1. Introduction
Spontaneous sourdough is a complex biological system obtained after the fermentation of cereal flour and water by means of bacteria and yeast, mostly deriving from the raw ingredients, the bread-making environment, and the bakers [
1]. The microorganisms in sourdough mainly belong to lactic acid bacteria (LAB) and yeast, which ferment the carbohydrates in flour, producing the carbon dioxide responsible for the bread dough rise and other metabolites such as organic acids and alcohols, which are responsible for the organoleptic properties of bread (flavor, texture and shelf life). Since ancient times, sourdough has been used as a natural leavening agent in bread production and shared among artisanal bakers and home-baking communities. Commonly, sourdoughs are classified into four types [
2]:
Type I sourdough is derived from a spontaneous fermentation process, which is followed by a daily backslopping, consisting in a cyclic reinoculation of a so-called “mother dough” using a newly prepared batch of flour and water. Fermentation temperature is set between 20–30 °C and the fermentation time ranges from 5 to 24 h. This sourdough can be refrigerated at regular intervals or at occurrence, and it is most commonly used in artisanal bakeries. The dough yield (ratio between the dough obtained and the flour used) is not exceeding 200.
Type II sourdough is obtained when a flour–water mixture is inoculated with LAB and yeast. Fermentation is conducted for one or more days and temperature is set above 30 °C. Sometimes, yeast is added at the final stage of the fermentation process. Sourdough type II is a semiliquid product with a dough yield between 200–300, and for this reason it is usually employed at an industrial scale.
When sourdough type II is commercialized in the form of a semidried product, it is called sourdough type III.
Type IV sourdough is a combination of type II and type I; in fact, the sourdough is obtained by inoculating selected microorganisms in a mixture of flour and water, and thereafter it is maintained by a backslopping procedure according to the type I sourdough method.
The fermentation of sourdough by spontaneous microflora can lead to the unpredictable growth of various microorganisms. The mutation of the sourdough microbial community over time is conditioned by the non-sterile and open-batch conditions of sourdough and by the use of ingredients potential microbial sources, as the flour, the water or the devices which are used in the backslopping process. Moreover, the development and the behavior of microbial species and the competitiveness between species depend on a multitude of factors, such as temperature, dough yield, and microbial metabolites [
3]. The use of spontaneously fermented sourdough can result in unstable product quality.
The use of selected LAB and yeasts as a starter in sourdough technology has become a common practice, mainly in industrial production. Starter microorganisms can be used as flavor carriers and texture improvers, or for their antifungal or health-promoting properties, in order to improve the performances and the properties of sourdough [
4,
5].
When starter microorganisms are inoculated, as in sourdough type IV, there is a competition between the starter microorganisms and the spontaneously growing microorganisms, and if the starter cultures cannot adapt to the sourdough substrate, and to the acidic conditions, then the spontaneously growing microorganisms can dominate [
6]. Moreover, De Vuyst and Neysen [
7] observed that the persistence of a microbial association over time is dependent on several factors, such as the process parameters (temperature, dough yield, time of fermentation) or microbial competition (bacteriocin production).
Until now, about 70 different species of LAB [
8] and 40 species of yeast have been identified in the sourdough environment. Among LAB,
L.
plantarum,
F. sanfranciscensis and
L. brevis were the most isolated species in worldwide sourdoughs [
4]. Young sourdoughs can harbor a consistent number of spontaneous microorganisms, but they were largely dominated by
L. plantarum and
L. brevis, while
F. sanfranciscensis is considered predominant in traditionally prepared and older sourdoughs [
7,
9]; in fact, when the sourdough becomes mature, the diversity of microflora decreases, mostly because the organic acids produced by LAB select for acid-tolerant microorganisms [
2].
This study describes the dynamics of the microbiota in three liquid sourdoughs that were propagated for 22 days in a laboratory. One of them was also studied after transferring to and propagation in a bakery. The sourdoughs were triggered using the same microbial starter, composed of L. plantarum, S. cerevisiae and C. lambica, with the purpose to guide the fermentation and dominate the mature sourdoughs. The starter strains were inoculated using different methods and sourdoughs were propagated under two different fermentation temperatures, 25 °C and 20 °C, in order to study the behavior of starter strains. To investigate the dynamics of the microbial population, culture-dependent and metataxonomic methods were applied.
2. Materials and Methods
2.1. Microorganisms and Growth Conditions
The strains of
L. plantarum PCC1034,
S. cerevisiae PCC1662 and
C. lambica PCC1649, isolated from Italian sourdoughs and belonging to the culture collection of Porto Conte Ricerche, were used as starters to inoculate a mixture of flour and water. The strains were identified with the instrument MALDI Biotyper (MicroFlex™, Bruker Daltonik GmbH, Bremen, Germany), using the software MBT Compass
® 4.1 and the attached libraries (Bruker Daltonik GmbH, Bremen, Germany). The
L. plantarum PCC1034 was selected among other strains, belonging to the same species, for its acidification capacity at different growth temperatures. Microbial strains were stored at −80 °C. The LAB strain was routinely propagated in MRS liquid medium (Oxoid, Basingstoke, Hampshire, UK), modified [
10] with the addition of fresh yeast extract (5%,
v/
v) and 1% maltose at a final pH of 5.5 (mMRS), and incubated at 28 °C in anaerobic conditions. The yeast strains were propagated in YEPD liquid medium [
10] and incubated at 28 °C under stirring conditions.
