Abstract
Dahi and chhurpi are the homemade, mildly acidic and mouthfeel fermented dairy products of Sikkim in India. Since yeasts co-exist among traditional fermented dairy foods, we believe that some species of yeasts may have some probiotic properties. Hence, the present study is aimed at screening some probiotic yeasts from dahi and chhurpi. A total of 3438 yeasts were isolated from 40 samples of dahi (1779 isolates) and 40 chhurpi (1659 isolates) and were preliminarily screened for probiotic properties on the basis of survival in low pH, resistance to bile salts and the percentage of hydrophobicity, out of which only 20 yeasts were selected for in vitro and genetic screening of probiotic properties. Saccharomyces cerevisiae DJT-2 and Debaryomyces prosopidis CPA-55 showed the highest hydrophobicity of 97.54% and 98.33%, respectively. S. cerevisiae DRC-42 and S. cerevisiae CGI-29 showed 93.88% and 91.69% auto-aggregation, respectively. All yeasts showed co-aggregation properties against pathogenic bacteria. Kluyveromyces marxianus DPA-41 and Pichia kudriavzevii CNT-3 showed excellent deconjugation activities. Probiotic genes for acid tolerance, bile tolerance, adhesion and antimicrobial activity were detected in S. cerevisiae DAO-17, K. marxianus DPA-41, S. cerevisiae CKL-10 and P. kudriavzevii CNT-3. Based on the results of in vitro and genetic screening of probiotic yeasts strains, S. cerevisiae DAO-17 (dahi), S. cerevisiae CKL-10 (chhurpi), P. kudriavzevii CNT-3 (chhurpi) and K. marxianus DPA-41(dahi) were selected as the potential probiotic yeasts.
1. Introduction
The souring of animal milk, by natural or back-sloping processes, is one of the oldest inventions of humans for prolonging the shelf life of perishable animal milk. Among the fermented dairy products, cheese and yogurt are the most popular products around the world with several health claims including having probiotic properties [1,2,3,4], followed by kefir, a viscous and slightly fizzy product obtained by the fermentation of milk and kefir grains [5], as a probiotic milk product [6,7]. The majority of populaces in the world cannot afford to buy the commercial probiotic milk products; however, they traditionally prepare various artisanal, naturally fermented dairy products from domesticated animals such as cow, buffalo, yak, camel, mare, sheep and donkey at household levels in different regions of the world [8,9,10,11,12,13,14], which may or may not have probiotics properties.
The probiotic properties of some region-specific and artisan-fermented dairy products of a few countries have also been reported, such as the lait caillé of Senegal [15], tarag and airag of Mongolia [16], amasi of South Africa [17], dadih of Indonesia [18], dahi of India [19,20] and nunu and wara of West Africa [21]. However, most of these probiotics properties in fermented dairy products have been shown by species of lactic acid bacteria (LAB) of Lacticaseibacillus, Lactiplantibacillus Levilactobacillus, Limosilactobacillus and Lactobacillus [22,23,24], and non-lactic acid bacteria such as Propionibacterium [25] and Bifidobacterium [26]. The probiotic properties of yeasts in fermented dairy products are barely reported [27,28,29]. Saccharomyces cerevisiae var. boulardii is the only clinically claimed probiotic yeast that is commercially available for human use [30], and it has also been reported from few fermented dairy products [31,32,33].
Ethnic people of the Himalayan regions of Sikkim state in India consume a diverse varieties of homemade fermented food products including animal milk products [34,35]. Dahi, fermented cow-milk, a slimy and viscous savory beverage (Figure 1a), and chhurpi, an artisan-fermented milk product similar to cottage cheese (Figure 1b), are the most popular homemade traditional dairy products in Sikkim [8]. Bacteria, mostly LAB, are the predominant microorganisms in the naturally fermented Himalayan milk products [8,36,37] with co-existence of several species of yeasts [34,38,39]. Probiotic bacteria have been isolated and screened for their probiotic properties in the dahi and chhurpi of Sikkim [20,40,41], also delineating some bio-functional properties such as angiotensin-converting enzyme (ACE) inhibitory and antioxidant activity [42,43]. Since yeasts co-exist in the traditional fermented dairy foods of the Himalayas, we believe that some species of yeasts present may show probiotic attributes. Hence, the present study is aimed at isolating the culturable yeasts from homemade samples of dahi and chhurpi of Sikkim, India and to identify by amplification of the D1/D2 domains of a large ribosomal subunit. It is also aimed at screening some probiotic properties by an in vitro method (survival in low pH, resistance to bile salts, percentage of hydrophobicity, auto-aggregation and co-aggregation, antagonistic activity, bile salt hydrolase (BSH) activity and lysozyme tolerance) and at genetic screening for probiotic traits.
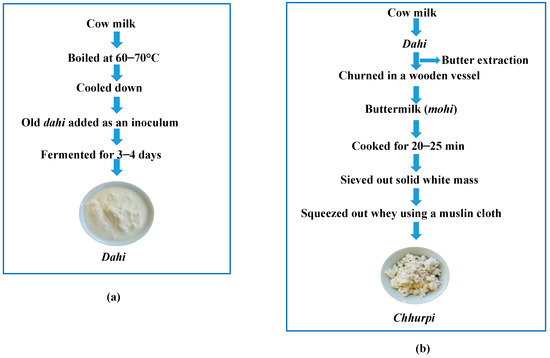
Figure 1.
Traditional method of preparation of (a) dahi and (b) chhurpi in Sikkim.
2. Materials and Methods
2.1. Collection of Samples
A total of 40 samples of dahi and 40 samples of chhurpi were collected from different places of four districts of Sikkim in India, viz. East, West, South and North. All samples were collected in presterilized containers and transported to the laboratory in an ice-box cooler and stored at 4 °C for immediate microbiological analysis.
2.2. Analysis of pH
One gram of each sample (dahi and chhurpi) was dissolved in 10 mL sterilized physiological saline (0.85% NaCl) and the pH was determined using a pH meter (GeNeiTM, Bangalore, India) calibrated with standard buffers [36].
2.3. Titratable Acidity
The titratable acidity of the samples was calculated by titrating the filtrates of a well-blended 10 g sample in 90 mL carbon-dioxide-free distilled water with 0.1 N sodium hydroxide to the end point of phenolphthalein (0.1% w/v in 95% ethanol) [44].
2.4. Moisture Content
Moisture content of the sample was analyzed by the simple weight difference method. The sample was kept in the oven at 105 °C for 4–6 h and moisture content was calculated based on initial and final weight difference [45]. Moisture was calculated as a percentage using the formula:
Moisture (%) = Fresh Weight − Dried weight/Fresh weight × 100
2.5. Viscosity
Apparent viscosity was measured according to the method described by Ali et al. [46]. The apparent viscosity was measured using a viscometer (DV1MRVTJ0, Brookfield AMETEK, Middleboro, MA, USA) in triplicate. The spindle used (LV-SC4-34 spindle at 4 rpm) in 150 mL of the sample was allowed to rotate for 1 min at 20 °C [46].
2.6. Microbiological Analysis
Samples were homogenized in a stomacher (400, Seward, London, UK) using stomacher bags in a ratio of 10:100 w/v dissolved in physiological solution (0.85% NaCl), and serial dilution (10−1 to 10−8) was made. One milliliter of the homogenized mixture was transferred into yeast malt (YM) agar (M424, HiMedia, Mumbai, India) plates under aerobic condition by the pour plate method in triplicate [39]. Colonies that appeared in the YM plates were selected randomly, or all were sampled if the plate contained less than 10 colonies, as according to Dewan and Tamang [39]. The number of colonies was counted as the colony-forming unit (cfu)/mL was represented as the log values for dahi, and cfu/g for chhurpi. The purity of the isolates was checked by streaking again on fresh YM plates, followed by microscopic examinations in a phase-contrast microscope (Olympus, CKX41, Tokyo, Japan) and stored in 20% glycerol at −80 °C for further analysis.
2.7. Preliminary Screening of Probiotic Isolates
Acid tolerance test: An acid tolerance test of all yeast isolates from 40 dahi and 40 chhurpi samples was conducted according to the method described by Greppi et al. [47] with slight modifications. Yeast malt (YM) broth (M425, HiMedia, Mumbai, India) inoculated with yeast cultures was incubated at 28 °C for 24 h, after which 1% (v/v) of the fresh cultures were inoculated in acidified (pH 2.0) YM broth. YM broth without inoculation was used as a control. Optical density of the inoculated broth was measured at 600 nm wavelength at 0 h and 24 h. Absorbance at 600 nm was measured at 0 h and 24 h of incubation, respectively, at 28 °C. The growth after 24 h of incubation at ∆OD600 ≥ 0.500 was considered as a threshold for selection of acid tolerance (pH 2.0) [48]).
