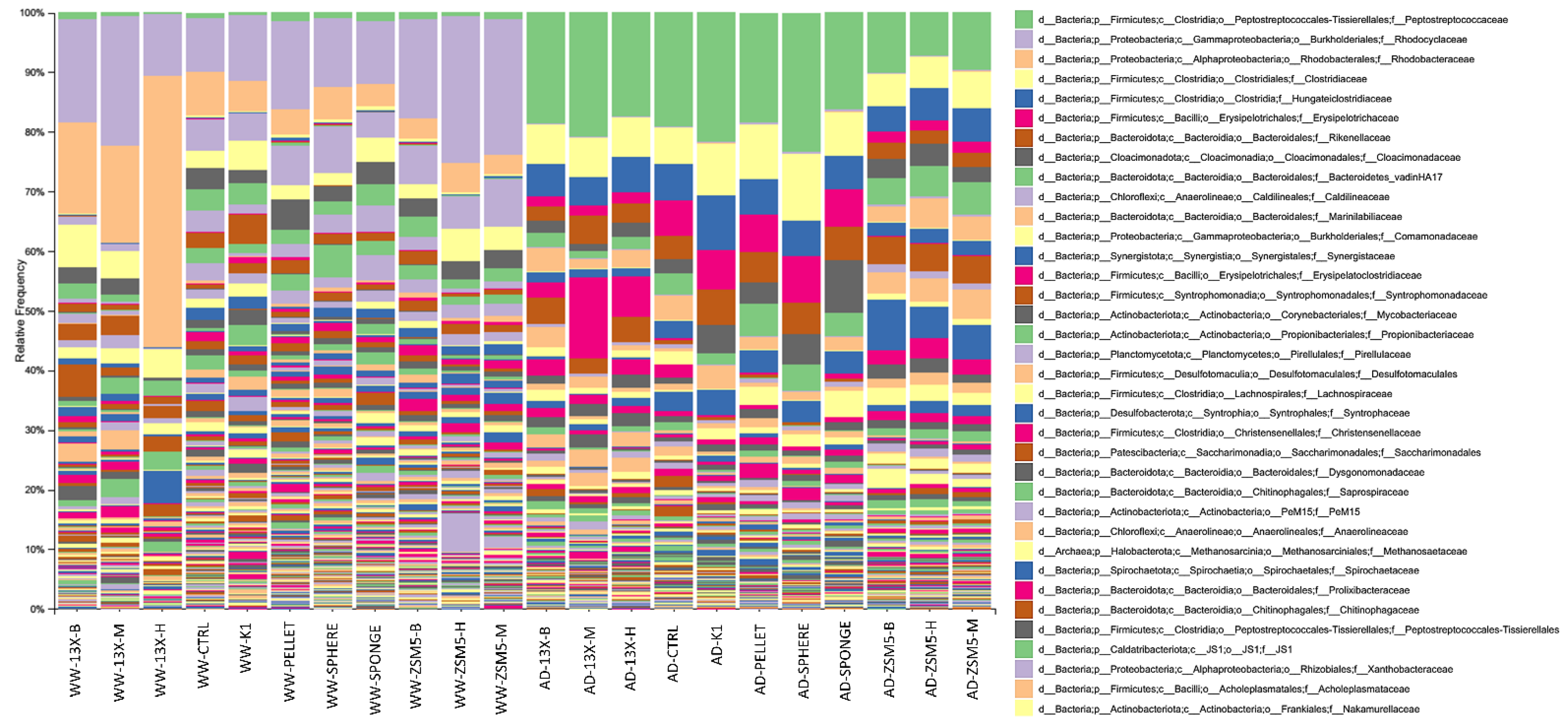
3.4. The Effect of the Novel 3D-Printed Biocarriers on Biofilm Composition and Formation
3.5. 16S rRNA Microbiome Analysis of Biofilms Developed on Biocarriers during Anaerobic Digestion
3.6. 16S rRNA Microbiome Analysis of Biofilms Developed on Biocarriers during Aerobic Digestion
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Al-Ghusain, I.; Hamoda, M.F.; Abd El-Ghany, M. Nitrogen transformations during aerobic/anoxic sludge digestion. Bioresour. Technol. 2002, 85, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, A.; Kumar, P.S.; Nhung, T.C.; Ramesh, B.; Srinivasan, S.; Rangasamy, G. A review on biological methodologies in municipal solid waste management and landfilling: Resource and energy recovery. Chemosphere 2022, 309, 136630. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Tashiro, Y.; Ishida, N.; Sakai, K. Application of autothermal thermophilic aerobic digestion as a sustainable recycling process of organic liquid waste: Recent advances and prospects. Sci. Total Environ. 2022, 828, 154187. [Google Scholar] [CrossRef]
- Pasalari, H.; Gholami, M.; Rezaee, A.; Esrafili, A.; Farzadkia, M. Perspectives on microbial community in anaerobic digestion with emphasis on environmental parameters: A systematic review. Chemosphere 2021, 270, 128618. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, D.; Huang, W.; Yang, Y.; Ji, M.; Nghiem, L.D.; Trinh, Q.T.; Tran, N.H. Insights into biofilm carriers for biological wastewater treatment processes: Current state-of-the-art, challenges, and opportunities. Bioresour. Technol. 2019, 288, 121619. [Google Scholar] [CrossRef]
- Raj Deena, S.; Kumar, G.; Vickram, A.S.; Rani Singhania, R.; Dong, C.D.; Rohini, K.; Anbarasu, K.; Thanigaivel, S.; Ponnusamy, V.K. Efficiency of various biofilm carriers and microbial interactions with substrate in moving bed-biofilm reactor for environmental wastewater treatment. Bioresour. Technol. 2022, 359, 127421. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; He, Z.; Liang, T.; Sheng, T.; Zhang, F.; Wu, D.; Ma, F. Colonization of biofilm in wastewater treatment: A review. Environ. Pollut. 2022, 293, 118514. [Google Scholar] [CrossRef]
- Yang, D.; Zhang, X.; Zhou, Y.; Xiu, Z. The Principle and Method of Wastewater Treatment in Biofilm Technology. J. Comput. Theor. Nanosci. 2015, 12, 2630–2638. [Google Scholar] [CrossRef]
- Sfetsas, T.; Patsatzis, S.; Chioti, A. A review of 3D printing techniques for bio-carrier fabrication. J. Clean. Prod. 2021, 318, 128469. [Google Scholar] [CrossRef]