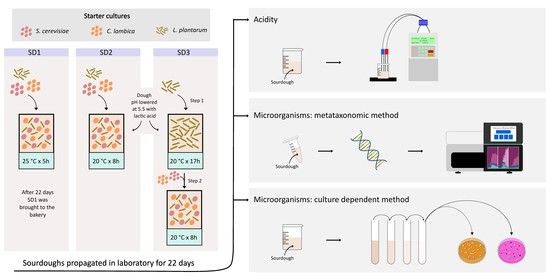
2.2. Preparation of Liquid Sourdough and Laboratory Propagation
In order to prepare the liquid sourdough, the selected strains of bacteria and yeast were inoculated in mMRS and YEPD liquid media, respectively, and incubated at 28 °C for 24 h. The next day, 400 μL of the bacterial culture and 200 μL of each yeast culture were inoculated in 40 mL of fresh mMRS and in 20 mL of fresh YEPD, respectively, and incubated at 28 °C. After 24 h, the cells were harvested (6076×
g for 10 min at 4 °C), resuspended in 10 mL of physiological solution and used to inoculate a mixture of 1.5 kg re-milled semolina (Mulino Brundu, Torralba, Italy) and 1.5 kg of sterile water. Liquid sourdough was managed in the laboratory using the Automatic Fermenter AFT5 (SITEP S.r.l., Voghiera, Italy). With regard to the cell number of inoculated strains, it was detected in each dough after inoculum. For all samples, the cell concentration was of the order of 10
7 UFC g
−1 for bacteria and 10
5 UFC g
−1 for yeast strains. Values were reported in
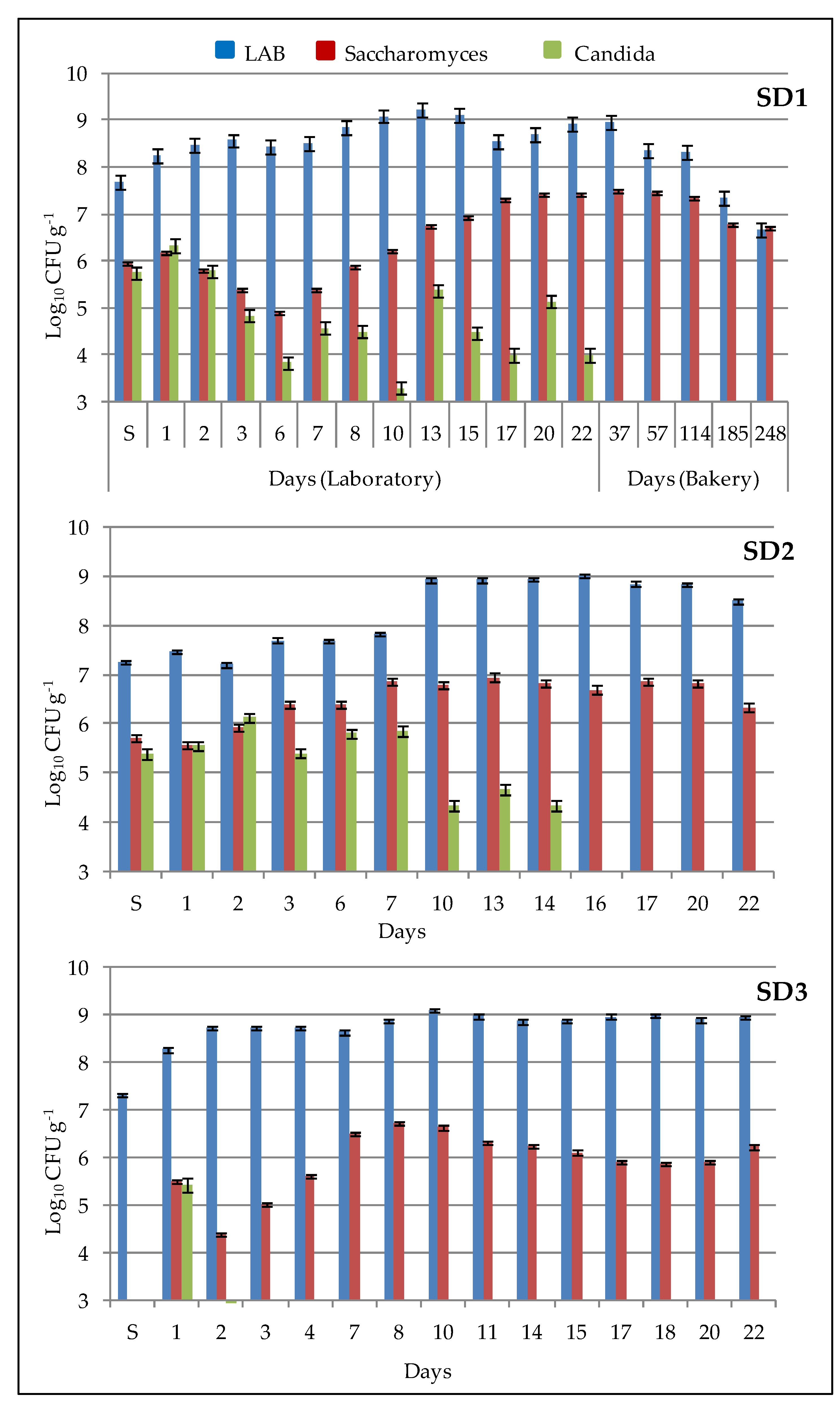
Figure 1 and indicated as “S”. The sourdoughs were refreshed over 22 days from Monday to Friday by a daily backslopping procedure, mixing an equal amount (ratio 1:1:1) of mother sourdough, fresh re-milled semolina and water, which was previously autoclaved 15 min at 120 °C. After the fermentation process, the sourdough was stored at a low temperature (5 ± 2 °C). The value of dough yield (DY) was 200.
Three different sourdoughs were prepared, called SD1, SD2 and SD3, using different methods of inoculum for the starter strains and different fermentation conditions, as follows:
For SD1, the starter strains were inoculated together. The pH of the dough was about 6.5 before starter addition. Fermentation was carried out at 25 °C for 5 h.
For SD2, the starter strains were inoculated together. The pH of the dough was lowered before starter addition at value 5.5, using lactic acid 90% (Sigma-Aldrich, Milan, Italy). Fermentation was carried out at 20 °C for 8 h.
