Heterologous Expression of Thermotolerant α-Glucosidase in Bacillus subtilis 168 and Improving Its Thermal Stability by Constructing Cyclized Proteins
Abstract
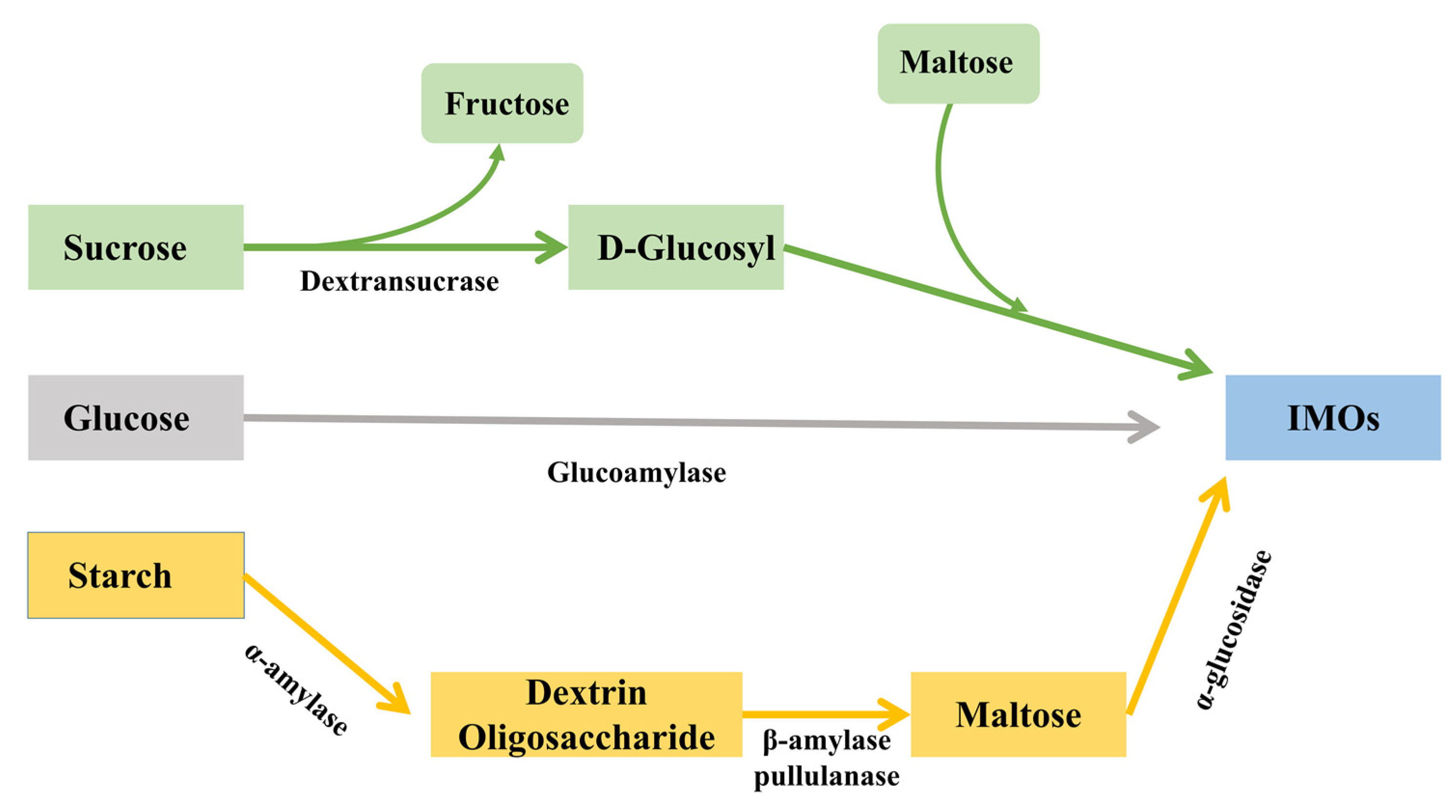
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Heterologous Expression of α-Glucosidase in B. subtilis 168
2.2.2. Expression Analysis and Purification of Recombinant α-Glucosidase
2.2.3. Determination of Recombinant α-Glucosidase Activity
2.2.4. Determination of Transglycosylation Activity
2.2.5. Construction of the Cyclized α-Glucosidase
2.2.6. Characterization of the Enzymatic Properties
2.2.7. Molecular Dynamics Simulation of Cyclized α-Glucosidase
3. Result and Discussion
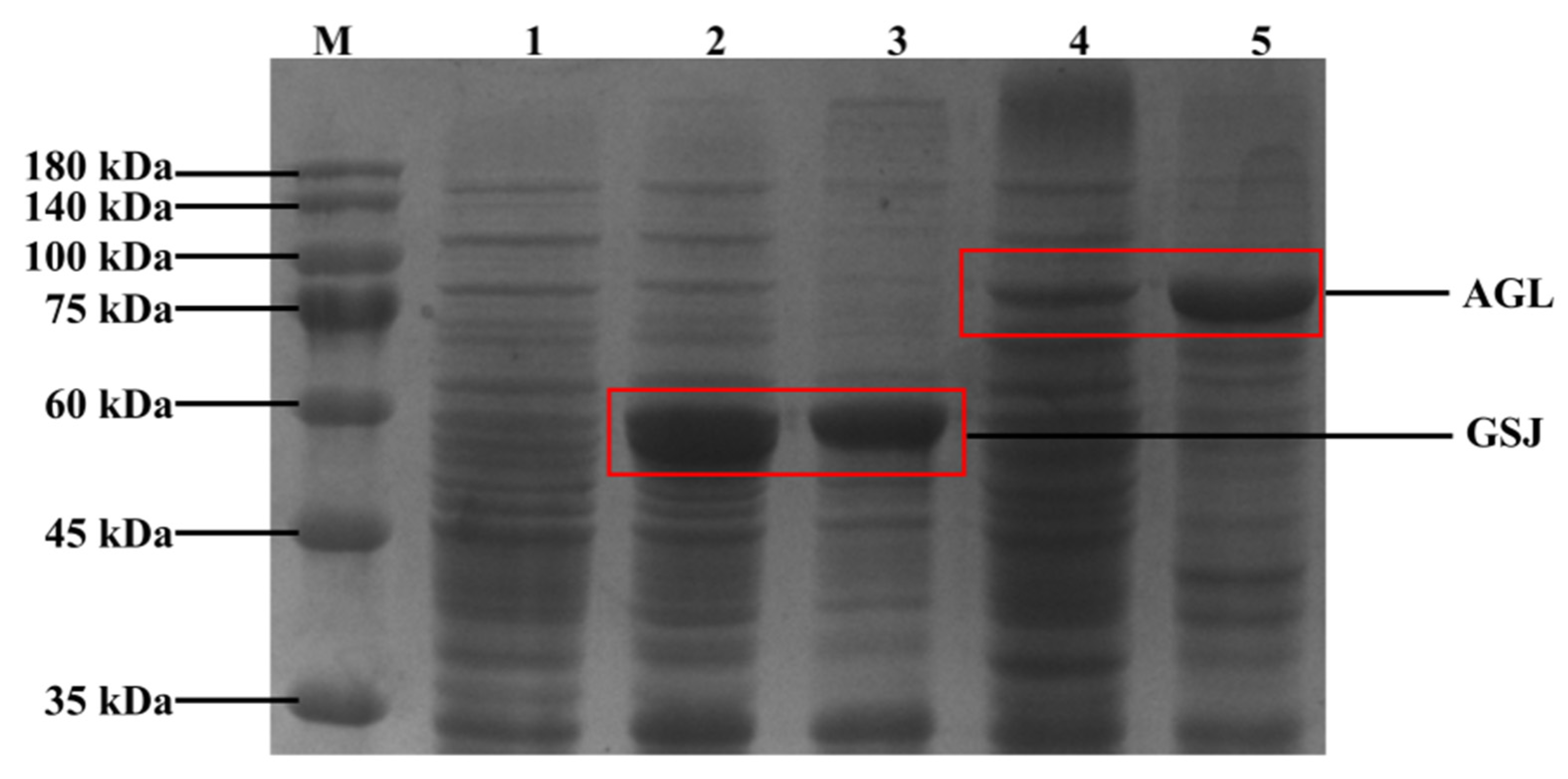
3.1. Heterologous Expression of Thermotolerant α-Glucosidase in B. Subtilis 168
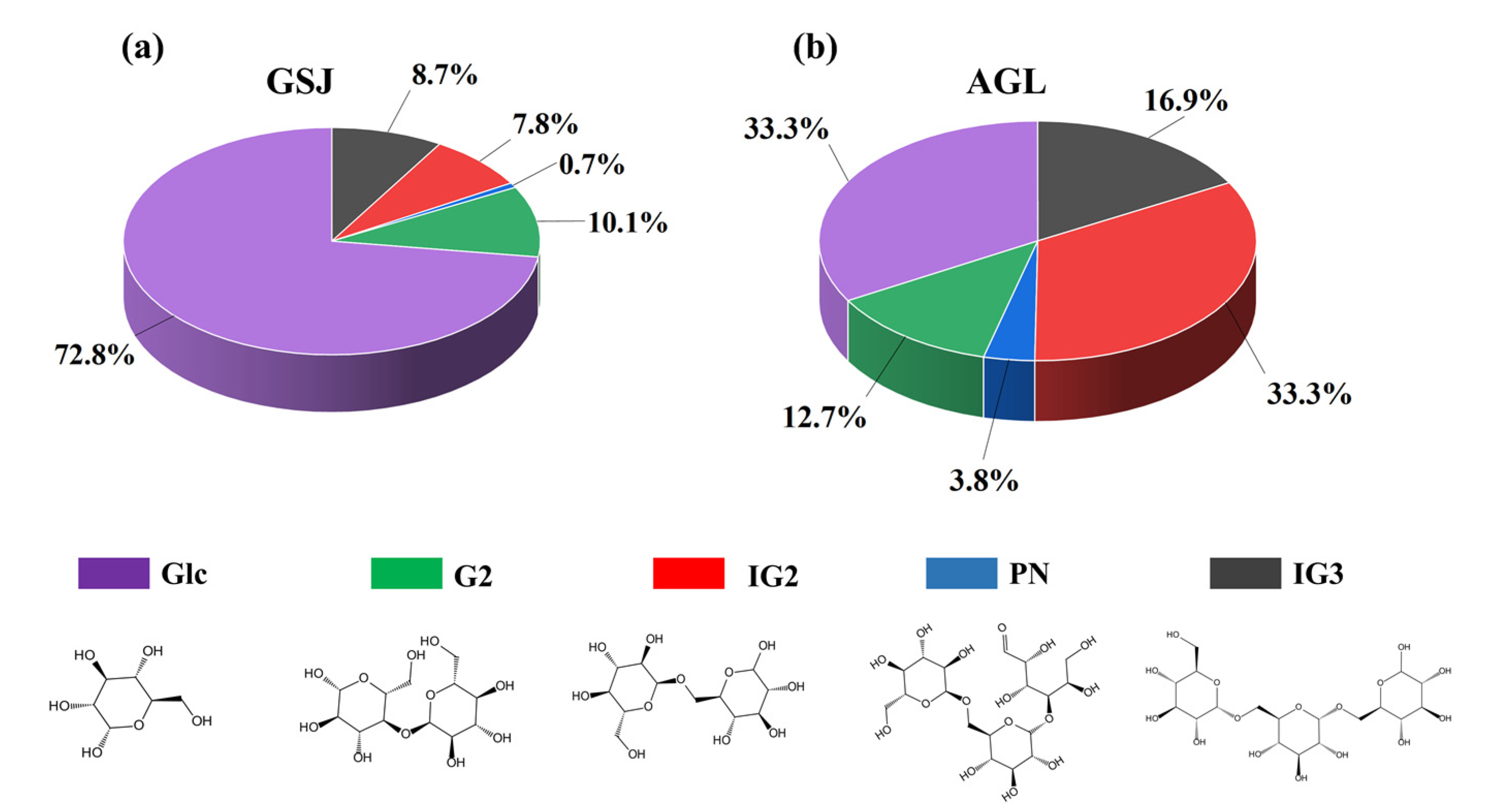
3.2. Transglucosylation Activity of Recombinant α-Glucosidase
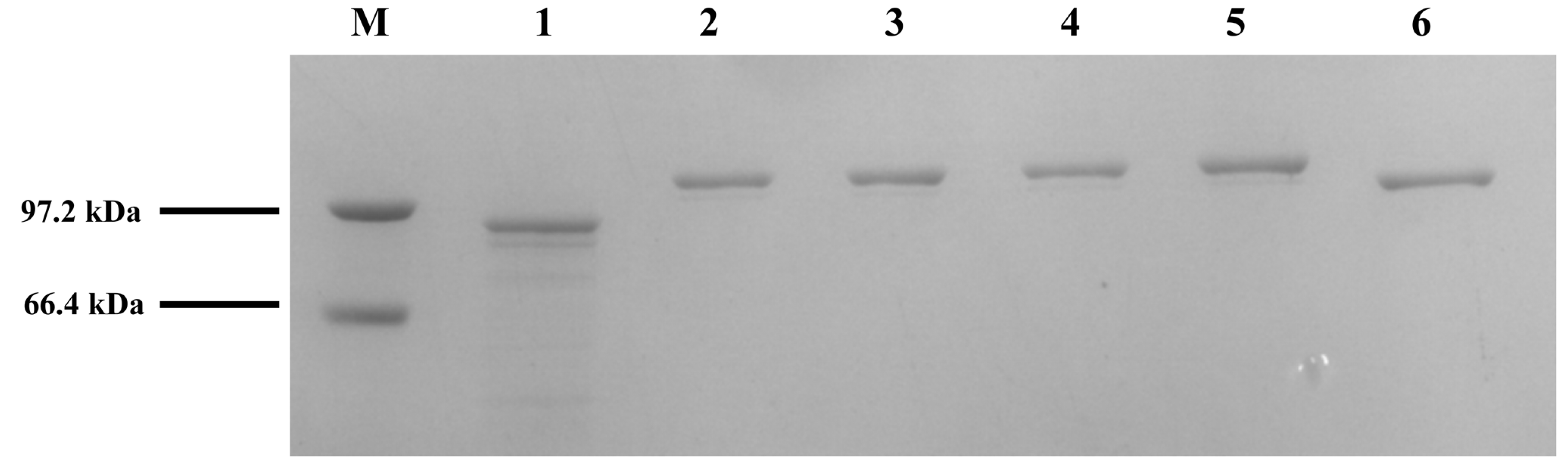
3.3. Construction of Cyclized AGL to Improve the Thermal Stability of Recombinant α-Glucosidase
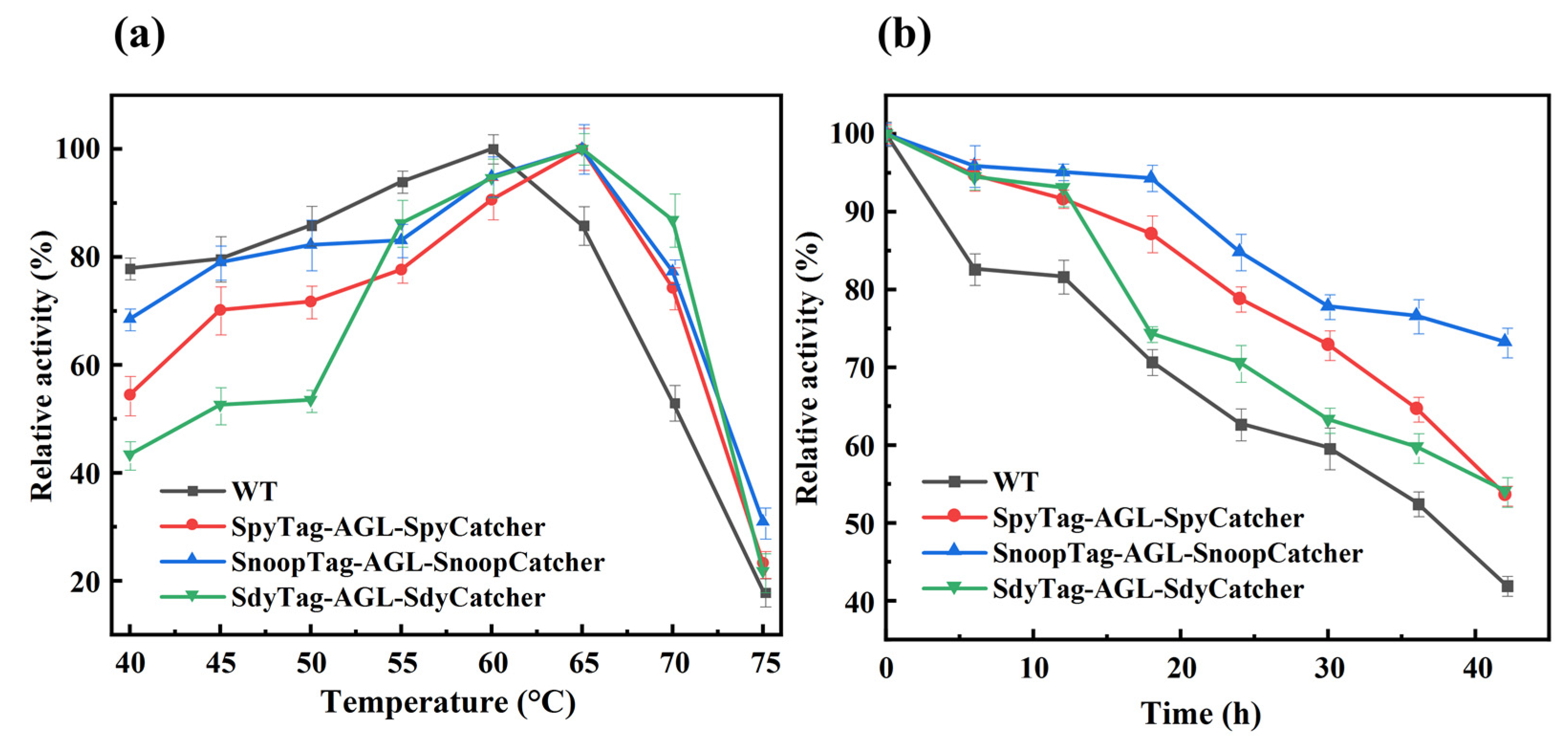
3.4. Optimal Temperature and Thermal Stability of Cyclized AGL
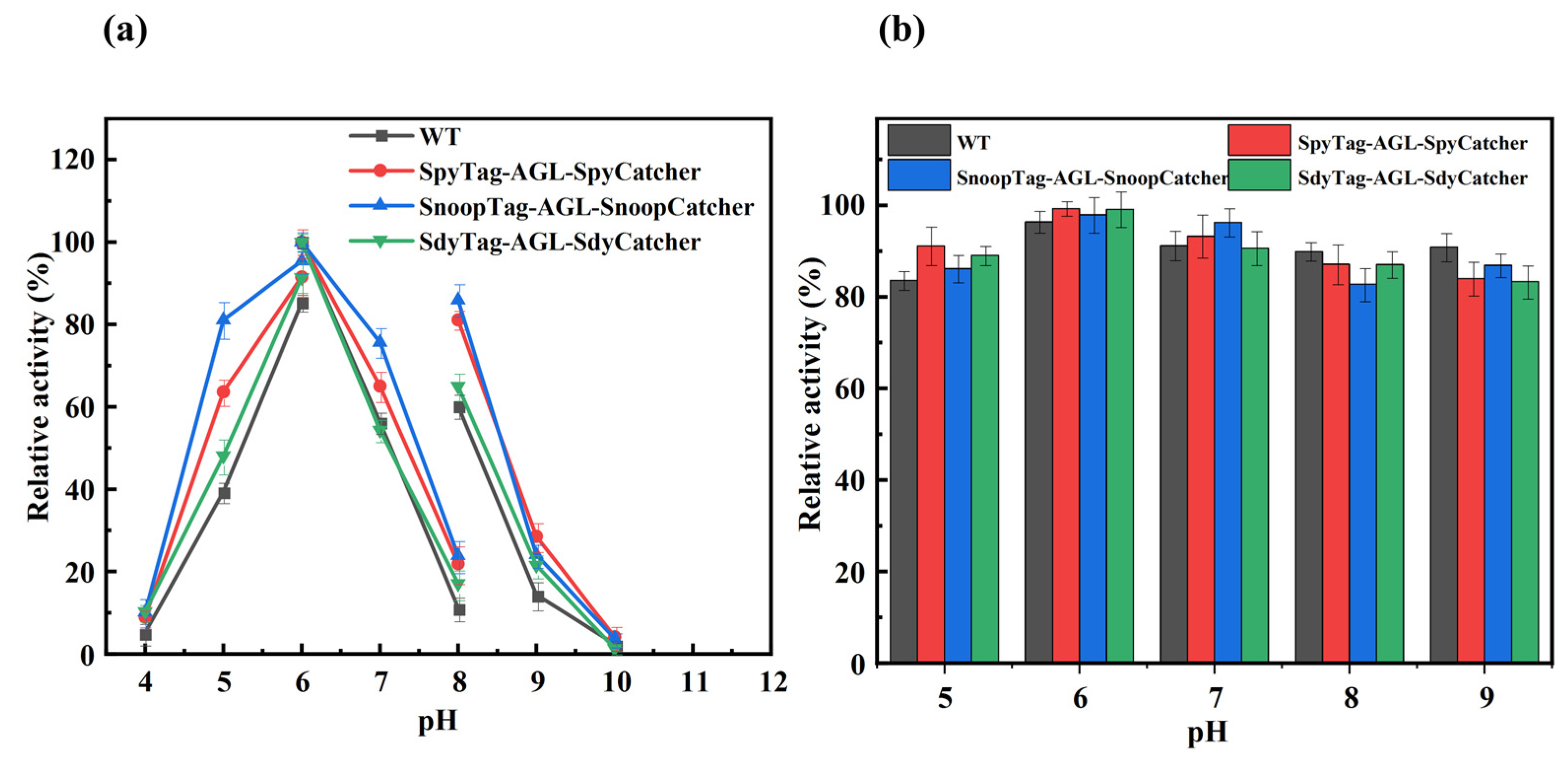
3.5. Optimal pH and pH Stability of Cyclized AGL
3.6. Kinetic Analysis of Recombinant α-Glucosidase
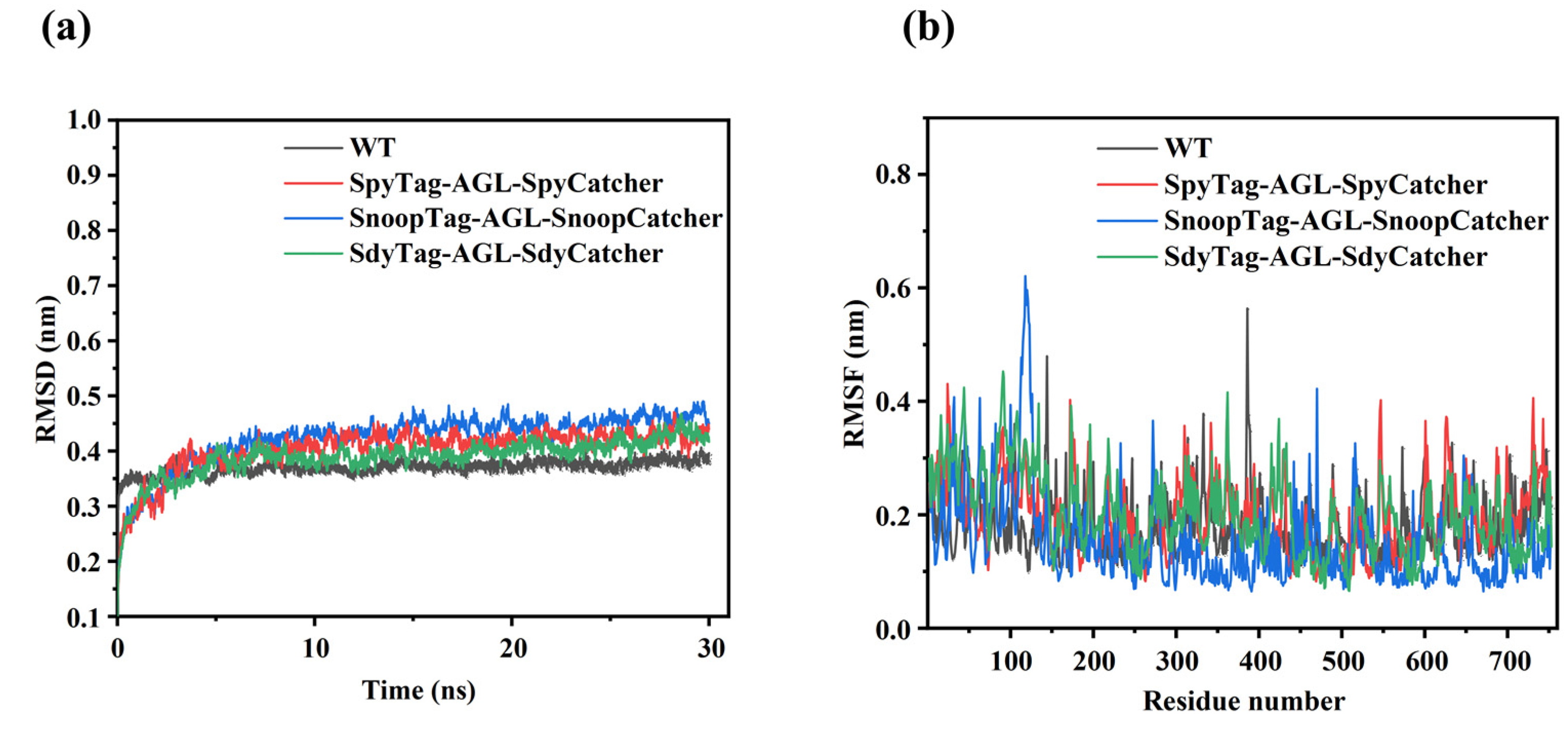
3.7. Molecular Dynamics Simulation Analysis of Cyclized AGL
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, Z.; Li, Z.; Su, Y.; Zhu, Y.; Zeng, W.; Chen, G.; Liang, Z. Continuous Production of Isomalto-oligosaccharides by Thermo-inactivated Cells of Aspergillus niger J2 with Coarse Perlite as an Immobilizing Material. Appl. Biochem. Biotechnol. 