- Da Silva, C.; Astals, S.; Peces, M.; Campos, J.L.; Guerrero, L. Biochemical methane potential (BMP) tests: Reducing test time by early parameter estimation. Waste Manag. 2018, 71, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Christensen, G.D.; Simpson, W.A.; Younger, J.J.; Baddour, L.M.; Barrett, F.F.; Melton, D.M.; Beachey, E.H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: A quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985, 22, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Abbott, S.L. 16S rRNA Gene Sequencing for Bacterial Identification in the Diagnostic Laboratory: Pluses, Perils, and Pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef] [PubMed]
- Mandakhalikar, K.D.; Rahmat, J.N.; Chiong, E.; Neoh, K.G.; Shen, L.; Tambyah, P.A. Extraction and quantification of biofilm bacteria: Method optimized for urinary catheters. Sci. Rep. 2018, 8, 8069. [Google Scholar] [CrossRef] [PubMed]
- Rice, E.W.; Baird, R.B.; Eaton, A.D. Standard Methods for the Examination of Water and Wastewater, 23rd ed.; American Public Health Association, American Water Works Association, Water Environment Federation: Alexandria, VA, USA, 2017. [Google Scholar]
- Hwang, B.K.; Kim, J.H.; Ahn, C.H.; Lee, C.H.; Song, J.Y.; Ra, Y.H. Effect of disintegrated sludge recycling on membrane permeability in a membrane bioreactor combined with a turbulent jet flow ozone contactor. Water Res. 2010, 44, 1833–1840. [Google Scholar] [CrossRef] [PubMed]
- Banti, D.C.; Karayannakidis, P.D.; Samaras, P.; Mitrakas, M.G. An innovative bioreactor set-up that reduces membrane fouling by adjusting the filamentous bacterial population. J. Membr. Sci. 2017, 542, 430–438. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Hartree, E. Determination of protein: A modification of the lowry method that gives a linear photometric response. Anal. Biochem. 1972, 48, 422–427. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Duan, L.; Jiang, W.; Song, Y.H.; Xia, S.Q.; Hermanowicz, S.W. The characteristics of extracellular polymeric substances and soluble microbial products in moving bed biofilm reactor-membrane bioreactor. Bioresour. Technol. 2013, 148, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Nobu, M.K.; Narihiro, T.; Mei, R.; Kamagata, Y.; Lee, P.K.H.; Lee, P.H.; McInerney, M.J.; Liu, W.T. Catabolism and interactions of uncultured organisms shaped by eco-thermodynamics in methanogenic bioprocesses. Microbiome 2020, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- Ijoma, G.N.; Nkuna, R.; Mutungwazi, A.; Rashama, C.; Matambo, T.S. Applying PICRUSt and 16S rRNA functional characterisation to predicting co-digestion strategies of various animal manures for biogas production. Sci. Rep. 2021, 11, 19913. [Google Scholar] [CrossRef]