For SD3, the pH of the dough was lowered before starter addition, as in SD2. The starter strains were inoculated separately. At first, the L. plantarum was inoculated, and it was left to ferment at 20 °C for 17 h. The following day, the yeast strains were inoculated throughout the refreshment step, and fermentation was carried out at 20 °C for 8 h.
The pH was lowered in order to support the growth of L. plantarum. The fermentation times were selected based on experience, as such values allowed sourdough pH values in between 4.0 and 4.5.
Samples of sourdoughs used for microbiological, chemical and metataxonomic analyses were collected about 24 h after the refreshment step, during the low-temperature phase, whereas the sample collected on Monday refers to the sourdough refreshed on Friday.
2.3. Refreshment of Liquid Sourdough in the Bakery
After 22 days, the liquid sourdough SD1 was transferred to an artisanal bakery (MFM Sunalle, Fonni, Italy), referred to as SD1-bak, and used for baking purposes. The sourdough was refreshed twice a week with the aid of the automatic bioreactor Fermentolevain FL80 (Esmach, Italy), mixing sourdough, semolina and water in a ratio of 1:1:1. Fermentation was carried out at 25 °C for 5 h, then the sourdough was cooled to 5 °C. Approximately every 2 months, and up to 8 months, one sample of sourdough was transferred to the Porto Conte Ricerche laboratory and analyzed.
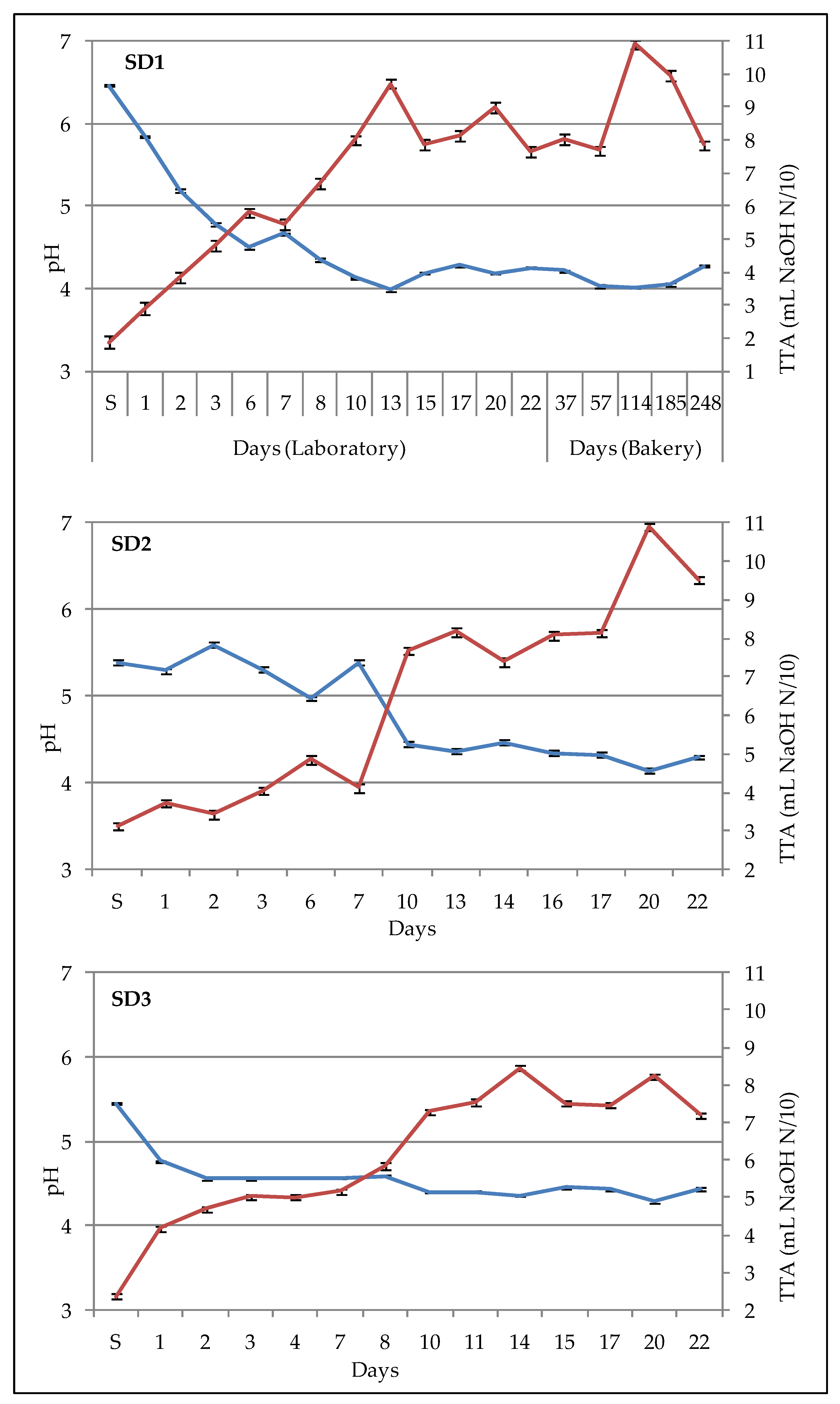
2.4. Determination of pH and Total Titratable Acidity
Ten grams of liquid sourdough were mixed with 90 mL of distillated water and stirred for 30 min, and then an automatic titrator (pH-Matic 23, Crison Instruments, Alella, Spain) was used to measure pH values and total titratable acidity (TTA), the latter was reported as the amount (mL) of NaOH N/10 to achieve pH 8.5 in 10 g of sample. Analyses were done in triplicate.