Bile tolerance Test: A bile tolerance test of all yeast isolates from 40 dahi and 40 chhurpi samples was performed following the method of Greppi et al. [47] with slight modifications. The YM broth inoculated with yeast cultures was incubated at 28 °C for 24 h, after which 1% (v/v) of the fresh cultures were inoculated in YM broth containing 0.3% oxgall (bile) (CR010, HiMedia, Mumbai, India). YM broth without inoculation was used as a control. Optical density of the inoculated broth was measured after 24 h of incubation at 28 °C at the 600 nm wavelength at 0 h and 24 h, respectively. The growth after 24 h of incubation at ∆OD600 ≥ 0.500 was considered as a threshold for survival tendency of isolates at 0.3% bile [48].
Hydrophobicity (%) test: A cell-surface hydrophobicity test of those isolates that showed both low acid (pH 2) and 0.3 bile tolerances was performed following the method described by Fernandez-Pacheco et al. [49]. Hydrocarbons n-hexadecane and xylene were used as solvents in the experiment. The yeast cultures were grown in YM broth at 30 °C and centrifuged at 5000× g for 5 min at 5 °C. The culture pellets were then washed twice with PBS (pH 7.0), and cell suspension was then adjusted to an A600 nm value of approximately 1.0 by using the buffer designated as ‘Ainitial’. The 3 mL of the cell suspension was mixed with 1 mL of each of the hydrocarbons and vortexed for uniform mixing. The two phases were allowed to separate for 3 h at 30 °C without agitation. After incubation, 1 mL of the upper layer (aqueous phase) was carefully taken and optical density was measured at 600 nm. The reading was designated as ‘Afinal’, and the percentage of cell surface hydrophobicity was calculated as follows:
Hydrophobicity (%) = (1 − (Afinal/Ainitial) × 100)
More than 80% hydrophobicity was considered as the threshold for a high hydrophobic nature of the yeast isolates [50].
2.8. Phenotypic and Biochemical Characterization
Preliminarily selected probiotic yeasts, on the basis of acid and bile tolerances and >80% hydrophobicity, were phenotypically characterized. Colony morphology, cell morphology, growth at different temperatures (25 °C, 30 °C, 37 °C, 40 °C and 45 °C), pH (2.0, 3.0 and pH 4.0) of preliminarily selected probiotic yeasts grown in YM broth at 28 °C for 48–72 h were performed [51]. All preliminary selected probiotic yeasts were tested for nitrate reduction [52], H2S production [53] and for fermentation of sugars (lactose, maltose, glucose, galactose, arabinose, mannose, rhamnose, raffinose, ribose, xylose, sucrose, trehalose and melibiose) [51].
2.9. Genotypic Characterization
2.9.1. DNA Extraction
DNA extraction of yeast isolates was done following the method of Renshaw et al. [54] with some modifications. The 2 mL of 24 h culture, grown in yeast malt broth (M425, HiMedia, Mumbai, India) at 28 °C, was centrifuged at 12,000× g rpm for 10 min. The supernatant was discarded and the pellet was washed twice with sterile 0.5 M NaCl, followed by suspension in 400 µL lysis buffer (Tris-HCl pH 8.0, 5 M NaCl, 0.5 M EDTA pH 8.0 and 10% SDS). The 2 µL of RNase A solution (20 mg/mL) (DS0003, HiMedia, Mumbai, India) was added, followed by incubation at 65 °C for 30 min. The 5 µL of proteinase K (RM2957, HiMedia, Mumbai, India) was added and kept at 65 °C for 30 min, after which 100 µL of 5 M NaCl was added and incubated at −20 °C for 10 min. The suspension was centrifuged at 12,000× g rpm for 10 min and the supernatant was transferred to a fresh tube. Equal volume of a phenol:chloroform:isoamyl-alcohol mixture (25:24:1 v/v) (MB078, HiMedia, Mumbai, India) was added and centrifuged at 12,000× g rpm for 10 min. The upper aqueous layer was carefully removed and transferred to a fresh tube, after which a double volume of chilled isopropanol was added and kept overnight at −20 °C. The suspension was then centrifuged at 14,000× g rpm for 10 min and the supernatant was discarded. The pellet was washed with 100 µL of chilled 70% ethanol and centrifuged at 8000× g rpm for 5 min. The supernatant was discarded and the pellet was allowed to dry at room temperature. The pellet was then dissolved in 50 µL of nuclease-free water. The quality of the DNA was checked using an Eppendorf Bio-Spectrometer (Model 6135 000 009, Hamburg, Germany). The quantified DNA was stored at −20 °C until required and DNA purity of 1.8 to 2.2 was used for PCR reaction.
2.9.2. PCR Amplification
Identification of yeast isolates was carried out by amplification of the D1/D2 domains of the large ribosomal subunit [55]. The PCR reaction was performed in a 50 μL reaction volume containing GoTaq® Green Master Mix (M7122, Promega, Madison, WI, USA), primers NL1 5′-GCA TAT CAA TAA GCG GAG GAA AAG-3′ and NL4 (5′-GGT CCG TGT TTC AAG ACG G-3′) and about 10–20 ng of the DNA template. The PCR amplification was carried out with a SimpliAmp™ Thermal Cycler (Cat No. A24811, ThermoFisher Scientific, Carlsbad, CA, USA) using the following conditions: 94 °C initial denaturation for 1 min; 35 amplification cycles of 30 s at 94 °C, 30 s at 58 °C and 30 s at 72 °C; and final extension at 72 °C for 5 min. The presence of amplicons was confirmed by 1% agarose gel electrophoresis and was visualized using a Gel DocTM EZ.
2.10. In Vitro Screening of Probiotic Properties
Auto-Aggregation and Co-Aggregation
Auto-aggregation (%) and co-aggregation (%) properties of yeasts were evaluated by following the method described by Ogunremi et al. [56] with slight modifications. Overnight-grown yeast cultures were harvested by centrifugation at 5000× g for 10 min at 5 °C, washed twice with 10 mL of PBS (pH 7.0), resuspended in 3 mL PBS (pH 7.0) and vortexed for 10 s. The 1 mL of the suspension was carefully taken from the upper zone and OD600 was measured and designated as ‘Ainitial’. The mixture was then vortexed and incubated at 37 °C for 3 h without agitation. After 3 h of incubation (ATime), absorbance was measured and the percentage was calculated using the following formula:
Auto-aggregation (%) = (1 − (ATime/AInitial) × 100)
Furthermore, the yeast isolates were tested for their ability to adhere to other bacteria (co-aggregation), particularly pathogenic strains that included Escherichia coli KL96 MTCC (Microbial Type Culture Collection, Chandigarh, India) 1583, Salmonella enterica subsp. enterica ser. typhimurium MTCC 3223, Staphylococcus aureus subsp. Aureus MTCC 740 and Bacillus cereus MTCC 1272. Overnight-grown yeasts cultures and the tested pathogens were harvested by centrifugation at 5000× g for 10 min at 5 °C, washed twice with 10 mL of PBS (pH 7.0), resuspended in 10 mL of PBS and OD600 was adjusted to 0.1, denoted as AYeast and APathogen, respectively. Equal volumes (2 mL each) of the yeast and pathogen suspensions were mixed in a vortex, and the mixture was incubated for 3 h at 37 °C without agitation. After incubation, the absorbance (AMix) of the mixture was measured at 600 nm and the co-aggregation percentage was calculated as follows:
Co-aggregation (%) = ((AYeast + APathogen/2) − AMix/(AYeast + APathogen)/2) × 100
2.11. Antimicrobial Activity
The antagonistic activity of yeasts was performed by method of Fernandez-Pacheco et al. [48] with slight modifications. Escherichia coli MTCC 1583, Salmonella enteric subsp. enteric ser. typhimurium MTCC 3223, Staphylococcus aureus subsp. Aureus MTCC 740 and Bacillus cereus MTCC 1272 were used as target pathogens. Lawn culture of the freshly prepared suspensions (1.5 × 108 CFU/mL, OD600 0.08–0.1) of the pathogenic strains (100 µL) was prepared on Muller Hinton agar (M173, HiMedia, Mumbai, India). The wells were prepared with the help of a cork borer and filled with 100 µL of active culture (1.5 × 108 CFU/mL, OD600 0.08–0.1) of yeast strains. The plates were incubated at 37 °C for 48–72 h. Antimicrobial activity was detected by observing the zone of inhibition that appeared after the incubation period.