- Wagner, M.; Loy, A.; Nogueira, R.; Purkhold, U.; Lee, N.; Daims, H. Microbial community composition and function in wastewater treatment plants. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2002, 81, 665–680. [Google Scholar] [CrossRef] [PubMed]
- Tice, H.; Mayilraj, S.; Sims, D.; Lapidus, A.; Nolan, M.; Lucas, S.; Del Rio, T.G.; Copeland, A.; Cheng, J.F.; Meincke, L.; et al. Complete genome sequence of Nakamurella multipartita type strain (Y-104(T)). Stand. Genom. Sci. 2010, 2, 168–175. [Google Scholar] [CrossRef]
- Zorz, J.K.; Kozlowski, J.A.; Stein, L.Y.; Strous, M.; Kleiner, M. Comparative Proteomics of Three Species of Ammonia-Oxidizing Bacteria. Front. Microbiol. 2018, 9, 938. [Google Scholar] [CrossRef]
- Montalvo, S.; Huiliñir, C.; Borja, R.; Sánchez, E.; Herrmann, C. Application of zeolites for biological treatment processes of solid wastes and wastewaters—A review. Bioresour. Technol. 2020, 301, 122808. [Google Scholar] [CrossRef]
- Montalvo, S.; Guerrero, L.; Borja, R.; Sánchez, E.; Milán, Z.; Cortés, I.; De La La Rubia, M.A. Application of natural zeolites in anaerobic digestion processes: A review. Appl. Clay Sci. 2012, 58, 125–133. [Google Scholar] [CrossRef]
- Sudibyo, H.; Shabrina, Z.L.; Wondah, H.R.; Hastuti, R.T.; Purnomo, C.W.; Budhijanto, W. Anaerobic digestion of landfill leachate with natural zeolite and sugarcane bagasse fly ash as the microbial immobilization media in packed bed reactor. Acta Polytech. 2018, 58, 57–68. [Google Scholar] [CrossRef]
- Andalib, M.; Elbeshbishy, E.; Mustafa, N.; Hafez, H.; Nakhla, G.; Zhu, J. Performance of an anaerobic fluidized bed bioreactor (AnFBR) for digestion of primary municipal wastewater treatment biosolids and bioethanol thin stillage. Renew. Energy 2014, 71, 276–285. [Google Scholar] [CrossRef]
- Montalvo, S.; Gonzalez, P.; Mena, C.; Guerrero, L.; Borja, R. Influence of the food to microorganisms (F/M) ratio and temperature on batch anaerobic digestion processes with and without zeolite addition. J. Environ. Sci. Health Part A 2012, 47, 1785–1794. [Google Scholar] [CrossRef] [PubMed]
- Andalib, M.; Hafez, H.; Elbeshbishy, E.; Nakhla, G.; Zhu, J. Treatment of thin stillage in a high-rate anaerobic fluidized bed bioreactor (AFBR). Bioresour. Technol. 2012, 121, 411–418. [Google Scholar] [CrossRef]
