Fermentation of Dairy-Relevant Sugars by Saccharomyces, Kluyveromyces, and Brettanomyces: An Exploratory Study with Implications for the Utilization of Acid Whey, Part I
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Inocula
2.3. Fermentation Media
2.4. Inoculation and Fermentation
2.5. Data Collection
2.5.1. Density, pH, and Microbial Enumeration
2.5.2. Analyses of Ethanol, Organic Acids, and Sugars
2.6. Statistical Analysis
2.6.1. General Analysis
2.6.2. Non-Linear Modeling
3. Results and Discussion
3.1. Fermentation Characterization
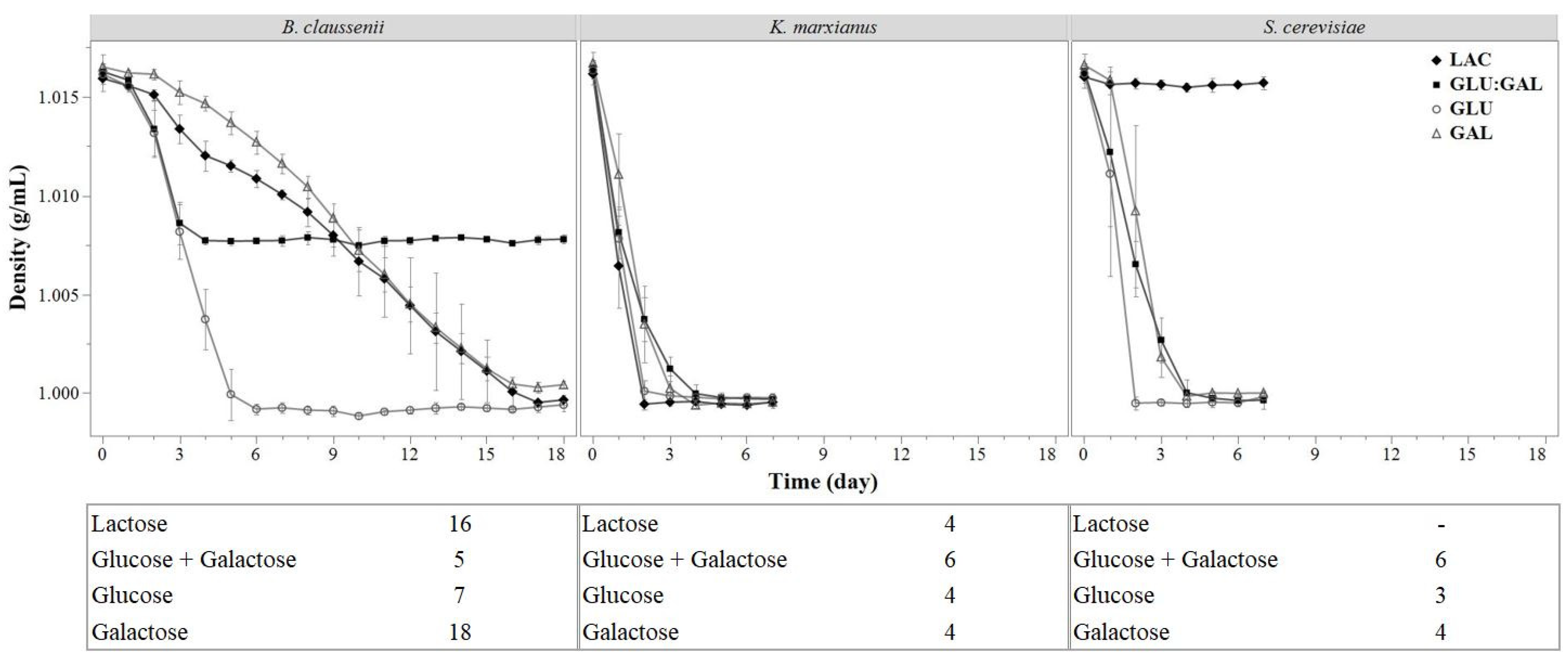
3.1.1. Density
3.1.1.1. Non-Linear Density Modeling
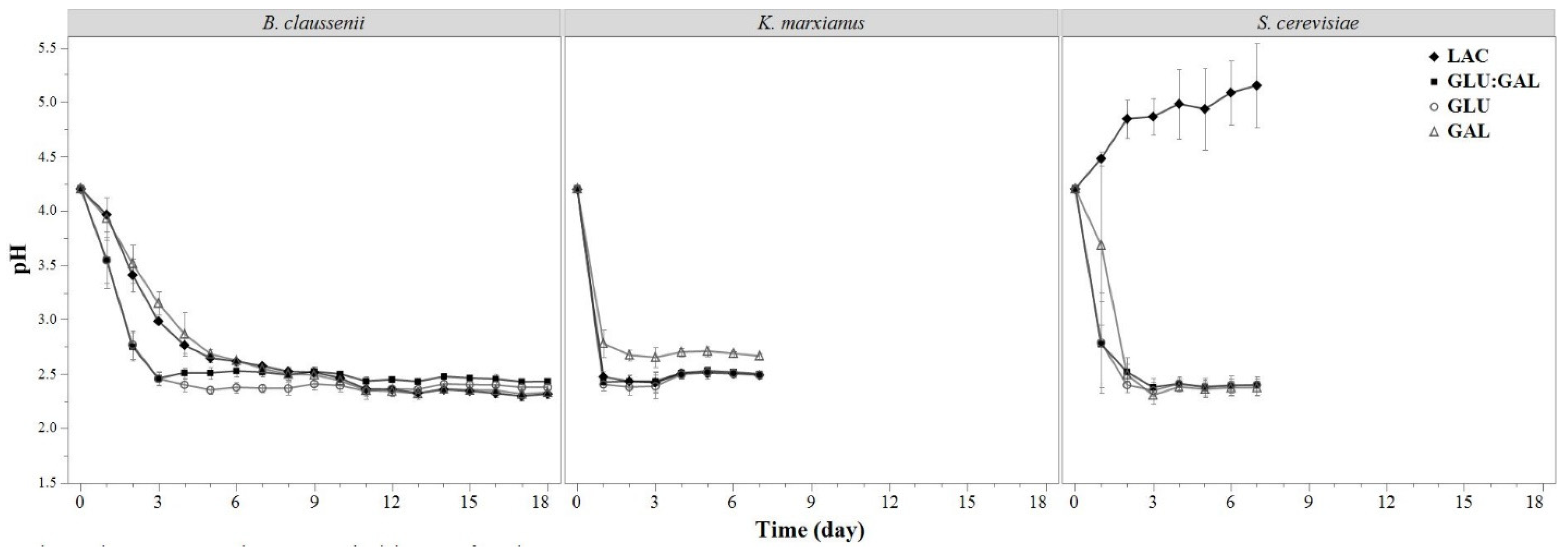
3.1.2. pH
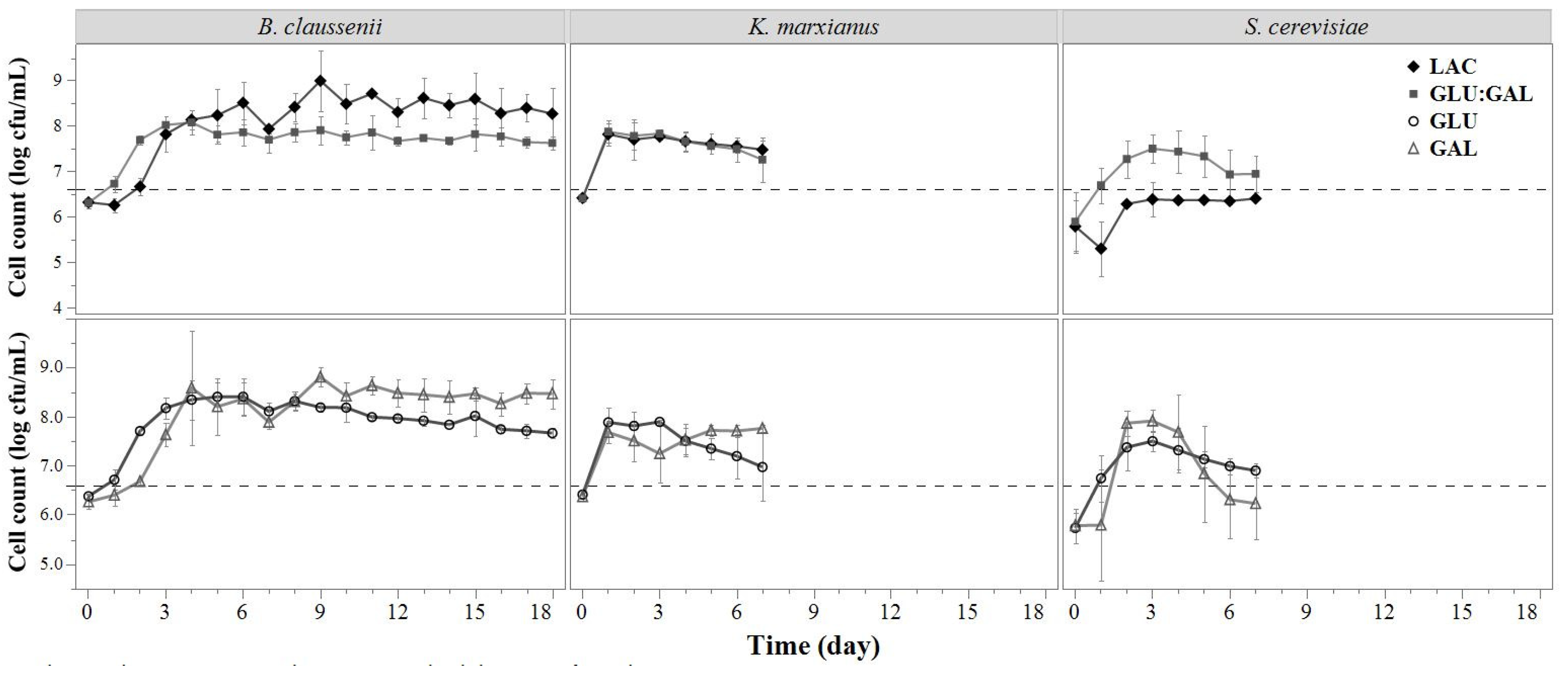
3.1.3. Microbial Concentration
3.2. Sugar Concentrations