2.5. Analysis of Sourdough Microorganisms—Culture Dependent Approach
The viable cell number of bacteria and yeast growing in sourdough was estimated by plate-count technique. Ten grams of sourdough were mixed for 2 min with 90 mL of sterile peptone solution (1 g/L of peptone in distilled water) in a sterile stomacher bag, using a Stomacher Lab blender 80 (VWR International PBI, Milano, Italy). Serial dilutions were performed and plated onto mMRS agar for bacterial enumeration and Rose-Bengal Chloramphenicol agar (Oxoid, Basingstoke, UK) for yeast enumeration. Plates of mMRS were incubated under anaerobiosis (AnaeroGen and AnaeroJar, Oxoid, Basingstoke, UK) at 28 °C for 48 h. Rose-Bengal Chloramphenicol agar plates (RB) were incubated at 28 °C for 72 h. The use of RB plates allowed for differentiating Saccharomyces from Candida based on colony morphology. Candida had white colonies with a rugged surface from which “feet” extended from the margins into the surrounding agar, and Saccharomyces colonies were circular in shape, violet and had a smooth surface.
2.6. Analysis of Sourdough Microorganisms—Metataxonomic Approach
2.6.1. Nucleic Acid Extraction and High-Throughput Sequencing Analysis
DNA was extracted from the samples collected in the three experiments, SD1, SD2 and SD3. Extraction was performed following the procedure reported in “Manual DNA Extraction from Food Samples” [
11] and using the ReliaPrep™ Blood gDNA Miniprep System (Promega, Milano, Italy). DNA quality and yield were evaluated via agarose gel and Qubit fluorometer (Life Technologies, Carlsbad, CA, USA). Libraries were constructed using Illumina’s recommendations as implemented in 16S Metagenomic Sequencing Library Preparation guide and Fungal Metagenomic Demonstrated Protocol. Two primers, Lac1 (5′-AGCAGTAGGGAATCTTCCA-3′) and Lac2 (5′-ATTYCACCGCTACACATG-3′), were used to amplify the variable regions 3 and 4 of the bacterial 16S rRNA gene [
12]. Among fungi, the gene-specific sequences used in this paper target the fungal ITS1 region between the 18S and 5.8S rRNA genes. They include the ITS1-F and ITS2 primers [
13], which are widely used for fungal barcoding studies. All primers were modified to contain adaptors for MiSeq sequencing.
Three separate gene-amplification reactions were performed for each sample, pooled together and cleaned up using AMPure XP (Beckman Coulter, Brea, CA, USA) magnetic beads. The next PCR attached dual index barcodes and sequencing adapters using the Illumina Nextera XT kit so that the PCR products may be pooled and sequenced directly. A final library size and quantification were conducted using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) and Qubit fluorometer, respectively. DNA sequencing was performed on the Illumina MiSeq platform using v3 chemistry according to the manufacturer’s specifications to generate paired-end reads of 251 bases of length in each direction.
2.6.2. Sequence Analyses of the 16S rRNA and ITS Amplicons