2.12. Deconjugation of Bile Salts (BSH Activity)
Bile salt hydrolase (BSH) activity was performed following the protocol described by Fadda et al. [57]. BSH activity was screened by spotting in duplicate 10 mL of cultures grown overnight in YM broth on the surface of YM agar plates supplemented with 0.5% (w/v) sodium taurocholate (RM011, HiMedia, Mumbai, India), 0.2% (w/v) sodium glycocholate (GRM8907, HiMedia, India) and sodium cholate (RM202, HiMedia, Mumbai, India) and 0.37 g L−1 of CaCl2 (GRM710, HiMedia, Mumbai, India). Plates were incubated at 30 °C for 72 h. The presence of halos around colonies, as well as white opaque colonies, indicated BSH activity.
2.13. Lysozyme Tolerance
The tolerance of the strains to lysozyme was checked as described by Vera-Pingitore et al. [58] with slight modifications. Overnight-grown yeast cultures were harvested by centrifugation at 5000× g for 10 min at 5 °C, washed twice with 10 mL of PBS (pH 7.0) and resuspended in 10 mL PBS (pH 7.0). Suspended cells were vortexed for 10 s, and 1 mL of the suspension was carefully taken, followed by measuring OD at the 600 nm wavelength as ‘Ainitial’. The suspended cells were vortexed again and treated with sterilized 100 μg/mL (100 mg L−1) of lysozyme (MB098, HiMedia, Mumbai, India) and the mixture was incubated for 1 h at 37 °C without agitation. After 1 h of incubation, OD600 was measured as ‘Afinal’ and the tolerance was calculated in percentage using the following formula: Lysozyme tolerance (%): Afinal/Ainitial × 100.
2.14. Genetic Screening for Probiotic Traits
The presence of genes (Table 1) in yeasts responsible for various probiotic traits was screened. Each reaction mixture for the PCR amplification of the probiotic genes was prepared by mixing 6 µL GoTaq® Green Master Mix (M7122, Promega, Madison, WI, USA), 0.6 µL forward primer, 0.6 µL reverse primer and 1 µL of template DNA, finally making a volume of 12 µL. The PCR products were run in 1% agarose gel for more than 500 bp amplicons and 2% agarose gel for less than 100 bp amplicons, then stained with ethidium bromide (RM813, HiMedia, Mumbai, India). The PCR conditions used were as follows: 1 cycle at 95 °C for 5 min; 40 cycles of 95 °C for 30 s, an annealing temperature for 10 s (depending on the primers listed Table 1) and at 72 °C for 15 s; and 1 cycle at 72 °C for 5 min.

Table 1.
Target genes, probiotic traits and primers used for detection of probiotic genes.
2.15. Bioinformatics Analysis
Raw sequence data were analyzed as per the method described by Palla et al. [55]. The quality of the raw sequences was initially checked using Sequence scanner v2.0 (ABI 3730XL Capillary Sequencers, Applied Biosystems, Foster City, CA, USA) and ChromasPro v1.34 (Technelysium Ltd., South Brisbane, Australia) to assemble the good quality sequences. Sequences were analyzed using BLAST (basic local alignment search tool) on the NCBI (National Center for Biotechnology Information) web (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) (accessed on 1 January 2020) [66]. ClustalW was used to align the identified sequences for analyzing phylogenetic relationship, and a phylogenetic tree was constructed using the neighbor-joining method based on the Kimura 2-parameter [67] by Molecular Evolutionary Genetics Analysis software, version 11 (MEGA v11.0.13) [66]. The sequences were submitted to the NCBI GenBank for accession numbers.
All experiments were performed in triplicate sets with mean ± SD values.
3. Results
3.1. Product Characteristics and Preliminary Screening of Probiotic Yeasts
Mean pH, moisture content, viscosity, titratable acidity and microbial loads of dahi were 4.32 ± 0.22, 94.05% ± 0.31, 367.14 cP ± 0.59, 0.87% ± 0.03 and 6.28 cfu/mL ± 0.84, respectively and chhurpi were 4.48 ± 0.33, 59.03% ± 0.34, 138.43 cP ± 0.43, 0.80% ± 0.06 and 6.28 cfu/g ± 0.70, respectively (Table 2).

Table 2.
Product characteristics, pH, moisture content, viscosity, titratable acidity and microbial load of dahi and chhurpi samples.
Preliminary screening for probiotic properties of a total of 3438 yeast isolates (1779 yeasts from samples of dahi and 1659 yeasts from chhurpi) was conducted on the basis of survival in low acid and bile tolerances based on a threshold value of ∆OD600 ≥ 0.500 (Figure 2). A total of 115 yeasts (54 isolates from dahi and 61 isolates from chhurpi) that survived in low pH and low bile salts were further screened for their hydrophobic nature or percentage of hydrophobicity. Out of these, 20 yeasts (8 isolates from dahi and 12 isolates from chhurpi) showed ≥80% hydrophobicity, which is considered as the threshold for selection of probiotic yeasts [50].
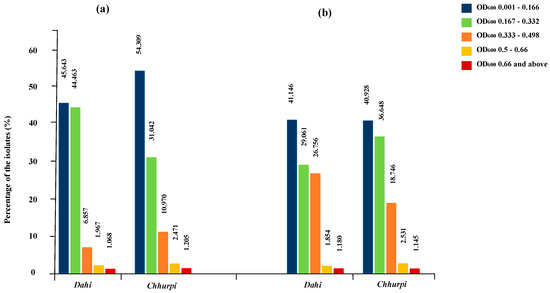
Figure 2.
Screening of survival percentages of years isolated from Dahi and Chhurpi intended as (a) low acid tolerance (pH = 2), and (b) bile tolerance (0.3% w/v).
Twenty screened probiotic yeasts were phenotypically tested and were tentatively identified as up to four major genera as Saccharomyces, Pichia, Kluyveromyces and Debaryomyces (Table 3).

Table 3.
Phenotypic characterization of probiotics yeasts isolated from dahi and chhurpi of Sikkim.
Further confirmation of their identities were performed by sequence analysis of the D1/D2 domain of large ribosomal RNA and a phylogenetic tree was constructed using the neighbor-joining method based on the Kimura 2-parameter (Figure 3).
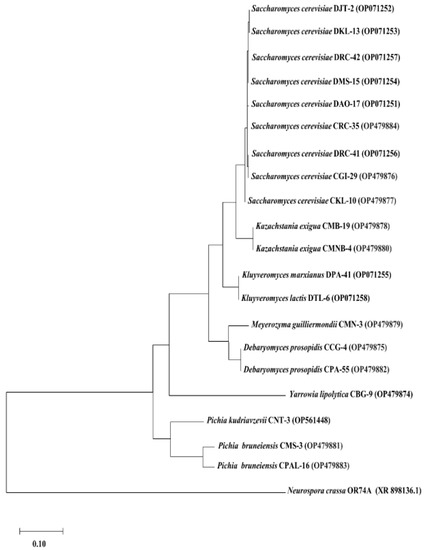
Figure 3.
Molecular phylogenetic analysis of 8 yeasts from dahi and 12 yeasts from chhurpi based on sequencing of the D1/D2 domain of a large ribosomal subunit, by the neighbor-joining method using MEGA v11.0.13 with Neurospora crassa OR74A as the out group.
Saccharomyces cerevisiae was the dominant probiotic yeast in the dahi samples, followed by Kluyveromyces marxianus and K. lactis, whereas S. cerevisiae, Debaryomyces prosopidis, Kazachstania exigua, Pichia bruneiensis, P. kudriavzevii, Meyerozyma guilliermondii and Yarrowia lipolytica were detected in the chhurpi samples (Table 4).

Table 4.
Molecular identification of probiotic yeasts strains isolated from dahi and chhurpi of Sikkim, based on sequencing of the D1/D2 domain of a large ribosomal subunit.
3.2. In Vitro Probiotic Screening
Saccharomyces cerevisiae DJT-2 (dahi) and Debaryomyces prosopidis CPA-55 (chhurpi) showed the highest hydrophobicity of 97.54% and 98.33%, respectively (Table 5). S. cerevisiae DRC-42 from dahi and S. cerevisiae CGI-29 from chhurpi showed 93.88% and 91.69% auto-aggregation, respectively (Table 5). All isolates showed co-aggregation properties against Escherichia coli KL96 MTCC 1583, Salmonella enterica subsp. enterica ser. typhimurium MTCC 3223, Staphylococcus aureus subsp. aureus MTCC 740 and Bacillus cereus MTCC 1272 (Table 5).

Table 5.