2018, 185, 1088–1099. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Hou, Y.; Juvonen, M.; Tuomainen, P.; Kajala, I.; Shukla, S.; Goyal, A.; Maaheimo, H.; Katina, K.; Tenkanen, M. Optimization of Isomaltooligosaccharide Size Distribution by Acceptor Reaction of Weissella confusa Dextransucrase and Characterization of Novel alpha-(1 -> 2)-Branched Isomaltooligosaccharides. J. Agric. Food Chem. 2016, 64, 3276–3286. [Google Scholar] [CrossRef] [PubMed]
- Ojha, S.; Mishra, S.; Chand, S. Production of isomalto-oligosaccharides by cell bound alpha-glucosidase of Microbacterium sp. Lwt-Food Sci. Technol. 2015, 60, 486–494. [Google Scholar] [CrossRef]
- Soto-Maldonado, C.; Concha-Olmos, J.; Caceres-Escobar, G.; Menses-Gomez, P. Sensory evaluation and glycaemic index of a food developed with flour from whole (pulp and peel) overripe banana (Musa cavendishii) discards. Lwt-Food Sci. Technol. 2018, 92, 569–575. [Google Scholar] [CrossRef]
- Wu, Q.; Pi, X.e.; Liu, W.; Chen, H.; Yin, Y.; Yu, H.D.; Wang, X.; Zhu, L. Fermentation properties of isomaltooligosaccharides are affected by human fecal enterotypes. Anaerobe 2017, 48, 206–214. [Google Scholar] [CrossRef]
- Madsen, L.R., II; Stanley, S.; Swann, P.; Oswald, J. A Survey of Commercially Available Isomaltooligosaccharide-Based Food Ingredients. J. Food Sci. 2017, 82, 401–408. [Google Scholar] [CrossRef]
- Gu, X.L.; Li, H.; Song, Z.H.; Ding, Y.N.; He, X.; Fan, Z.Y. Effects of isomaltooligosaccharide and Bacillus supplementation on sow performance, serum metabolites, and serum and placental oxidative status. Anim. Reprod. Sci. 2019, 207, 52–60. [Google Scholar] [CrossRef]