3.3. Production of Ethanol and Organic Acids
3.3.1. Ethanol
3.3.2. Organic Acids
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Statista Dossier on Greek Yogurt in the U.S. Available online: https://www-statista-com.proxy.library.cornell.edu/study/25622/greek-yogurt-statista-dossier/ (accessed on 4 August 2020).
- Erickson, B. Acid Whey: Is the Waste Product an Untapped Goldmine? Available online: https://cen.acs.org/articles/95/i6/Acid-whey-waste-product-untapped.html (accessed on 8 February 2019).
- Menchik, P.; Zuber, T.; Zuber, A.; Moraru, C.I. Short communication: Composition of coproduct streams from dairy processing: Acid whey and milk permeate. J. Dairy Sci. 2019, 102, 3978–3984. [Google Scholar] [CrossRef] [PubMed]
- Jelen, P. Whey Processing|Utilization and Products. In Encyclopedia of Dairy Sciences, 2nd ed.; Fuquay, J.W., Ed.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 731–737. [Google Scholar]
- Ketterings, Q.; Czymmek, K.; Gami, S.; Godwin, G.; Ganoe, K. Guidelines for Land Application of Acid Whey; Department of Animal Science Publication Series; Cornell University: Ithaca, NY, USA, 2017. [Google Scholar]
- Mano, J.; Liu, N.; Hammond, J.H.; Currie, D.H.; Stephanopoulos, G. Engineering Yarrowia lipolytica for the utilization of acid whey. Metab. Eng. 2020, 57, 43–50. [Google Scholar] [CrossRef] [PubMed]
- United States Environmental Protection Agency. Food Recovery Hierarchy. Available online: https://www.epa.gov/sustainable-management-food/food-recovery-hierarchy#about (accessed on 7 July 2021).