Probiotic characteristics of yeasts isolated from dahi and chhurpi of Sikkim.
Some isolates showed BSH activity by hydrolyzing sodium cholate (by three isolates); sodium taurocholates by (three isolates); and sodium glycocholate (by three isolates). Antimicrobial activity against Escherichia coli KL96 MTCC 1583, Salmonella enterica subsp. enterica ser. typhimurium MTCC 3223, Staphylococcus aureus subsp. aureus MTCC 740 and Bacillus cereus MTCC 1272 was shown by many yeast isolates (Table 5). All isolates showed remarkable resistance (75% to 95%) against lysozyme, among which S. cerevisiae CGI-29 (chhurpi) showed the highest value of 95% (Table 5).
3.3. Detection of Probiotic Genes
Based on the results of in vitro probiotic screening results, further detection of probiotic genes was performed using target genes, probiotic traits and primers (Table 1). The acid tolerance genes, viz. TPS1, HSP150 and SED1, were screened; out of 20 isolates, 19 yeasts showed the presence of the TPS1 gene (Figure 4a) and HSP150 gene, and 10 yeasts showed the presence of the SED1 gene (Figure 4b). The genes for bile tolerance, viz. YIM1, PDR1, YOR1, ERG3 and EPA1, were also screened. Out of 20 isolates, 19 yeasts showed the presence of the YIM1 gene (Figure 4c) and 2 isolates showed the presence of PDR1, YOR1, ERG3 (Figure 4d) and EPA1. Genes screened for adhesion were FLO1, FLO5, FLO10, FLO11 and AGA1. Out of 20 isolates, 9 yeasts showed the presence of the FLO1 gene (Figure 4e), 10 isolates for FLO5, 16 isolates for FLO10, 8 isolates for FLO11 and 6 isolates for AGA1 (Figure 4f). Three genes, viz. Apid, khs and pela, were screened for antimicrobial activity. Out of 20 isolates, 3 yeasts (S. cerevisiae DAO-17 (dahi), K. marxianus DPA-41 (dahi), S. cerevisiae CKL-10 (chhurpi)) showed S. cerevisiae DAO-17 (dahi), S. cerevisiae CKL-10 (chhurpi), Pichia kudriavzevii CNT-3 (chhurpi) and the presence of Apid gene (Figure 4g) and the other 3 yeasts [S. cerevisiae DAO-17 (dahi), S. cerevisiae CKL-10 (chhurpi), Pichia kudriavzevii CNT-3 (chhurpi)] showed the presence of pelA genes (Figure 4h). The gene khs for antimicrobial activity was not detected in any of the isolates.
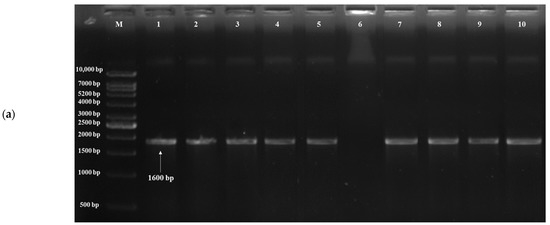
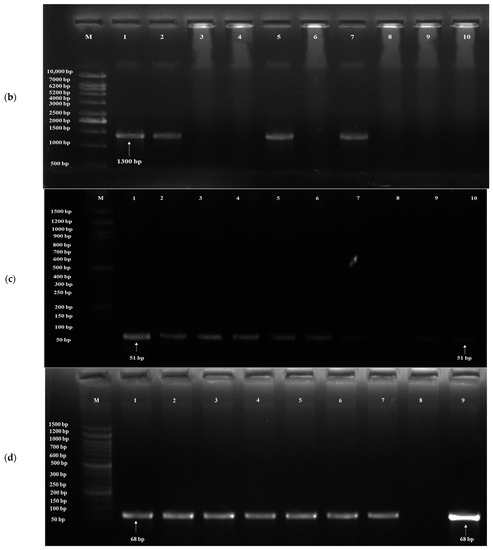
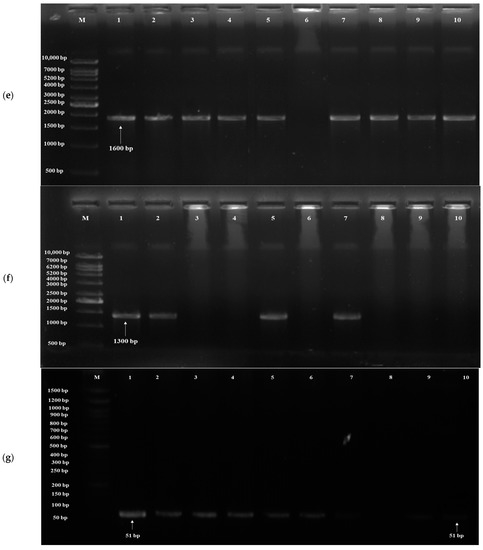
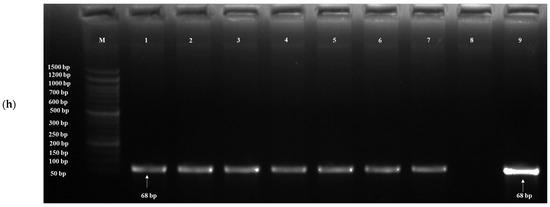
Figure 4.
(a) Agarose gel electrophoresis showing PCR amplification of TPS1 gene for acid tolerance. M: 1 kb DNA ladder; (1) Saccharomyces cerevisiae DAO-17; (2) S. cerevisiae DJT-2; (3) S. cerevisiae DKL-13; (4) S. cerevisiae DMS-15; (5) Kluyveromyces marxianus DPA-41; (6) Pichia bruneiensis CMS-3; (7) Debaryomyces prosopidis CPA-55; (8) Kazachstania exigua CMNB-4; (9) Meyerozyma guilliermondii CMN-3; (10) Yarrowia lipolytica CBG-9. (b) Agarose gel electrophoresis showing PCR amplification of SED1 gene for acid tolerance. M: 1 kb DNA ladder; (1) S. cerevisiae DAO-17; (2) K. marxianus DPA-41; (3) S. cerevisiae DJT-2; (4) S. cerevisiae DKL-13; (5) S. cerevisiae DRC-41; (6) S. cerevisiae DMS-15; (7) D. prosopidis CPA-55; (8) K. exigua CMB-19; (9) M. guilliermondii CMN-3; (10) Pichia bruneiensis CMS-3. (c) Agarose gel electrophoresis showing PCR amplification of YIM1 gene for bile tolerance. M: 50 bp DNA ladder; (1) S. cerevisiae DAO-17; (2) K. marxianus DPA-41; (3) S. cerevisiae DJT-2; (4) K. lactis DTL-6; (5) S. cerevisiae DRC-41; (6) S. cerevisiae DMS-15; (7) D. prosopidis CPA-55; (8) S. cerevisiae DKL-13; (9) M. guilliermondii CMN-3; (10) P. bruneiensis CMS-3. (d) Agarose gel electrophoresis showing PCR amplification of ERG3 gene for bile tolerance. M: 50 bp DNA ladder; (1) S. cerevisiae DAO-17; (2) S. cerevisiae DJT-2; (3) S. cerevisiae DMS-15; (4) M. guilliermondii CMN-3; (5) K. lactis DTL-6; (6) S. cerevisiae CGI-29; (7) D. prosopidis CPA-55; (8) S. cerevisiae DKL-13; (9) K. exigua CMNB-4. (e) Agarose gel electrophoresis showing PCR amplification of FLO1 gene for adhesion. M: 1 kb DNA ladder; (1) S. cerevisiae DAO-17; (2) S. cerevisiae DJT-2; (3) S. cerevisiae DKL-13; (4) K. marxianus DPA-41; (5) K. lactis DTL-6; (6) S. cerevisiae CGI-29; (7) D. prosopidis CPA-55; (8) S. cerevisiae CRC-35; (9) K. exigua CMNB-4; (10) Y. lipolytica CBG-9. (f) Agarose gel electrophoresis showing PCR amplification of AGA1 gene for adhesion. (1) Saccharomyces cerevisiae DAO-17; (2) S. cerevisiae DMS-15; (3) S. cerevisiae DJT-2; (4) S. cerevisiae DKL-13; (5) K. lactis DTL-6; (6) S. cerevisiae DRC-42; (7) M. guilliermondii CMN-3; (8) S. cerevisiae CRC-35; (9) P. kudriavzevii CNT-3; (10) Y. lipolytica CBG-9; M: 100 bp DNA ladder. (g) Agarose gel electrophoresis showing PCR amplification of Apid gene for antimicrobial activity. M: 1 kb DNA ladder; (1) S. cerevisiae DKL-13; (2) S. cerevisiae DJT-2; (3) S. cerevisiae CKL-10; (4) S. cerevisiae DMS-15; (5) K. lactis DTL-6; (6) Pichia bruneiensis CMS-3; (7) D. prosopidis CPA-55; (8) K. exigua CMNB-4; (9) M. guilliermondii CMN-3; (10) Y. lipolytica CBG-9. (h) Agarose gel electrophoresis showing PCR amplification of pelA gene for antimicrobial activity. M: 1 kb DNA ladder; (1) S. cerevisiae DKL-13; (2) S. cerevisiae DJT-2; (3) K. lactis DTL-6; (4) S. cerevisiae DMS-15; (5) S. cerevisiae CKL-10; (6) P. bruneiensis CMS-3; (7) D. prosopidis CPA-55; (8) Saccharomyces cerevisiae CGI-29; (9) M. guilliermondii CMN-3; (10) S. cerevisiae CRC-35.