- Nakakuki, T. Present status and future of functional oligosaccharide development in Japan. Pure Appl. Chem. 2002, 74, 1245–1251. [Google Scholar] [CrossRef]
- Bivolarski, V.; Vasileva, T.; Bozov, P.; Iliev, I. INFLUENCE OF DIFFERENT ACCEPTORS ON SYNTHESIS OF GLUCOOLIGOSACCHARIDES BY PURIFIED DEXTRANSUCRASE FROM LEUCONOSTOC MESENTEROIDES URE 13. Comptes Rendus De L Acad. Bulg. Des. Sci. 2013, 66, 1405–1412. [Google Scholar]
- Sorndech, W.; Sagnelli, D.; Blennow, A.; Tongta, S. Combination of amylase and transferase catalysis to improve IMO compositions and productivity. Lwt-Food Sci. Technol. 2017, 79, 479–486. [Google Scholar] [CrossRef]
- Rudeekulthamrong, P.; Sawasdee, K.; Kaulpiboon, J. Production of long-chain isomaltooligosaccharides from maltotriose using the thermostable amylomaltase and transglucosidase enzymes. Biotechnol. Bioprocess Eng. 2013, 18, 778–786. [Google Scholar] [CrossRef]
- Ma, M.; Okuyama, M.; Sato, M.; Tagami, T.; Klahan, P.; Kumagai, Y.; Mori, H.; Kimura, A. Effects of mutation of Asn694 in Aspergillus niger alpha-glucosidase on hydrolysis and transglucosylation. Appl. Microbiol. Biotechnol. 2017, 101, 6399–6408. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-L.; Tong, X.; Chen, S.-W.; Chen, S.; Wu, D.; Fang, S.-G.; Wu, J.; Chen, J. Heterologous Expression and Biochemical Characterization of alpha-Glucosidase from Aspergillus niger by Pichia pastroris. J. Agric. Food Chem. 2010, 58, 4819–4824. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, W.; Bah, F.B.M.; Song, C.; Zhou, Y.; Ji, L.; Yuan, Y. Heterologous Expression of a Thermostable alpha-Glucosidase from Geobacillus sp. Strain HTA-462 by Escherichia coli and Its Potential Application for Isomaltose-Oligosaccharide Synthesis. Molecules 2019, 24, 1413. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Xue, Y.; Ma, Y. Evaluation and directed evolution for thermostability improvement of a GH 13 thermostable alpha-glucosidase from Thermus thermophilus TC11. BMC Biotechnol. 2015, 15, 97. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-E.; Park, S.R.; Woo, J.Y.; Hwang, H.S.; Cha, J.; Lee, H. Enzymatic Properties of a Thermostable alpha-Glucosidase from Acidothermophilic Crenarchaeon Sulfolobus tokodaii Strain 7. J. Microbiol. Biotechnol. 2013, 23, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Hung, V.S.; Hatada, Y.; Goda, S.; Lu, J.; Hidaka, Y.; Li, Z.J.; Akita, M.; Ohta, Y.; Watanabe, K.; Matsui, H.; et al. alpha-Glucosidase from a strain of deep-sea Geobacillus: A potential enzyme for the biosynthesis of complex carbohydrates. Appl. Microbiol. Biotechnol. 2005, 68, 757–765. [Google Scholar] [CrossRef]
- Zhou, C.; Xue, Y.; Ma, Y. Enhancing the thermostability of alpha-glucosidase from Thermoanaerobacter tengcongensis MB4 by single proline substitution. J. Biosci. Bioeng. 2010, 110, 12–17. [Google Scholar] [CrossRef]
- Zakeri, B.; Howarth, M. Spontaneous Intermolecular Amide Bond Formation between Side Chains for Irreversible Peptide Targeting. J. Am. Chem. Soc. 2010, 132. [Google Scholar] [CrossRef]
- Zakeri, B.; Fierer, J.O.; Celik, E.; Chittock, E.C.; Schwarz-Linek, U.; Moy, V.T.; Howarth, M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA 2012, 109, E690–E697. [Google Scholar] [CrossRef]