- Li, R.; Liu, D.; Zhang, Y.; Zhou, J.; Tsang, Y.F.; Liu, Z.; Duan, N.; Zhang, Y. Improved methane production and energy recovery of post-hydrothermal liquefaction waste water via integration of zeolite adsorption and anaerobic digestion. Sci. Total Environ. 2019, 651, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Wijesinghe, D.T.N.; Dassanayake, K.B.; Sommer, S.G.; Scales, P.; Chen, D. Biogas improvement by adding Australian zeolite during the anaerobic digestion of C: N ratio adjusted swine manure. Waste Biomass Valorization 2019, 10, 1883–1887. [Google Scholar] [CrossRef]
- Kotobuki, M. Properties of Al2O3 Pastes Using Inorganic Na2SiO3 Binder and Organic Binder for Direct Ink Writing. Phys. Status Solidi B-Basic Solid State Phys. 2022, 259, 2100520. [Google Scholar] [CrossRef]
- Schwarz, J.A.; Contescu, C.; Contescu, A. Methods for Preperation of Catalyctic Materials. Chem. Rev. 1995, 95, 477–510. [Google Scholar] [CrossRef]
- Chen, S.J.; Zhu, M.; Fu, Y.; Huang, Y.X.; Tao, Z.C.; Li, W.L. Using 13X, LiX, and LiPdAgX zeolites for CO2 capture from post-combustion flue gas. Appl. Energy 2017, 191, 87–98. [Google Scholar] [CrossRef]
- Bernholc, J.; Brenner, D.; Nardelli, M.B.; Meunier, V.; Roland, C. Mechanical and electrical properties of nanotubes. Annu. Rev. Mater. Res. 2002, 32, 347. [Google Scholar] [CrossRef]
- Aranzabal, A.; Iturbe, D.; Romero-Sáez, M.; González-Marcos, M.P.; González-Velasco, J.R.; González-Marcos, J.A. Optimization of process parameters on the extrusion of honeycomb shaped monolith of H-ZSM-5 zeolite. Chem. Eng. J. 2010, 162, 415–423. [Google Scholar] [CrossRef]
- Jiang, Z.; An, N.; Chu, Y.; Cao, B.; Wu, F.; Zhang, Y.; Zhang, Y.; Li, Y.; Zhang, Y. Growth, biofilm formation and atrazine degrading gene (trzN) expression of Arthrobacter sp. DNS10 cultured with montmorillonite, kaolinite and goethite. Chemosphere 2022, 307, 135904. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, C.; Chen, Z.; Song, Y.; Li, H.; Han, Y.; Hou, Y.; Guo, J. Multifaceted synergistic electron transfer mechanism for enhancing denitrification by clay minerals. Sci. Total Environ. 2022, 812, 152222. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Luo, X.; Liu, S.; Wan, W.; Huang, Q.; Chen, W. Synergistic effect of biofilm growth and cadmium adsorption via compositional changes of extracellular matrix in montmorillonite system. Bioresour. Technol. 2020, 315, 123742. [Google Scholar] [CrossRef] [PubMed]
- Cai, P.; Liu, X.; Ji, D.; Yang, S.; Walker, S.L.; Wu, Y.; Gao, C.; Huang, Q. Impact of soil clay minerals on growth, biofilm formation, and virulence gene expression of Escherichia coli O157:H7. Environ. Pollut. 2018, 243, 953–960. [Google Scholar] [CrossRef]