4. Discussion
Probiotic properties in fermented diary products have several clinically tested health benefits that mostly originated from some lactic acid bacteria and few non-lactic acid bacteria [68,69]. In comparison to bacterial probiotics, the potentiality of yeasts as probiotics is inadequately studied, although yeasts are very important for sustaining the balance of the GI tract such as antagonistic interactions against noxious microbiota [3]. However, it has been reported that along with bacteria, some yeasts that originated from fermented dairy foods also have potential probiotic properties [29,70]. Several studies showed that yeasts could positively interact with probiotic bacteria by enhancing their survival and stimulating their growth [28,70,71,72]. Both bacteria and yeasts are reported in homemade fermented dairy products of Sikkim [36,38,39]. Dahi and chhurpi are the most common dietary items in the local gastronomy of Sikkim in India for lactose-intolerant ethnic consumers, which supplement nutritional value and digestibility, as well as desirable organoleptic taste. Dahi and chhurpi are acidic in nature with high apparent viscosity. High apparent viscosity improves the texture, quality and mouthfeel of fermented dairy products [73]. Earlier we reported few probiotic lactic acid bacteria from dahi and chhurpi of Sikkim [20,40]. Since yeasts are also co-existing in these dairy products, we screened some probiotic yeasts and studied their probiotic properties in these artisan-fermented dairy products.
Those yeasts, which resist low pH and low bile salts in human GI tracts, are apparently considered to possess the potential probiotics characteristics [50,74]. Moreover, survival in low pH and resistance to low bile salts are considered as the preliminary screening criteria for probable probiotic characters of yeasts [75]. We choose the resistances to low pH and low bile salts as preliminary screening parameters for possible probiotic attributes in the samples. On the basis of growth survival in low pH and low bile salts, 115 isolates were preliminarily screened and were further screened for their high hydrophobic characteristics. The ability of microorganisms to adhere to the epithelial cells and mucosal surfaces is critical for probiotic selection [76]. Hence, hydrophobicity is considered as one the important criteria for the selection of yeasts as probiotics [49]. However, although ≥70% hydrophobicity is considered to be hydrophobic in nature [77], in this experiment we selected ≥80% hydrophobicity as the threshold for the selection of high hydrophobicity [50,78]. Finally, 20 yeasts were selected as potential probiotic yeasts, which included Saccharomyces cerevisiae, Kluyveromyces marxianus, K. lactis, Debaryomyces prosopidis, Kazachstania exigua, Pichia bruneiensis, P. kudriavzevii, Meyerozyma guilliermondii and Yarrowia lipolytica. These yeasts are normally present in naturally fermented dairy products [28,79,80,81].
Auto-aggregation of probiotic strains is necessary for their adhesion to intestinal epithelial cells [82]. It has been reported that an auto-aggregation ability above 80% is considered as a strong adhesion ability [83,84]. In our study, we observed that Saccharomyces cerevisiae DRC-42 (dahi) and S. cerevisiae CGI-29 (chhurpi) showed 93.88% and 91.69% auto-aggregation, respectively, indicating their strong adherence abilities as the probiotic character. Some yeasts from dahi and chhurpi showed efficient co-aggregation with Escherichia coli, Salmonella enterica subsp. enterica ser. typhimurium, Staphylococcus aureus subsp. aureus and Bacillus cereus, possibly preventing bacterial colonization and secreting antimicrobial substances [85]. One of the most desirable properties of probiotic yeasts is their antibacterial activity against pathogens that penetrate various mucosal sites [71,86]. Many yeasts from dahi and chhurpi show antimicrobial activity against some pathogenic bacteria. Some yeasts strains from dahi and chhurpi differed in their bile salt hydrolase (BSH) substrate preference and activity. Kluyveromyces marxianus DPA-41 (dahi) and Pichia kudriavzevii CNT-3 (chhurpi) showed excellent deconjugation activities for sodium cholate, sodium taurocholates and sodium glycocholate. BSH activity is a relevant property for probiotic strains to survive the toxic effects of conjugated bile salts in the duodenum [57] and is also correlated with the ability to lower serum cholesterol levels in hypercholesterolemic patients [87]. Saccharomyces cerevisiae CGI-29 (chhurpi) showed the highest value of lysozyme resistance, which is considered as a promising criterion for the selection of new probiotic strains [29,70]. In vitro screening of probiotic yeasts Saccharomyces cerevisiae DAO-17 (dahi), S. cerevisiae CKL-10 (chhurpi) and Pichia kudriavzevii CNT-3 (chhurpi) showed maximum probiotic properties, which were also reported in other fermented dairy products as probiotic yeasts [47,49,50,75]. Though in vitro screening of probable probiotic yeasts is easy and common, the reliability of their probiotic abilities is not fully confirmed [88]. To get more reliable results, the genes responsible for major probiotic characteristics are detected by the PCR method using different primers [89,90]. However, genetic screening for probiotic yeasts is very limited compared to that for probiotic bacteria. We attempted to perform the limited gene detections for probiotic properties in yeasts isolated from dahi and chhurpi based on the target genes, probiotic traits and primers used [60,61,62,64,65]. Probiotic genes for acid tolerance, bile tolerance, adhesion and antimicrobial activity were detected in S. cerevisiae DAO-17, K. marxianus DPA-41, S. cerevisiae CKL-10 and P. kudriavzevii CNT-3. Genes involved in the acid shock condition are the TPS1, HSP150 and SED1 genes. However, the SED1 gene also induced under other stress conditions [91]. Finally, four yeast strains (Saccharomyces cerevisiae DAO-17 (dahi), S. cerevisiae CKL-10 (chhurpi), Pichia kudriavzevii CNT-3 (chhurpi) and Kluyveromyces marxianus DPA-41(dahi)) were selected as the potential probiotic yeasts based on results of in vitro and genetic screening.
5. Conclusions
This study mainly focused on the isolation of potential probiotic yeasts from popular homemade fermented dairy products of Sikkim, viz. dahi and chhurpi. Out of the 3438 yeasts isolated from these samples, only 20 yeasts were selected for in vitro and genetic screening of probiotic properties. Though our selection of probiotic candidates was based on limited in vitro and genetic screening for probiotic traits, some cultural yeast strains, viz. Saccharomyces cerevisiae DAO-17 (dahi), S. cerevisiae CKL-10 (chhurpi), Pichia kudriavzevii CNT-3 (chhurpi) and Kluyveromyces marxianus DPA-41(dahi), showed potential probiotic properties, which may be developed as probiotic yeast starter culture(s) for the production of functional dairy products. Further studies need to be done to study their functional properties and the whole genome analysis of potential strains.
Author Contributions
Conceptualization, J.P.T.; methodology, S.L.; software, S.L. and J.P.T.; investigation, S.L.; resources, J.P.T.; data curation, J.P.T.; writing—original draft preparation, S.L.; writing—review and editing, J.P.T.; visualization, S.L.; supervision, J.P.T.; project administration, J.P.T.; funding acquisition, J.P.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
All identified gene sequences based on the D1/D2 domain of large ribosomal RNA were deposited at GenBank-NCBI under the accession number: OP071251-OP071258, OP479874-OP479884 and OP561448.