- Veggiani, G.; Nakamura, T.; Brenner, M.D.; Gayet, R.V.; Yan, J.; Robinson, C.V.; Howarth, M. Programmable polyproteams built using twin peptide superglues. Proc. Natl. Acad. Sci. USA 2016, 113, 1202–1207. [Google Scholar] [CrossRef]
- Fierer, J.O.; Veggiani, G.; Howarth, M. SpyLigase peptide-peptide ligation polymerizes affibodies to enhance magnetic cancer cell capture. Proc. Natl. Acad. Sci. USA 2014, 111, E1176–E1181. [Google Scholar] [CrossRef]
- Wong, J.X.; Gonzalez-Miro, M.; Sutherland-Smith, A.J.; Rehm, B.H.A. Covalent Functionalization of Bioengineered Polyhydroxyalkanoate Spheres Directed by Specific Protein-Protein Interactions. Front. Bioeng. Biotechnol. 2020, 8, 44. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhao, Y.; Zhou, X.; Liu, N.; Ming, D.; Zhu, L.; Jiang, L. Improving the thermostability of trehalose synthase from Thermomonospora curvata by covalent cyclization using peptide tags and investigation of the underlying molecular mechanism. Int. J. Biol. Macromol. 2021, 168, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Y.; Wang, X.; Zhang, D.; Wu, S.; Zhang, G. Enhanced thermal stability of lichenase from Bacillus subtilis 168 by SpyTag/SpyCatcher-mediated spontaneous cyclization. Biotechnol. Biofuels 2016, 9, 79. [Google Scholar] [CrossRef]
- Schoene, C.; Bennett, S.P.; Howarth, M. SpyRing interrogation: Analyzing how enzyme resilience can be achieved with phytase and distinct cyclization chemistries. Sci. Rep. 2016, 6, 21151. [Google Scholar] [CrossRef] [PubMed]