For 16S rDNA gene sequencing, data quality control and analyses were performed using the QIIME pipeline package v.1.9.1 [
14]. The overlapping paired-end reads were merged using the script join_paired_ends.py inside the QIIME package. Only Illumina reads with a length >200 bp were retained for further analysis. Operational taxonomic units (OTUs) were generated using a pipeline based on USEARCH’s OTU clustering recommendations (
http://www.drive5.com/usearch/manual/otu_clustering.html (accessed on 26 October 2021)) using the closed-reference OTU picking to allow clustering of 16S sequences, as previously described [
14]. Reads were clustered at 97% identity using UCLUST to produce OTUs. The taxonomy classification was determined in accordance with the Greengenes 13_8 database. In addition, taxonomic attribution was completed by searching in the NCBI 16S ribosomal RNA sequences database through the Nucleotide BLASTdatabase [
15].
For ITS gene sequencing, data quality control and analyses were performed using the BaseSpace ITS Metagenomics App by Illumina. The ITS Metagenomics workflow performs a taxonomic classification using the UNITE database. Taxonomic identification of strains was completed by comparing the sequences of each sample with those reported in the Nucleotide BLAST database [
15].
2.6.3. Data Availability
The sequence data have been deposited in the Sequence Read Archive of the NCBI database (BioProject ID: PRJNA886648).
2.7. Statistical Analysis and Graph Generation
For each sourdough sample, a standard ANOVA procedure was applied to the dataset of acidity and to the cell count of each microbial group. The means were separated by LSD test at a
p = 0.05 significance level using the Statgraphics Centurion 18 software package (version 18, Statpont Technologies Inc., Warrenton, VA, USA). Some data were subjected to non-parametric statistical analysis by determining the median value. Metataxonomic count data were uploaded to the web application MicrobiomeAnalyst (
http://www.microbiomeanalyst.ca (accessed on 26 October 2021)) to assess different statistics through comparative analysis. The relative proportion of read counts was used as a quantitative estimation of the abundance of each taxon of the three sourdoughs. The diversity within each sample (alpha diversity) was estimated with the Shannon diversity index, which takes into account the number of species and their frequency in each sourdough sample. Statistical significance testing between samples was considered and differences were assigned as statistically significant at
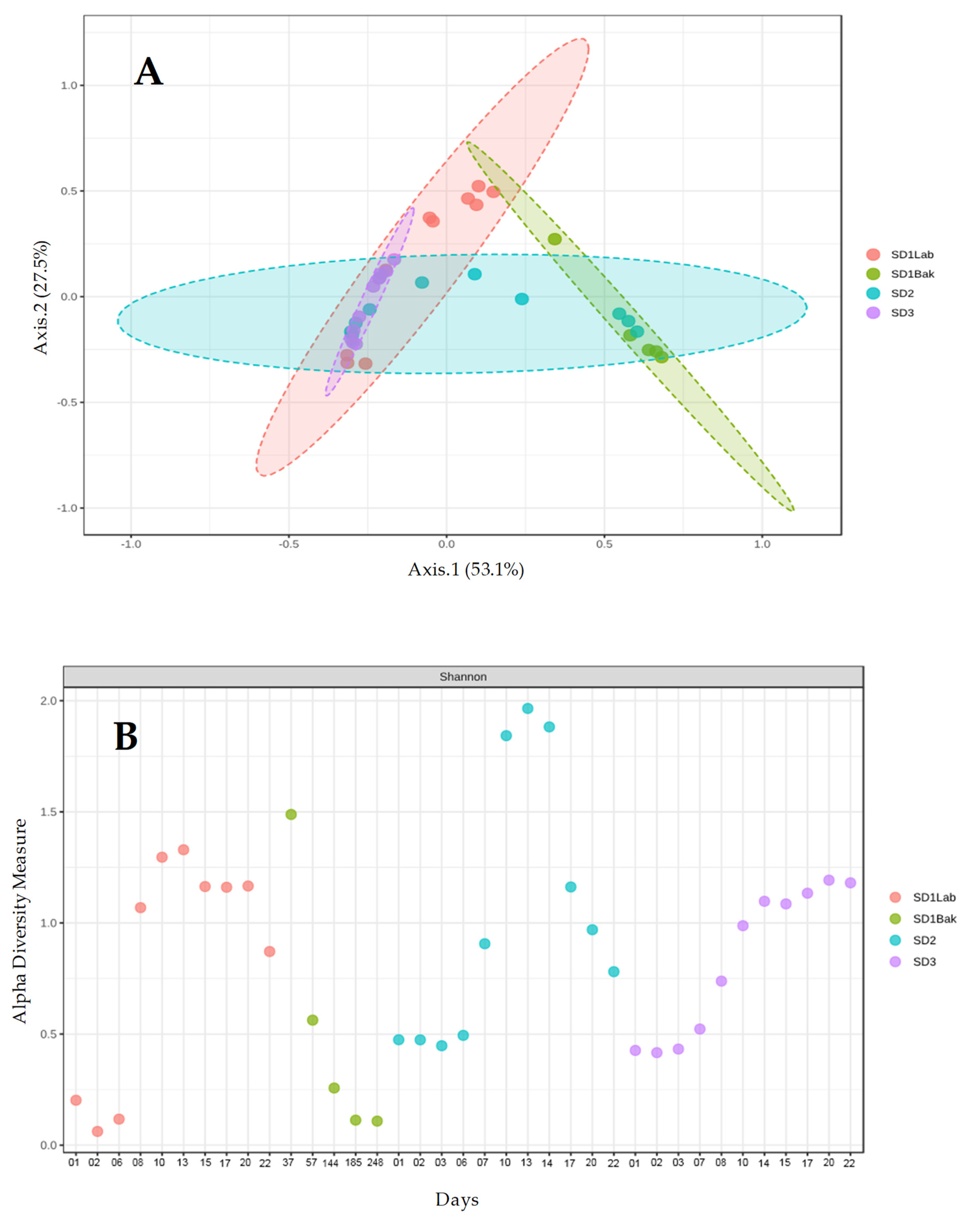
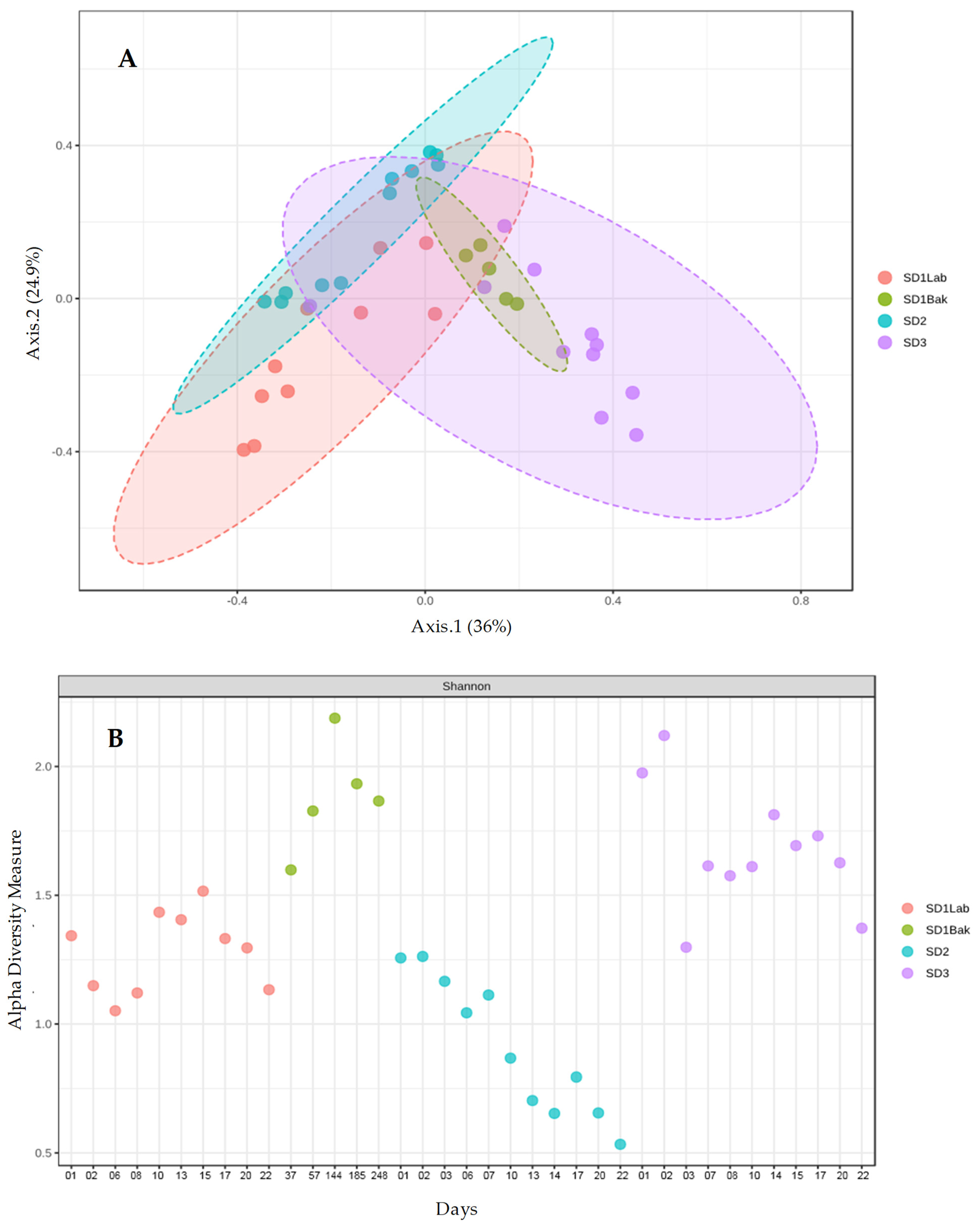
p < 0.05. The differences between microbial samples (beta-diversity) were calculated and visualized as Principal Coordinate Analysis (PCoA). The statistical significance of group clustering was calculated through a permutational multivariate analysis (PERMANOVA) on taxonomic data.