- Song, Z.; Zhang, X.; Ngo, H.H.; Guo, W.; Song, P.; Zhang, Y.; Wen, H.; Guo, J. Zeolite powder based polyurethane sponges as biocarriers in moving bed biofilm reactor for improving nitrogen removal of municipal wastewater. Sci. Total Environ. 2019, 651, 1078–1086. [Google Scholar] [CrossRef]
- Nguyen, T.M.; Ha, P.T.; Le, T.T.H.; Phan, K.S.; Mai, T.T.T.; Hoang, P.H. Modification of expanded clay carrier for enhancing the immobilization and nitrogen removal capacity of nitrifying and denitrifying bacteria in the aquaculture system. J. Biosci. Bioeng. 2022, 134, 41–47. [Google Scholar] [CrossRef]
- Saidulu, D.; Srivastava, A.; Gupta, A.K. Enhancement of wastewater treatment performance using 3D printed structures: A major focus on material composition, performance, challenges, and sustainable assessment. J. Environ. Manag. 2022, 306, 114461. [Google Scholar] [CrossRef] [PubMed]
- Wingender, J.; Neu, T.R.; Flemming, H.-C. What are Bacterial Extracellular Polymeric Substances? In Microbial Extracellular Polymeric Substances; Springer: Berlin/Heidelberg, Germany, 1999; pp. 1–19. [Google Scholar]
- Lin, H.; Zhang, M.; Wang, F.; Meng, F.; Liao, B.-Q.; Hong, H.; Chen, J.; Gao, W. A critical review of extracellular polymeric substances (EPSs) in membrane bioreactors: Characteristics, roles in membrane fouling and control strategies. J. Membr. Sci. 2014, 460, 110–125. [Google Scholar] [CrossRef]
- Laspidou, C.S.; Rittmann, B.E. A unified theory for extracellular polymeric substances, soluble microbial products, and active and inert biomass. Water Res. 2002, 36, 2711–2720. [Google Scholar] [CrossRef]
- Liu, D.; Li, C.; Guo, H.; Kong, X.; Lan, L.; Xu, H.; Zhu, S.; Ye, Z. Start-up evaluations and biocarriers transfer from a trickling filter to a moving bed bioreactor for synthetic mariculture wastewater treatment. Chemosphere 2019, 218, 696–704. [Google Scholar] [CrossRef]
- Lin, H.; Sun, S.; Lin, Z.; Chen, M.; Fang, L.; Ma, R.; Lin, J.; Luo, J. Bio-carrier-enhanced aerobic granulation: Effects on the extracellular polymeric substances production and microorganism community. Chemosphere 2021, 280, 130756. [Google Scholar] [CrossRef]
- Wang, M.; Zhou, J.; Yuan, Y.-X.; Dai, Y.-M.; Li, D.; Li, Z.-D.; Liu, X.-F.; Zhang, X.-Y.; Yan, Z.-Y. Methane production characteristics and microbial community dynamics of mono-digestion and co-digestion using corn stalk and pig manure. Int. J. Hydrogen Energy 2016, 42, 4893–4901. [Google Scholar] [CrossRef]