- Si, M.; Xu, Q.; Jiang, L.; Huang, H. SpyTag/SpyCatcher Cyclization Enhances the Thermostability of Firefly Luciferase. PLoS ONE 2016, 11, e0162318. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Jiang, Y.; Duan, Z.-Y.; Shao, W.-L.; Li, H.-Z. Expression and characterization of an alpha-glucosidase from Thermoanaerobacter ethanolicus JW200 with potential for industrial application. Biologia 2009, 64, 1053–1057. [Google Scholar] [CrossRef]
- Kang, W.; Ma, T.; Liu, M.; Qu, J.; Liu, Z.; Zhang, H.; Shi, B.; Fu, S.; Ma, J.; Lai, L.T.F.; et al. Modular enzyme assembly for enhanced cascade biocatalysis and metabolic flux. Nat. Commun. 2019, 10, 4248. [Google Scholar] [CrossRef]
- You, J.; Yang, C.; Pan, X.; Hu, M.; Du, Y.; Osire, T.; Yang, T.; Rao, Z.J.B.T. Metabolic engineering of Bacillus subtilis for enhancing riboflavin production by alleviating dissolved oxygen limitation. Bioresour. Technol. 2021, 333, 125228. [Google Scholar] [CrossRef]
- Jung, J.H.; Seo, D.H.; Holden, J.F.; Kim, H.S.; Baik, M.Y.; Park, C.S. Broad substrate specificity of a hyperthermophilic alpha-glucosidase from Pyrobaculum arsenaticum. Food Sci. Biotechnol. 2016, 25, 1665–1669. [Google Scholar] [CrossRef] [PubMed]
- Anindyawati, T.; Ann, Y.-G.; Ito, K.; Iizuka, M.; Minamiura, N. Two kinds of novel α-glucosidases from Aspergillus awamori KT-11: Their purifications, properties and specificities. J. Ferment. Bioeng. 1998, 85, 465–469. [Google Scholar] [CrossRef]
- Shimba, N.; Shinagawa, M.; Hoshino, W.; Yamaguchi, H.; Yamada, N.; Suzuki, E.-i. Monitoring the hydrolysis and transglycosylation activity of alpha-glucosidase from Aspergillus niger by nuclear magnetic resonance spectroscopy and mass spectrometry. Anal. Biochem. 2009, 393, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Cascales, L.; Craik, D.J. Naturally occurring circular proteins: Distribution, biosynthesis and evolution. Org. Biomol. Chem. 2010, 8, 5035–5047. [Google Scholar] [CrossRef]