4. Discussion
The stability of sourdough microflora, in terms of microbial species, is an important factor from an industrial point of view, in order to obtain the standardization of baking processes and of products. The stability and the predominance of specific strains in a sourdough ecosystem is dependent on several factors, such as the metabolic properties of the strains, the microbial interactions, and the technological and ecological parameters [
2,
16]. In the current work, strains of
L. plantarum,
S. cerevisiae and
C. lambica, isolated from spontaneous sourdoughs, were used to ferment semolina doughs, with the aim of developing a liquid sourdough that had a stable microflora over time. Two fermentation temperatures were investigated and the impact on the microbial strains was observed. The metataxonomic analysis revealed the complexity of the fungi microflora over time, which was characterized by the presence of numerous species; most of them were pathogens of plants, likely suggesting the flour as the origin, and just a few of them were yeasts. With regard to the bacteria, most of them were lactic acid bacteria and their heterogeneity was very low; as a matter of fact, alongside the
L. plantarum species, inoculated as starter strain, few species developed in all the sourdoughs examined. Above all,
L. brevis and
Leuc.
citreum were commonly detected from day one to the end of the experiments, except when the
F. sanfranciscensis became dominant and all the other species disappeared.
The metataxonomic method used in this work does not allow for the identification of a specific microbial strain, so we cannot assert with absolute certainty the persistence of the starter strains during sourdough propagation. However, we could reasonably believe that they were present due to the following reasons: (a) the number of cells inoculated with the starter strains, about 10
7 CFU g
−1 for bacteria and 10
5 CFU g
−1 for yeasts, (
Figure 1) were close to the values found for the dominant microflora in a mature sourdough (10
6–10
9 CFU g
−1 for bacteria, 10
5–10
8 CFU g
−1 for yeasts) [
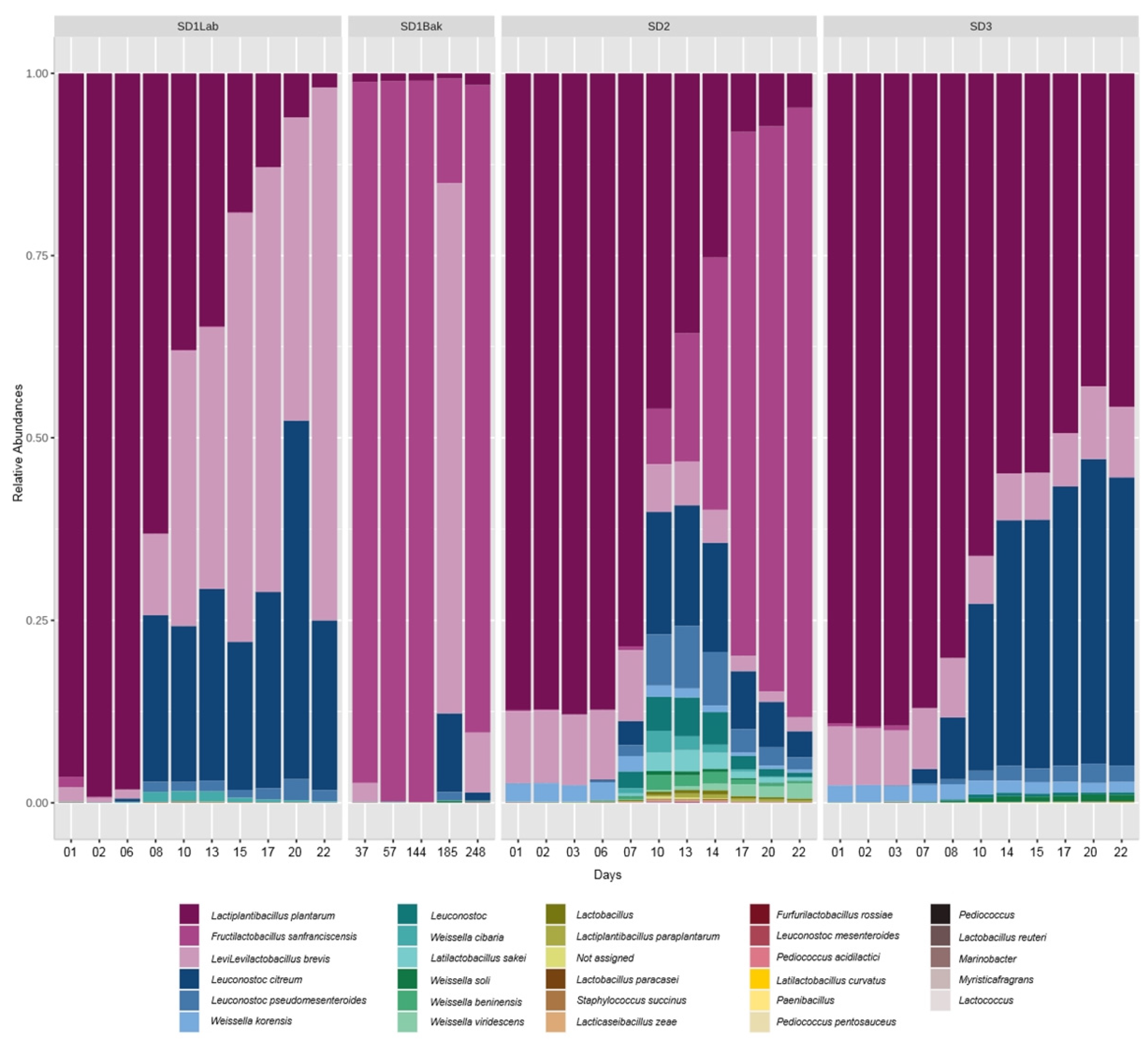
16]; (b) the relative abundance of
L.