- Wang, H.; Tolvanen, K.; Lehtomäki, A.; Puhakka, J.; Rintala, J. Microbial community structure in anaerobic co-digestion of grass silage and cow manure in a laboratory continuously stirred tank reactor. Biogeochemistry 2009, 21, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Venkiteshwaran, K.; Milferstedt, K.; Hamelin, J.; Fujimoto, M.; Johnson, M.; Zitomer, D.H. Correlating methane production to microbiota in anaerobic digesters fed synthetic wastewater. Water Res. 2017, 110, 161–169. [Google Scholar] [CrossRef]
- Yan, L.; Gao, Y.; Wang, Y.; Liu, Q.; Sun, Z.; Fu, B.; Wen, X.; Cui, Z.; Wang, W. Diversity of a mesophilic lignocellulolytic microbial consortium which is useful for enhancement of biogas production. Bioresour. Technol. 2012, 111, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, X.; Zhou, J.; Yuan, Y.; Dai, Y.; Li, D.; Li, Z.; Liu, X.; Yan, Z. The dynamic changes and interactional networks of prokaryotic community between co-digestion and mono-digestions of corn stalk and pig manure. Bioresour. Technol. 2017, 225, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Ferry, J.G. Enzymology of one-carbon metabolism in methanogenic pathways. Fems Microbiol. Rev. 1999, 23, 13–38. [Google Scholar] [CrossRef]
- Gao, Y.; Yang, A.; Bao, J.; Ma, R.; Yan, L.; Wang, Y.; Wang, W. Bioreactor performance and microbial community dynamics in a production-scale biogas plant in northeastern China. Int. J. Agric. Biol. Eng. 2017, 10, 191–201. [Google Scholar] [CrossRef]
- Zhao, Y.; Xu, C.; Ai, S.; Wang, H.; Gao, Y.; Yan, L.; Mei, Z.; Wang, W. Biological pretreatment enhances the activity of functional microorganisms and the ability of methanogenesis during anaerobic digestion. Bioresour. Technol. 2019, 290, 121660. [Google Scholar] [CrossRef]
- Wang, H.; Lim, T.T.; Duong, C.; Zhang, W.; Xu, C.; Yan, L.; Mei, Z.; Wang, W. Long-Term Mesophilic Anaerobic Co-Digestion of Swine Manure with Corn Stover and Microbial Community Analysis. Microorganisms 2020, 8, 188. [Google Scholar] [CrossRef]
- Stepanov, V.G.; Xiao, Y.; Tran, Q.; Rojas, M.; Willson, R.C.; Fofanov, Y.; E Fox, G.; Roberts, D.J. The presence of nitrate dramatically changed the predominant microbial community in perchlorate degrading cultures under saline conditions. BMC Microbiol. 2014, 14, 225. [Google Scholar] [CrossRef]
- Oren, A. The Family Rhodocyclaceae. In The Prokaryotes: Alphaproteobacteria and Betaproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 975–998. [Google Scholar]