- Boecker, J.K.; Friedel, K.; Matern, J.C.J.; Bachmann, A.-L.; Mootz, H.D. Generation of a Genetically Encoded, Photoactivatable Intein for the Controlled Production of Cyclic Peptides. Angew. Chem. Int. Ed. 2015, 54, 2116–2120. [Google Scholar] [CrossRef]
- De Rosa, L.; Cortajarena, A.L.; Romanelli, A.; Regan, L.; D’Andrea, L.D. Site-specific protein double labeling by expressed protein ligation: Applications to repeat proteins. Org. Biomol. Chem. 2012, 10, 273–280. [Google Scholar] [CrossRef]
- Agwa, A.J.; Craik, D.J.; Schroeder, C.I. Cyclizing Disulfide-Rich Peptides Using Sortase A. Methods Mol. Biol. (Clifton N.J.) 2019, 2012, 29–41. [Google Scholar] [CrossRef]
- Minato, Y.; Ueda, T.; Machiyama, A.; Shimada, I.; Iwai, H. Segmental isotopic labeling of a 140 kDa dimeric multi-domain protein CheA from Escherichia coli by expressed protein ligation and protein trans-splicing. J. Biomol. NMR 2012, 53, 191–207. [Google Scholar] [CrossRef]
- Lin, Y.; Jin, W.; Wang, J.; Cai, Z.; Wu, S.; Zhang, G. A novel method for simultaneous purification and immobilization of a xylanase-lichenase chimera via SpyTag/SpyCatcher spontaneous reaction. Enzym. Microb. Technol. 2018, 115, 29–36. [Google Scholar] [CrossRef]

| Enzyme | Specific Enzyme Activity (U·mg−1) | Relative Enzyme Activity (100%) |
|---|---|---|
| WT | 27.91 ± 0.5 | 100.0 ± 1.7 |
| RIAD-AGL-RIDD | 12.01 ± 0.7 | 43.5 ± 2.5 |
| SpyTag-AGL-SpyCatcher | 26.43 ± 0.3 | 94.7 ± 1.1 |
| SnoopTag-AGL-SnoopCatcher | 28.55 ± 0.6 | 102.3 ± 2.1 |
| SdyTag-AGL-SdyCatcher | 27.18 ± 0.3 | 97.4 ± 1.1 |