plantarum after one day was ca. 90% (
Figure 4) in all sourdoughs, whereas it was quite low for
S. cerevisiae, which tend to increase over time (
Figure 5); (c) the first source of sourdough contamination is the flour, where the number of bacteria ranged from 10
4 to 10
6 CFU g
−1 [
18], a value lower than the number of starter cells. The
S. cerevisiae is one of the most encountered yeast species in spontaneous sourdough [
19] whereas
C. lambica, synonymous with
Pichia fermentans, is a maltose-negative microorganism that is not considered a typical sourdough microorganism, but it was isolated to a lesser extent from sourdough [
20,
21,
22].
Candida humilis (syn.
Candida milleri) and
Candida krusei are the most frequently isolated
Candida species in sourdough [
5]. In this work, more than 10
5 CFU g
−1 of
S. cerevisiae and of
C. lambica were inoculated as starters in sourdoughs. The cells of
C.
lambica decreased over time in all sourdoughs, as showed in
Figure 1 and
Figure 5, but to a lesser extent in SD1, where the relative abundance was the highest (34.6%) and the decrease over time was slow. In fact, the relative abundance was 1.5% at day 22, while in SD2 it was 1.4% at day 17 and in SD3 it was 1.8% at day 10. The
S. cerevisiae was found at high relative abundance both in SD1 and SD2 (
Table 1), and the increase of the abundance seems to be associated with the decrease of
L. plantarum and the growth of
F. sanfranciscensis in both samples, as observed in
Figure 4 and
Figure 5. The
L.
plantarum is the most employed LAB species in sourdough prepared with starter strains [
4], and, despite this, not so many papers studied the permanence in sourdough of
L.
plantarum used as starter. Minervini et al. [
23], studying the robustness of seven strains of
L. plantarum, showed that five of them maintained an elevated number of cells during 10 days of sourdough propagation; nevertheless, new emerging strains were found. The
L. plantarum is considered a ubiquitous microorganism with a relatively large genome size that allows the expression of important metabolic functions. It has been isolated from different fermented foods including spontaneous sourdough, where it is frequently associated with
L. brevis [
24]. It is noticeable that
L. brevis was always present in SD1, SD2 and SD3. In SD1, the cell number increased toward the end of laboratory experiment, and in SD3 its relative abundance was quite constant, ranging from 6.4% to 10%. In SD2, the abundance decreased after 10 days, and the same was observed in bakery-propagated SD1, corresponding in both samples to the appearance of the
F. sanfranciscensis species (
Figure 4), which became dominant over the other strains, reaching about 80% of relative abundance at the end of the experiment. The growth of
F.
sanfranciscensis lowered the bacterial diversity, as stated by Comasio et al. [
25]. The predominance of
F.
sanfranciscensis in bakery-propagated SD1 is likely due to the power of the “house microbiota”, namely the microorganisms contaminating the setting and the equipment of the bakery; the
F. sanfranciscensis was probably the main bacterial strain in the bakery environment and this led to the replacement of the native strains in the sourdough. The
F. sanfranciscensis has the smallest genome and the highest density of ribosomal operons within the lactobacilli group, and this feature is retained to favor its predominance in sourdough substrate [
16]. Siragusa et al. [
26] investigated the predominance of nine strains of
F. sanfranciscensis inoculated as starters in different type I sourdoughs, and they observed that only three of them were able to dominate during 10 days of continuous propagation. Viiard et al. [
27] studied the LAB community of a rye sourdough used in a bakery that was initiated with a commercial starter containing
Limosilactobacillus pontis; after 28 months of refreshments, the analysis revealed the presence of
F. sanfranciscensis and
L. pontis.
As previously reported in the literature [
2,
28], the dynamics of the microbial community are influenced deeply by the fermentation temperature, but the other process parameters (fermentation time, number of refreshment steps, aeration, dough yield, etc.) are considered important as well. In this work, the effect of different fermentation temperatures (25 °C and 20 °C) on sourdough microflora cannot be easily accounted for. At 25 °C, the total number of viable cells (bacteria and yeast) seems to increase with respect to the sourdoughs fermented at 20 °C (
Figure 1); regarding the LAB, three LAB species contributed to increase the number of viable cells, as indicated by metataxonomic analysis in
Figure 4; therefore, the high temperature did not favor the growth of
L. plantarum. Presumptive
Candida grew up to the 22nd day and the number of presumptive
Saccharomyces cells is consistent in sourdough fermented at 25 °C. The low fermentation temperature is reported [
28] to favor the growth of yeast and heterofermentative LAB species in sourdoughs produced worldwide, whereas homofermentative and facultatively heterofermentative LABs were favored at a high fermentation temperature (>30 °C). In natural sourdoughs, dominated by heterofermentative LABs and the yeast
C. milleri, the LAB and yeast cells increased when the temperature was raised from 15 °C to 27 °C [
29].