- Urakami, T.; Araki, H.; Oyanagi, H.; Suzuki, K.-I.; Komagata, K. Paracoccus aminophilus sp. nov. and Paracoccus aminovorans sp. nov., Which Utilize N,N-Dimethylformamide. Int. J. Syst. Evol. Microbiol. 1990, 40, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zheng, J.W.; Liang, B.; Wang, C.H.; Cai, S.; Ni, Y.Y.; He, J.; Li, S.P. Biodegradation of Chloroacetamide Herbicides by Paracoccus sp. FLY-8 in Vitro. J. Agric. Food Chem. 2011, 59, 4614–4621. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, D.; Richard, M.G.; Daigger, G.T. Manual on the Causes and Control of Activated Sludge Bulking, Foaming and Other Solids Separation Problems; IWA Publishing: London, UK, 2003. [Google Scholar]
- Dubinina, G.; Savvichev, A.; Orlova, M.; Gavrish, E.; Verbarg, S.; Grabovich, M.; Dubinina, G.; Savvichev, A.; Orlova, M.; Gavrish, E.; et al. Beggiatoa leptomitoformis sp. nov., the first freshwater member of the genus capable of chemolithoautotrophic growth. Int. J. Syst. Evol. Microbiol. 2017, 67, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Schwedt, A.; Kreutzmann, A.-C.; Polerecky, L.; Schulz-Vogt, H.N. Sulfur Respiration in a Marine Chemolithoautotrophic Beggiatoa Strain. Front. Microbiol. 2012, 2, 276. [Google Scholar] [CrossRef]
- Shintani, T.; Liu, W.T.; Hanada, S.; Kamagata, Y.; Miyaoka, S.; Suzuki, T.; Nakamura, K. Micropruina glycogenica gen. nov., sp, nov., a new Gram-positive glycogen-accumulating bacterium isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 2000, 50, 201–207. [Google Scholar] [CrossRef]
- Wang, R.; Peng, Y.; Cheng, Z.; Ren, N. Understanding the role of extracellular polymeric substances in an enhanced biological phosphorus removal granular sludge system. Bioresour. Technol. 2014, 169, 307–312. [Google Scholar] [CrossRef]
- Basuvaraj, M.; Fein, J.; Liss, S.N. Protein and polysaccharide content of tightly and loosely bound extracellular polymeric substances and the development of a granular activated sludge floc. Water Res. 2015, 82, 104–117. [Google Scholar] [CrossRef]




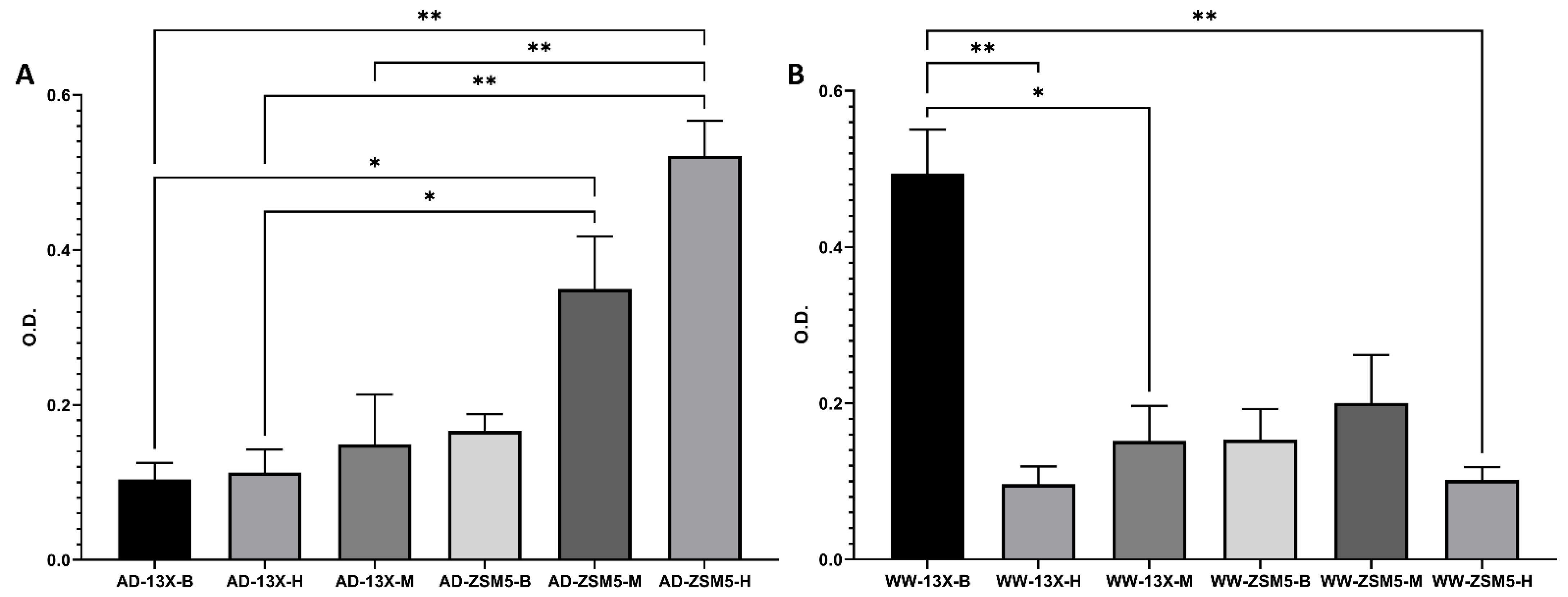
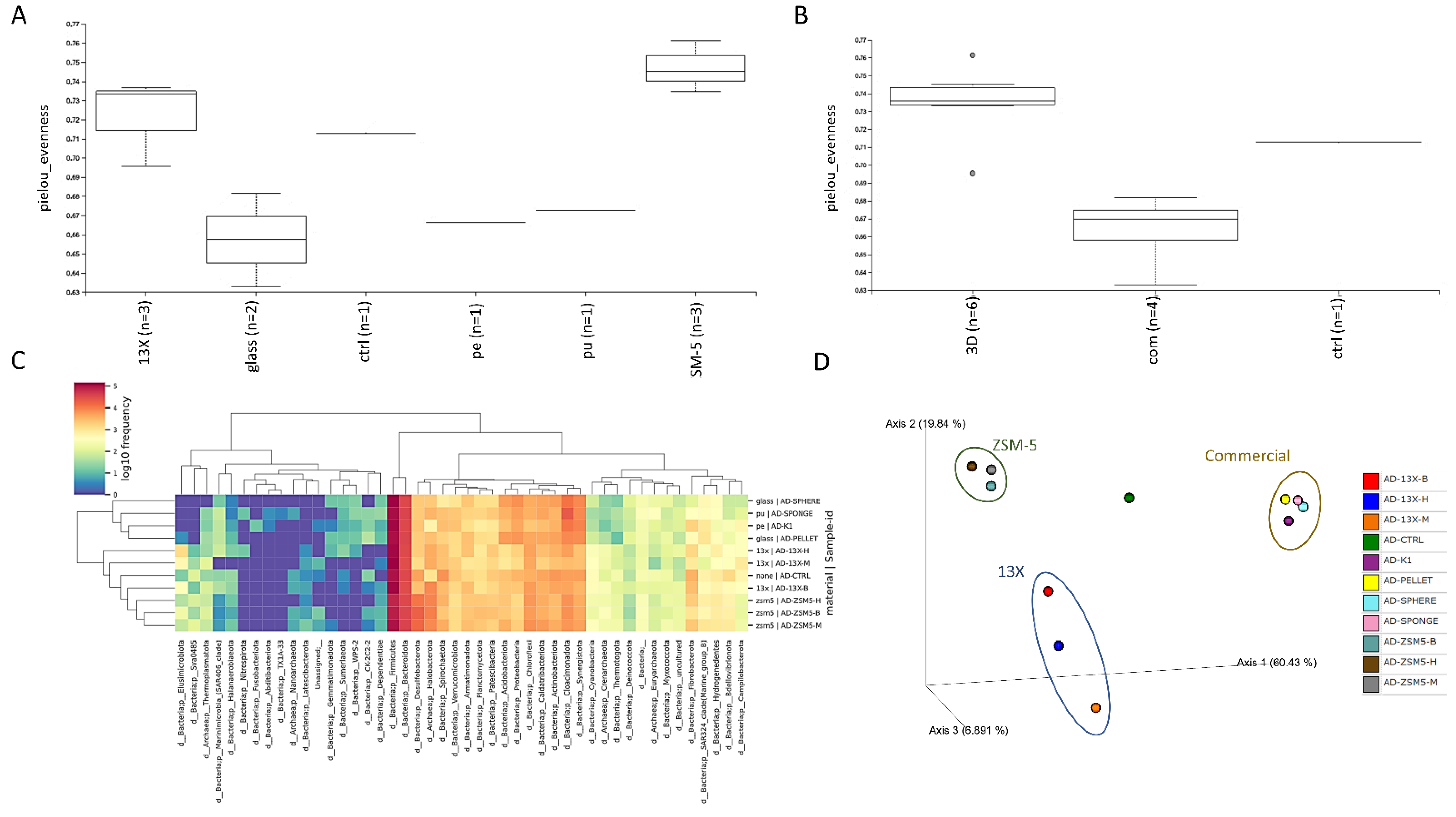
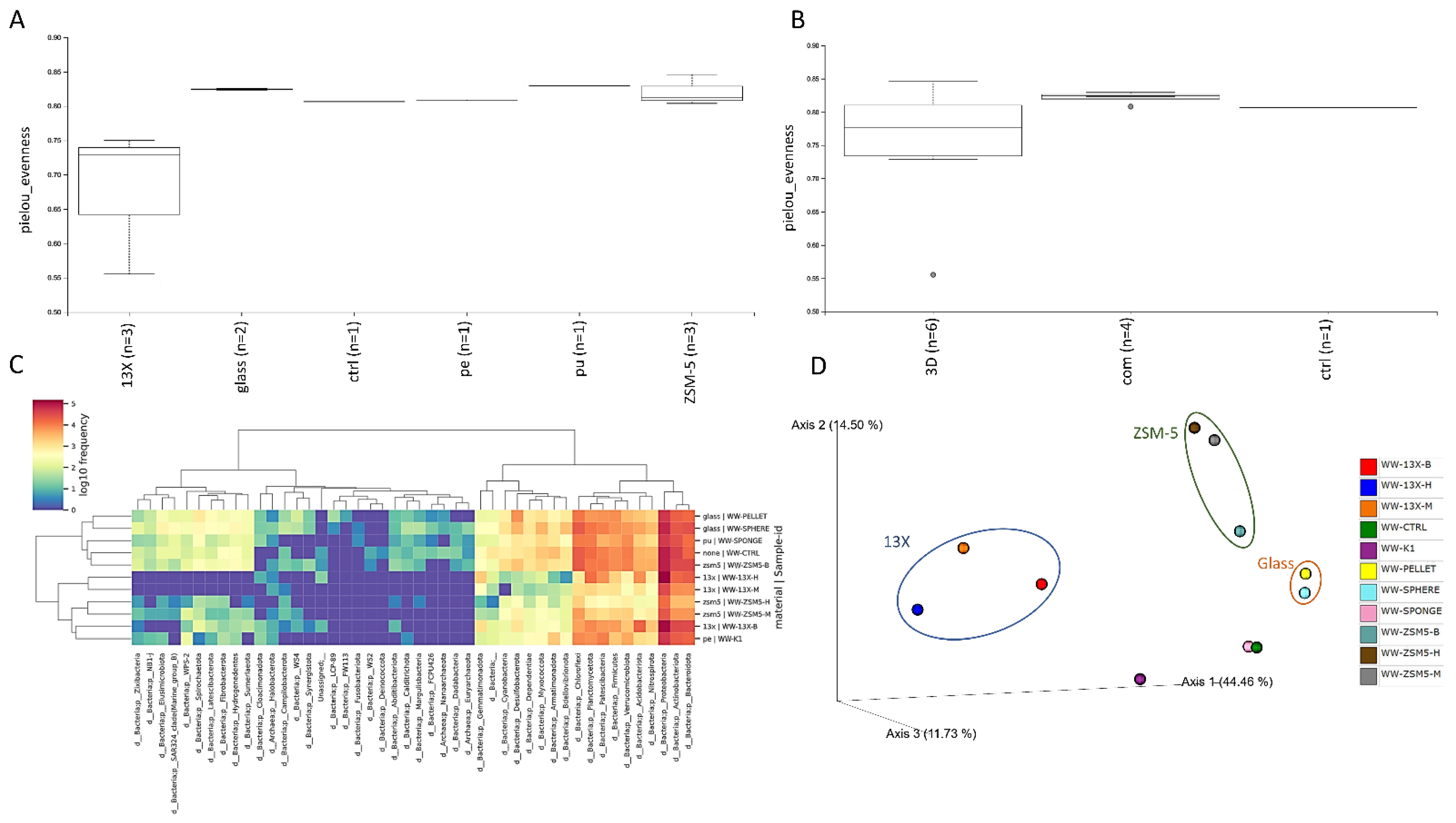
| Mean OD Values | Adherence | Biofilm Formation |
|---|---|---|
| <0.120 | None | None/weak |
| 0.120–0.240 | Moderate | Moderate |
| ≥0.240 | Strong | High |
| Specific Surface Area (m2/g) | Total Pore Volume (cc/g) | Micropore Volume (cc/g) | Mesopore Volume (cc/g) | Micropore Diameter (Angstrom) | Mesopore Diameter (Angstrom) | |
|---|---|---|---|---|---|---|
| 13X | 688 | 0.35 | 0.29 | 0.06 | 10.2 | 55 |
| ZSM5 | 549 | 0.57 | 0.21 | 0.36 | 5 | 42 |
| Bentonite | 52 | 0.15 | 0.02 | 0.13 | na | 55 |
| Montmorillonite | 32 | 0.16 | 0.01 | 0.15 | na | 55 |
| Halloysite Nanotubes | 89 | 0.32 | 0.03 | 0.29 | na | 55 and 116 |
| 13X/Bentonite | 590 | 0.42 | 0.24 | 0.18 | 9 | 42, 116 |