Acknowledgments
Sonam Lama is grateful for the National Fellowship and Scholarship for Higher Education for ST students, Ministry of Tribal Affairs, Govt. of India, for financial support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mukhtar, H.; Yaqub, S.; ul Haq, I. Production of probiotic Mozzarella cheese by incorporating locally isolated Lactobacillus acidophilus. Ann. Microbiol. 2020, 70, 56. [Google Scholar] [CrossRef]
- Savaiano, D.A.; Hutkins, R.W. Yogurt, cultured fermented milk, and health: A systematic review. Nutr. Rev. 2021, 79, 599–614. [Google Scholar] [CrossRef]
- Zahoor, F.; Sooklim, C.; Songdech, P.; Duangpakdee, O.; Soontorngun, N. Selection of potential yeast probiotics and a cell factory for xylitol or acid production from honeybee samples. Metabolites 2021, 11, 312. [Google Scholar] [CrossRef] [PubMed]
- Hadjimbei, E.; Botsaris, G.; Chrysostomou, S. Beneficial effects of yoghurts and probiotic fermented milks and their functional food potential. Foods 2022, 11, 2691. [Google Scholar] [CrossRef] [PubMed]
- Lynch, K.M.; Wilkinson, S.; Daenen, L.; Arendt, E.K. An update on water kefir: Microbiology, composition and production. Int. J. Food Microbiol. 2021, 345, 109128. [Google Scholar] [CrossRef]
- Castellone, V.; Bancalari, E.; Rubert, J.; Gatti, M.; Neviani, E.; Bottari, B. Eating fermented: Health benefits of LAB-fermented foods. Foods 2021, 10, 2639. [Google Scholar] [CrossRef]
- Yilmaz, B.; Elibol, E.; Shangpliang, H.N.J.; Ozogul, F.; Tamang, J.P. Microbial communities in home-made and commercial kefir and their hypoglycemic properties. Fermentation 2022, 8, 590. [Google Scholar] [CrossRef]
- Rai, R.; Shangpliang, H.N.J.; Tamang, J.P. Naturally fermented milk products of the Eastern Himalayas. J. Ethn. Foods 2016, 3, 270–275. [Google Scholar] [CrossRef]
- Teneva-Angelova, T.; Balabanova, T.; Boyanova, P.; Beshkova, D. Traditional Balkan fermented milk products. Eng. Life Sci. 2018, 18, 807–819. [Google Scholar] [CrossRef] [PubMed]
- Agyei, D.; Owusu-Kwarteng, J.; Akabanda, F.; Akomea-Frempong, S. Indigenous African fermented dairy products: Processing technology, microbiology and health benefits. Crit. Rev. Food Sci. Nutr. 2020, 60, 991–1006. [Google Scholar] [CrossRef]
- Faccia, M.; D’Alessandro, A.; Summer, A.; Hailu, Y. Milk products from minor dairy species: A review. Animals 2020, 10, 1260. [Google Scholar] [CrossRef] [PubMed]
- Ganzorig, K.; Urashima, T.; Fukuda, K. Exploring potential bioactive peptides in fermented bactrian camel’s milk and mare’s milk made by Mongolian nomads. Foods 2020, 9, 1817. [Google Scholar] [CrossRef]
- Tamang, J.P.; Cotter, P.; Endo, A.; Han, N.S.; Kort, R.; Liu, S.Q.; Mayo, B.; Westerik, N.; Hutkins, R. Fermented foods in a global age: East meets west. Compr. Rev. Food Sci. Food Saf. 2020, 19, 184–217. [Google Scholar] [CrossRef]
- Tamang, J.P. “Ethno-Microbiology” of ethnic Indian fermented foods and alcoholic beverages. J. Appl. Microbiol. 2022, 133, 145–161. [Google Scholar] [CrossRef]
- Parker, M.; Zobrist, S.; Donahue, C.; Edick, C.; Mansen, K.; Hassan, Z.N.M.; Heerikhuisen, M.; Sybesma, W.; Molenaar, D.; Diallo, A.M.; et al. Naturally fermented milk from northern senegal: Bacterial community composition and probiotic enrichment with Lactobacillus rhamnosus. Front. Microbiol. 2018, 9, 2218. [Google Scholar] [CrossRef]
- Takeda, S.; Yamasaki, K.; Takeshita, M.; Kikuchi, Y.; Tsend-Ayush, C.; Dashnyam, B.; Ahhmed, A.M.; Kawahara, S.; Muguruma, M. The investigation of probiotic potential of lactic acid bacteria isolated from traditional Mongolian dairy products. Anim. Sci. J. 2011, 82, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Maleke, M.S.; Adefisoye, M.A.; Doorsamy, W.; Adebo, O.A. Processing, nutritional composition and microbiology of amasi: A Southern African fermented milk product. Sci. Afr. 2021, 12, e00795. [Google Scholar] [CrossRef]
- Venema, K.; Surono, I.S. Microbiota composition of dadih—A traditional fermented buffalo milk of West Sumatra. Lett. Appl. Microbiol. 2019, 68, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, R.; Chandragunasekaran, A.S.; Chellappan, G.; Rajaram, K.; Ramamoorthi, G.; Ramakrishna, B.S. Probiotic potential of lactic acid bacteria present in home made curd in southern India. Indian J. Med. Res. 2014, 140, 345–355. [Google Scholar]
- Rai, R.; Tamang, J.P. In vitro and genetic screening of probiotic properties of lactic acid bacteria isolated from naturally fermented cow-milk and yak-milk products of Sikkim, India. World J. Microbiol. Biotechnol. 2022, 38, 25. [Google Scholar] [CrossRef]
- Adegboye, B.; Banwo, K.; Ogunremi, O.; Sanni, A. Probiotic potentials of yeasts isolated from nunu (African fermented milk) and wara (African soft cheese). Adv. Food Sci. 2014, 36, 115–124. [Google Scholar]
- Tamang, J.P.; Watanabe, K.; Holzapfel, W.H. Diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 2016, 7, 377. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Ağagündüz, D.; Yılmaz, B.; Şahin, T.Ö.; Güneşliol, B.E.; Ayten, Ş.; Russo, P.; Spano, G.; Rocha, J.M.; Bartkiene, E.; Özogul, F. Dairy lactic acid bacteria and their potential function in dietetics: The food–gut-health axis. Foods 2021, 10, 3099. [Google Scholar] [CrossRef]
- Gaucher, F.; Kponouglo, K.; Rabah, H.; Bonnassie, S.; Ossemond, J.; Pottier, S.; Jardin, J.; Briard-Bion, V.; Marchand, P.; Blanc, P.; et al. Propionibacterium freudenreichii CIRM-BIA 129 osmoadaptation coupled to acid-adaptation increases its viability during freeze-drying. Front. Microbiol. 2019, 10, 2324. [Google Scholar] [CrossRef]
- Wong, C.B.; Odamaki, T.; Xiao, J.Z. Beneficial effects of Bifidobacterium longum subsp. longum BB536 on human health: Modulation of gut microbiome as the principal action. J. Funct. Foods 2019, 54, 506–519. [Google Scholar] [CrossRef]
- Gut, A.M.; Vasiljevic, T.; Yeager, T.; Donkor, O.N. Characterization of yeasts isolated from traditional kefir grains for potential probiotic properties. J. Funct. Foods 2019, 58, 56–66. [Google Scholar] [CrossRef]
- Galli, V.; Venturi, M.; Mari, E.; Guerrini, S.; Granchi, L. Selection of yeast and lactic acid bacteria strains, isolated from spontaneous raw milk fermentation, for the production of a potential probiotic fermented milk. Fermentation 2022, 8, 407. [Google Scholar] [CrossRef]
- Tamang, J.P.; Lama, S. Probiotic properties of yeasts in traditional fermented foods and beverages. J. Appl. Microbiol. 2022, 132, 3533–3542. [Google Scholar] [CrossRef]
- Roohvand, F.; Ehsani, P.; Abdollahpour-Alitappeh, M.; Shokri, M.; Kossari, N. Biomedical applications of yeasts—A patent view, part two: Era of humanized yeasts and expanded applications. Expert Opin. Ther. Pat. 2020, 30, 609–631. [Google Scholar] [CrossRef]
- Karaolis, C.; Botsaris, G.; Pantelides, I.; Tsaltas, D. Potential application of Saccharomyces boulardii as a probiotic in goat’s yoghurt: Survival and organoleptic effects. Int. J. Food Sci. Technol. 2013, 48, 1445–1452. [Google Scholar] [CrossRef]
- Sarwar, A.; Aziz, T.; Al-Dalali, S.; Zhao, X.; Zhang, J.; ud Din, J.; Chen, C.; Cao, Y.; Yang, Z. Physicochemical and microbiological properties of synbiotic yogurt made with probiotic yeast Saccharomyces boulardii in Combination with inulin. Foods 2019, 8, 468. [Google Scholar] [CrossRef]