The values of pH and TTA observed in sourdough samples were consistent with values found in other papers [
4]. The
L. plantarum species, which is the most abundant and important bacterial species in the laboratory-propagated sourdoughs, showed a similar total relative abundance in SD1, SD2 and SD3 (
Table 1), but the decrease over time was more pronounced in SD1 and less pronounced in SD3 (
Figure 4). Anyway, the acidifying activity in the first week was greater in SD1 than in SD2 and SD3, and the lowest pH values were observed in SD1. Therefore, the higher fermentation temperature in SD1 could have favored the metabolism of
L.
plantarum; in the first week thereafter, the development of
L.
brevis, a heterofermentative species, could have contributed to the production of organic acids and to the pH decrease [
5].
Concerning the sourdough samples fermented at the same temperature (SD2 and SD3) during the first week, the acidification was faster in SD3 than in SD2, likely because of the different method used to inoculate the
L.
plantarum. In fact, the starter strains were inoculated all together in SD2, whereas in SD3 the
L. plantarum was inoculated alone and left to ferment for 17 h, and then the yeast strains were added in the subsequent backslopping. The analysis of the data reported in
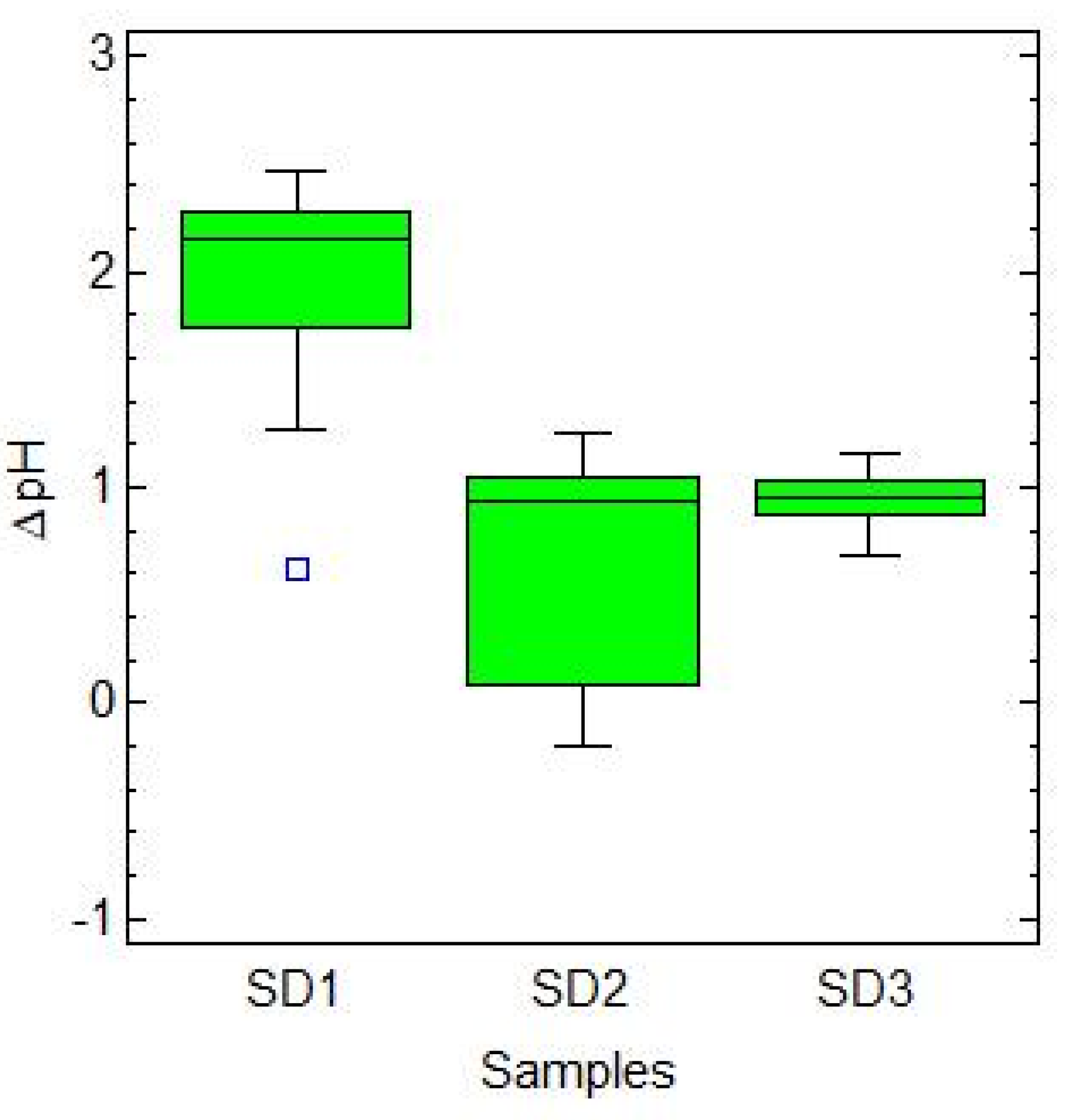
Figure 3 indicates that the ∆pH values were more homogeneous in SD3 compared to SD2, and the same phenomenon can be observed for the ∆TTA and bacterial cell density values reported in
Figures S2 and S4, respectively. Therefore, in SD3, the growth of lactic acid bacteria and the acidification over the propagation period was more uniform and stable compared to the other samples. Regarding the evolution of starter yeasts, differences can be observed in SD2 and SD3 (
Figure 5), unless the fermentation was conducted at the same temperature (20 °C). Indeed, both the
S.
cerevisiae and the
C.
lambica grew well in SD2, where the lowering of dough pH before starter addiction could have favored the growth of yeast, according with that reported by Minervini et al. [
16] and to the yeast growth in SD1, where the dough pH was not lowered. On the contrary, in SD3, the lowering of pH did not favor the growth of yeast strains, probably because their growth was conditioned by the rapid growth of
L.
plantarum in the first days. The different methods used for starter addition seem to have affected the behavior of starter strains and the acidification process in sourdoughs.