| 13X/Halloysite Nanotubes | 711 | 0.49 | 0.29 | 0.20 | 9 | 34, 55, 97 |
| 13X/Montmorillonite | 687 | 0.42 | 0.28 | 0.14 | 9 | 39, 54, 94 |
| ZSM5/Bentonite | 527 | 0.56 | 0.21 | 0.35 | 9,4 | 35, 51, 116 |
| ZSM5/Montmorillonite | na | na | na | na | na | na |
| ZSM5/Halloysite Nanotubes | 383 | 0.37 | 0.15 | 0,22 | na | na |
| Source | Material | Sample | d__Bacteria | d__Archaea | Unassigned | ||
|---|---|---|---|---|---|---|---|
| ASVs | (%) | ASVs | (%) | ||||
| 3D | 13X | AD-13X-B | 205,558 | 96.02 | 8517 | 3.98 | 0 |
| AD-13X-H | 195,934 | 97.82 | 4371 | 2.18 | 0 | ||
| AD-13X-M | 202,854 | 97.83 | 4486 | 2.16 | 8 | ||
| ZSM-5 | AD-ZSM5-B | 178,934 | 94.59 | 10,223 | 5.40 | 15 | |
| AD-ZSM5-H | 160,829 | 94.74 | 8936 | 5.26 | 0 | ||
| AD-ZSM5-M | 207,491 | 96.11 | 8391 | 3.89 | 6 | ||
| com | glass | AD-PELLET | 199,781 | 98.62 | 2805 | 1.38 | 0 |
| AD-SPHERE | 221,908 | 99.01 | 2219 | 0.99 | 0 | ||
| pe | AD-K1 | 215,865 | 99.64 | 772 | 0.36 | 2 | |
| pu | AD-SPONGE | 214,017 | 98.26 | 3784 | 1.74 | 0 | |
| none | none | AD-CTRL | 191,480 | 99.26 | 1423 | 0.74 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chioti, A.G.; Tsioni, V.; Patsatzis, S.; Filidou, E.; Banti, D.; Samaras, P.; Economou, E.A.; Kostopoulou, E.; Sfetsas, T. Characterization of Biofilm Microbiome Formation Developed on Novel 3D-Printed Zeolite Biocarriers during Aerobic and Anaerobic Digestion Processes. Fermentation 2022, 8, 746. https://doi.org/10.3390/fermentation8120746
Chioti AG, Tsioni V, Patsatzis S, Filidou E, Banti D, Samaras P, Economou EA, Kostopoulou E, Sfetsas T. Characterization of Biofilm Microbiome Formation Developed on Novel 3D-Printed Zeolite Biocarriers during Aerobic and Anaerobic Digestion Processes. Fermentation. 2022; 8(12):746. https://doi.org/10.3390/fermentation8120746
Chicago/Turabian StyleChioti, Afroditi G., Vasiliki Tsioni, Stefanos Patsatzis, Eirini Filidou, Dimitra Banti, Petros Samaras, Eleni Anna Economou, Eleni Kostopoulou, and Themistoklis Sfetsas. 2022. "Characterization of Biofilm Microbiome Formation Developed on Novel 3D-Printed Zeolite Biocarriers during Aerobic and Anaerobic Digestion Processes" Fermentation 8, no. 12: 746. https://doi.org/10.3390/fermentation8120746
APA StyleChioti, A. G., Tsioni, V., Patsatzis, S., Filidou, E., Banti, D., Samaras, P., Economou, E. A., Kostopoulou, E., & Sfetsas, T. (2022). Characterization of Biofilm Microbiome Formation Developed on Novel 3D-Printed Zeolite Biocarriers during Aerobic and Anaerobic Digestion Processes. Fermentation, 8(12), 746. https://doi.org/10.3390/fermentation8120746