- Ansari, F.; Alian Samakkhah, S.; Bahadori, A.; Jafari, S.M.; Ziaee, M.; Khodayari, M.T.; Pourjafar, H. Health-promoting properties of Saccharomyces cerevisiae var. boulardii as a probiotic; characteristics, isolation, and applications in dairy products. Crit. Rev. Food Sci. Nutr. 2021, 13, 1949577. [Google Scholar] [CrossRef] [PubMed]
- Tamang, J.P.; Jeyaram, K.; Rai, A.K.; Mukherjee, P.K. Diversity of beneficial microorganisms and their functionalities in community-specific ethnic fermented foods of the Eastern Himalayas. Food Res. Int. 2021, 148, 110633. [Google Scholar] [CrossRef]
- Tamang, J.P. Dietary culture and antiquity of the Himalayan fermented foods and alcoholic fermented beverages. J. Ethn. Foods 2022, 9, 30. [Google Scholar] [CrossRef]
- Shangpliang, H.; Rai, R.; Keisam, S.; Jeyaram, K.; Tamang, J.P. Bacterial community in naturally fermented milk products of Arunachal Pradesh and Sikkim of India analysed by high-throughput amplicon sequencing. Sci. Rep. 2018, 8, 1532. [Google Scholar] [CrossRef] [PubMed]
- Shangpliang, H.N.J.; Tamang, J.P. Phenotypic and genotypic characterisation of lactic acid bacteria isolated from exotic naturally fermented milk (cow and yak) products of Arunachal Pradesh, India. Int. Dairy J. 2021, 118, 105038. [Google Scholar] [CrossRef]
- Dewan, S.; Tamang, J.P. Microbial and analytical characterization of Chhu-A traditional fermented milk product of Sikkim Himalayas. J. Sci. Ind. Res. 2006, 65, 747–752. [Google Scholar]
- Dewan, S.; Tamang, J.P. Dominant lactic acid bacteria and their technological properties isolated from the Himalayan ethnic fermented milk products. Antonie Leeuwenhoek 2007, 92, 343–352. [Google Scholar] [CrossRef]
- Tamang, J.P.; Dewan, S.; Thapa, S.; Olasupo, N.A.; Schillinger, U.; Wijaya, A.; Holzapfel, W.H. Identification and enzymatic profiles of the predominant lactic acid bacteria isolated from soft-variety Chhurpi, a traditional cheese typical of the Sikkim Himalayas. Food Biotechnol. 2000, 14, 99–112. [Google Scholar] [CrossRef]
- Goel, A.; Halami, P.M.; Tamang, J.P. Genome analysis of Lactobacillus plantarum isolated from some Indian fermented foods for bacteriocin production and probiotic marker genes. Front. Microbiol. 2020, 11, 40. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.K.; Kumari, R.; Sanjukta, S.; Sahoo, D. Production of bioactive protein hydrolysate using the yeasts isolated from soft chhurpi. Bioresour. Technol. 2016, 1, 239–245. [Google Scholar] [CrossRef]
- Abedin, A.M.; Chourasia, R.; Phukon, L.C.; Singh, S.P.; Rai, A.K. Characterization of ACE inhibitory and antioxidant peptides in yak and cow milk hard chhurpi cheese of the Sikkim Himalayan region. Food Chem. 2022, 13, 100231. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis, 20th ed.; Association of Official Analytical Chemists: Rockville, MD, USA, 2016. [Google Scholar]
- Pratap, D.; Maurya, V.K.; Kumar, N.; Singh, R.; Upadhyay, A. Studies on physico-chemical and organoleptic properties of soymilk blended dahi (curd) with toned milk (cattle milk). Curr. Nutr. Food Sci. 2018, 14, 61–67. [Google Scholar] [CrossRef]
- Ali, K.; Mehmood, M.H.; Iqbal, M.A.; Masud, T.; Qazalbash, M.; Saleem, S.; Ahmed, S.; Tariq, M.R.; Safdar, W.; Nasir, M.A.; et al. Isolation and characterization of exopolysaccharide-producing strains of Lactobacillus bulgaricus from curd. Food Sci. Nutr. 2019, 7, 1207–1213. [Google Scholar] [CrossRef]
- Greppi, A.; Saubade, F.; Botta, C.; Humblot, C.; Guyot, J.P.; Cocolin, L. Potential probiotic Pichia kudriavzevii strains and their ability to enhance folate content of traditional cereal-based African fermented food. Food Microbiol. 2017, 62, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Zhang, Y.; Zhang, Y.; Liu, Y.; Wang, S.; Dong, X.; Wang, Y.; Zhang, H. Screening of potential probiotic properties of Lactobacillus fermentum isolated from traditional dairy products. Food Control 2010, 21, 695–701. [Google Scholar] [CrossRef]
- Fernandez-Pacheco, P.; Arévalo-Villena, M.; Bevilacqua, A.; Corbo, M.R.; Pérez, A.B. Probiotic characteristics in Saccharomyces cerevisiae strains: Properties for application in food industries. LWT-Food Sci. Technol. 2018, 97, 332–340. [Google Scholar] [CrossRef]
- Alkalbani, N.S.; Osaili, T.M.; Al-Nabulsi, A.A.; Obaid, R.S.; Olaimat, A.N.; Liu, S.Q.; Ayyash, M.M. In vitro characterization and identification of potential probiotic yeasts isolated from fermented dairy and non-dairy food products. J. Fungi 2022, 8, 544. [Google Scholar] [CrossRef]
- Umeh, S.O.; Agwuna, L.C.; Okafor, U.C. Yeasts from local sources: An alternative to the conventional brewer’s yeast. Int. J. Biotechnol. Food Sci. 2017, 30, 191–195. [Google Scholar]
- Kotzekidou, P. Identification of yeasts from black olives in rapid system microtitre plates. Food Microbiol. 1997, 14, 609–616. [Google Scholar] [CrossRef]
- Oliveira, V.A.; Vicente, M.A.; Fietto, L.G.; de Miranda Castro, I.; Coutrim, M.X.; Schüller, D.; Alves, H.; Casal, M.; de Oliveira Santos, J.; Araújo, L.D.; et al. Biochemical and molecular characterization of Saccharomyces cerevisiae strains obtained from sugar-cane juice fermentations and their impact in cachaca production. Appl. Environ. Microbiol. 2008, 74, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Renshaw, M.A.; Olds, B.P.; Jerde, C.L.; McVeigh, M.M.; Lodge, D.M. The room temperature preservation of filtered environmental DNA samples and assimilation into a phenol–chloroform–isoamyl alcohol DNA extraction. Mol. Ecol. Resour. 2015, 15, 168–176. [Google Scholar] [CrossRef]
- Palla, M.; Blandino, M.; Grassi, A.; Giordano, D.; Sgherri, C.; Quartacci, M.F.; Reyneri, A.; Agnolucci, M.; Giovannetti, M. Characterization and selection of functional yeast strains during sourdough fermentation of different cereal wholegrain flours. Sci. Rep. 2020, 10, 12856. [Google Scholar] [CrossRef] [PubMed]
- Ogunremi, O.; Sanni, A.; Agrawal, R. Probiotic potentials of yeasts isolated from some cereal-based Nigerian traditional fermented food products. J. Appl. Microbiol. 2015, 119, 797–808. [Google Scholar] [CrossRef] [PubMed]
- Fadda, M.E.; Mossa, V.; Deplano, M.; Pisano, M.B.; Cosentino, S. In vitro screening of Kluyveromyces strains isolated from Fiore Sardo cheese for potential use as probiotics. LWT-Food Sci. Technol. 2017, 75, 100–106. [Google Scholar] [CrossRef]
- Vera-Pingitore, E.; Jimenez, M.E.; Dallagnol, A.; Belfiore, C.; Fontana, C.; Fontana, P.; von Wright, A.; Vignolo, G.; Plumed-Ferrer, C. Screening and characterization of potential probiotic and starter bacteria for plant fermentations. LWT-Food Sci. Technol. 2016, 1, 288–294. [Google Scholar] [CrossRef]
- Wilson, R.A.; Jenkinson, J.M.; Gibson, R.P.; Littlechild, J.A.; Wang, Z.Y.; Talbot, N.J. Tps1 regulates the pentose phosphate pathway, nitrogen metabolism and fungal virulence. EMBO J. 2007, 26, 3673–3685. [Google Scholar] [CrossRef]
- Marinangeli, P.; Angelozzi, D.; Ciani, M.; Clementi, F.; Mannazzu, I. Minisatellites in Saccharomyces cerevisiae genes encoding cell wall proteins: A new way towards wine strain characterisation. FEMS Yeast Res. 2004, 4, 427–435. [Google Scholar] [CrossRef][Green Version]