| Enzyme | Km (mM) | Vmax (U mg−1) | Kcat (S−1) |
|---|---|---|---|
| WT | 1.83 ± 0.06 | 36.42 ± 0.12 | 282.31±2.1 |
| SpyTag-AGL-SpyCatcher | 1.86 ± 0.03 | 35.94 ± 0.07 | 276.38±7.2 |
| SnoopTag-AGL-SnoopCatcher | 1.82 ± 0.05 | 36.12 ± 0.15 | 278.92±6.8 |
| SdyTag-AGL-SdyCatcher | 1.86 ± 0.06 | 36.31 ± 0.19 | 281.59±6.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Hu, M.; Fang, M.; Wang, Q.; Lu, R.; Zhang, H.; Xu, M.; Zhang, X.; Rao, Z. Heterologous Expression of Thermotolerant α-Glucosidase in Bacillus subtilis 168 and Improving Its Thermal Stability by Constructing Cyclized Proteins. Fermentation 2022, 8, 498. https://doi.org/10.3390/fermentation8100498
Wang Z, Hu M, Fang M, Wang Q, Lu R, Zhang H, Xu M, Zhang X, Rao Z. Heterologous Expression of Thermotolerant α-Glucosidase in Bacillus subtilis 168 and Improving Its Thermal Stability by Constructing Cyclized Proteins. Fermentation. 2022; 8(10):498. https://doi.org/10.3390/fermentation8100498
Chicago/Turabian StyleWang, Zhi, Mengkai Hu, Ming Fang, Qiang Wang, Ruiqi Lu, Hengwei Zhang, Meijuan Xu, Xian Zhang, and Zhiming Rao. 2022. "Heterologous Expression of Thermotolerant α-Glucosidase in Bacillus subtilis 168 and Improving Its Thermal Stability by Constructing Cyclized Proteins" Fermentation 8, no. 10: 498. https://doi.org/10.3390/fermentation8100498
APA StyleWang, Z., Hu, M., Fang, M., Wang, Q., Lu, R., Zhang, H., Xu, M., Zhang, X., & Rao, Z. (2022). Heterologous Expression of Thermotolerant α-Glucosidase in Bacillus subtilis 168 and Improving Its Thermal Stability by Constructing Cyclized Proteins. Fermentation, 8(10), 498. https://doi.org/10.3390/fermentation8100498