- Mannazzu, I.; Simonetti, E.; Marinangeli, P.; Guerra, E.; Budroni, M.; Thangavelu, M.; Clementi, F. SED1 gene length and sequence polymorphisms in feral strains of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2002, 68, 5437–5444. [Google Scholar] [CrossRef]
- Caudle, K.E.; Barker, K.S.; Wiederhold, N.P.; Xu, L.; Homayouni, R.; Rogers, P.D. Genome wide expression profile analysis of the Candida glabrata Pdr1 regulon. Eukaryot. Cell 2011, 10, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.S.; Choo, Y.M.; Lee, K.S.; Yoon, H.J.; Kim, I.; Je, Y.H.; Sohn, H.D.; Jin, B.R. Cloning and expression profiling of four antibacterial peptide genes from the bumblebee Bombus ignitus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2008, 15, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Younis, G.; Awad, A.; Dawod, R.E.; Yousef, N.E. Antimicrobial activity of yeasts against some pathogenic bacteria. Vet. World. 2017, 10, 979. [Google Scholar] [CrossRef] [PubMed]
- Damas-Buenrostro, L.C.; Gracia-González, G.; Hernández-Luna, C.E.; Galán-Wong, L.J.; Pereyra-Alférez, B.; Sierra-Benavides, J.A. Detection of FLO genes in lager and wild yeast strains. J. Am. Soc. Brew. Chem. 2008, 66, 184–187. [Google Scholar] [CrossRef]
- Nutaratat, P.; Boontham, W.; Khunnamwong, P. A novel yeast genus and two novel species isolated from pineapple leaves in Thailand: Savitreella phatthalungensis gen. nov., sp. nov. and Goffeauzyma siamensis sp. nov. J. Fungi 2022, 8, 118. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- García-Burgos, M.; Moreno-Fernández, J.; Alférez, M.J.M.; Díaz-Castro, J.; López-Aliaga, I. New perspectives in fermented dairy products and their health relevance. J. Funct. Foods 2020, 72, 104059. [Google Scholar] [CrossRef]
- Widyastuti, Y.; Febrisiantosa, A.; Tidona, F. Health-Promoting properties of lactobacilli in fermented dairy products. Front. Microbiol. 2021, 12, 673890. [Google Scholar] [CrossRef]
- Staniszewski, A.; Kordowska-Wiater, M. Probiotic and potentially probiotic yeasts-characteristics and food application. Foods 2021, 10, 1306. [Google Scholar] [CrossRef]
- Hatoum, R.; Labrie, S.; Fliss, I. Antimicrobial and probiotic properties of yeasts: From fundamental to novel applications. Front. Microbiol. 2012, 3, 421. [Google Scholar] [CrossRef]
- Suharja, A.; Henriksson, A.; Liu, S.Q. Impact of Saccharomyces cerevisiae on Viability of probiotic Lactobacillus rhamnosus in fermented milk under ambient conditions. J. Food Process. Preserv. 2014, 38, 326–337. [Google Scholar] [CrossRef]
- Buldo, P.; Sokolowsky, M.; Hoegholm, T. The role of starter cultures on oral processing properties of different fermented milk products. Food Hydrocoll. 2021, 114, 106571. [Google Scholar] [CrossRef]
- Markowiak, P.; Śliżewska, K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients 2017, 9, 1021. [Google Scholar] [CrossRef]
- Moradi, R.; Nosrati, R.; Zare, H.; Tahmasebi, T.; Saderi, H.; Owlia, P. Screening and characterization of in-vitro probiotic criteria of Saccharomyces and Kluyveromyces strains. Iran J. Microbiol. 2018, 10, 123–131. [Google Scholar]
- Kechagia, M.; Basoulis, D.; Konstantopoulou, S.; Dimitriadi, D.; Gyftopoulou, K.; Skarmoutsou, N.; Fakiri, E.M. Health benefits of probiotics: A review. ISRN Nutr. 2013, 2013, 481651. [Google Scholar] [CrossRef] [PubMed]
- Nostro, A.; Cannatelli, M.A.; Crisafi, G.; Musolino, A.D.; Procopio, F.; Alonzo, V. Modifications of hydrophobicity, in vitro adherence and cellular aggregation of Streptococcus mutans by Helichrysum italicum extract. Lett. Appl. Microbiol. 2004, 38, 423–427. [Google Scholar] [CrossRef]
- de Oliveira Coelho, B.; Fiorda-Mello, F.; de Melo Pereira, G.V.; Thomaz-Soccol, V.; Rakshit, S.K.; de Carvalho, J.C.; Soccol, C.R. In vitro probiotic properties and dna protection activity of yeast and lactic acid bacteria isolated from a honey-based kefir beverage. Foods 2019, 8, 485. [Google Scholar] [CrossRef]
- Kumura, H.; Tanoue, Y.; Tsukahara, M.; Tanaka, T.; Shimazaki, K. Screening of dairy yeast strains for probiotic applications. J. Dairy Sci. 2004, 87, 4050–4056. [Google Scholar] [CrossRef]
- Cissé, H.; Kagambèga, B.; Sawadogo, A.; Tankoano, A.; Sangaré, G.; Traoré, Y.; Ouoba, I.I.L.; Savadogo, A. Molecular characterization of Bacillus, lactic acid bacteria and yeast as potential probiotic isolated from fermented food. Sci. Afr. 2019, 6, 00175. [Google Scholar] [CrossRef]
- Nahidul-Islam, S.M.; Kuda, T.; Takahashi, H.; Kimura, B. Bacterial and fungal microbiota in traditional Bangladeshi fermented milk products analysed by culture-dependent and culture-independent methods. Food Res. Int. 2018, 111, 431–437. [Google Scholar] [CrossRef]
- Pérez, P.F.; Minnaard, Y.; Disalvo, E.A.; De Antoni, G.L. Surface properties of bifidobacterial strains of human origin. Appl. Environ. Microbiol. 1998, 64, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Syal, P.; Vohra, A. Probiotic potential of yeasts isolated from traditional Indian fermented foods. Int. J. Microbiol. Res. 2013, 5, 390. [Google Scholar] [CrossRef]
- Bonatsou, S.; Karamouza, M.; Zoumpopoulou, G.; Mavrogonatou, E.; Kletsas, D.; Papadimitriou, K.; Tsakalidou, E.; Nychas, G.J.E.; Panagou, E.Ζ. Evaluating the probiotic potential and technological characteristics of yeasts implicated in cv. Kalamata natural black olive fermentation. Int. J. Food Microbiol. 2018, 271, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Suvarna, S.; Dsouza, J.; Ragavan, M.L.; Das, N. Potential probiotic characterization and effect of encapsulation of probiotic yeast strains on survival in simulated gastrointestinal tract condition. Food Sci. Biotechnol. 2018, 27, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Fakruddin, M.; Hossain, M.N.; Ahmed, M.M. Antimicrobial and antioxidant activities of Saccharomyces cerevisiae IFST062013, a potential probiotic. BMC Complement. Altern. Med. 2017, 17, 64. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Gómez, J.G.; López-Bonilla, A.; Trejo-Tapia, G.; Ávila-Reyes, S.V.; Jiménez-Aparicio, A.R.; Hernández-Sánchez, H. In vitro bile salt hydrolase (BSH) activity screening of different probiotic microorganisms. Foods 2021, 10, 674. [Google Scholar] [CrossRef] [PubMed]
- Papadimitriou, K.; Zoumpopoulou, G.; Foligné, B.; Alexandraki, V.; Kazou, M.; Pot, B.; Tsakalidou, E. Discovering probiotic microorganisms: In vitro, in vivo, genetic and omics approaches. Front. Microbiol. 2015, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Turpin, W.; Humblot, C.; Guyot, J.P. Genetic screening of functional properties of lactic acid bacteria in a fermented pearl millet slurry and in the metagenome of fermented starchy foods. Appl. Environ. Microbiol. 2011, 77, 8722–8734. [Google Scholar] [CrossRef] [PubMed]
- Turpin, W.; Humblot, C.; Noordine, M.L.; Thomas, M.; Guyot, J.P. Lactobacillaceae and cell adhesion: Genomic and functional screening. PLoS ONE 2012, 7, e38034. [Google Scholar] [CrossRef] [PubMed]
- Kawahata, M.; Masaki, K.; Fujii, T.; Iefuji, H. Yeast genes involved in response to lactic acid and acetic acid: Acidic conditions caused by the organic acids in Saccharomyces cerevisiae cultures induce expression of intracellular metal metabolism genes regulated by Aft1p. FEMS Yeast Res. 2006, 6, 924–936. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).