- Rocha-Mendoza, D.; Kosmerl, E.; Krentz, A.; Zhang, L.; Badiger, S.; Miyagusuku-Cruzado, G.; Mayta-Apaza, A.; Giusti, M.; Jiménez-Flores, R.; García-Cano, I. Invited review: Acid whey trends and health benefits. J. Dairy Sci. 2021, 104, 1262–1275. [Google Scholar] [CrossRef] [PubMed]
- Skryplonek, K.; Dmytrow, I.; Mituniewicz-Malek, A. Probiotic fermented beverages based on acid whey. J. Dairy Sci. 2019, 102, 7773–7780. [Google Scholar] [CrossRef] [PubMed]
- Kovtuna, K.; Martynova, J.; Krēķe, S.I.; Scherbaka, R.; Vigants, A. The sweet and acidic whey as substrates for probiotics biomass production. J. Biotechnol. 2019, 305, S53–S54. [Google Scholar] [CrossRef]
- Luo, S.; Demarsh, T.A.; deRiancho, D.; Stelick, A.; Alcaine, S.D. Characterization of the fermentation and sensory profiles of novel yeast-fermented acid whey beverages. Foods 2021, 10, 1204. [Google Scholar] [CrossRef] [PubMed]
- Fior Markets. Global Functional Beverages Market Is Expected to Reach USD 216.7 Billion by 2028: Fior Markets. Available online: https://www.globenewswire.com/en/news-release/2021/05/24/2234940/0/en/Global-Functional-Beverages-Market-Is-Expected-to-Reach-USD-216-7-billion-by-2028-Fior-Markets.html (accessed on 13 August 2021).
- Conway, J. Market Size of Non-Alcoholic Beer Worldwide from 2016 to 2024; Statista: Paris, France, 2020. [Google Scholar]
- Lawton, M.R.; deRiancho, D.L.; Alcaine, S.D. Lactose utilization by Brettanomyces claussenii expands potential for valorization of dairy by-products to functional beverages through fermentation. Curr. Opin. Food Sci. 2021, 42, 93–101. [Google Scholar] [CrossRef]
- Steensels, J.; Daenen, L.; Malcorps, P.; Derdelinckx, G.; Verachtert, H.; Verstrepen, K.J. Brettanomyces yeasts—From spoilage organisms to valuable contributors to industrial fermentations. Int. J. Food Microbiol. 2015, 206, 24–38. [Google Scholar] [CrossRef] [Green Version]
- Karim, A.; Gerliani, N.; Aïder, M. Kluyveromyces marxianus: An emerging yeast cell factory for applications in food and biotechnology. Int. J. Food Microbiol. 2020, 333, 108818. [Google Scholar] [CrossRef]
- Roman, W. Yeasts, 1st ed.; Junk: The Hague, The Netherlands, 1957. [Google Scholar]
- Rivera Flores, V.K.; Alcaine, S.D. Cornell University, Ithaca, NY, USA. Unpublished work. 2019. [Google Scholar]
- Salaün, F.; Mietton, B.; Gaucheron, F. Buffering capacity of dairy products. Int. Dairy J. 2005, 15, 95–109. [Google Scholar] [CrossRef]
- O′Connell, J.E.; Fox, P.F. Effect of beta-lactoglobulin and precipitation of calcium phosphate on the thermal coagulation of milk. J. Dairy Res. 2001, 68, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Chavez, S.A.; Arevalo-Gallegos, S.; Rascon-Cruz, Q. Lactoferrin: Structure, function and applications. Int. J. Antimicrob. Agents 2009, 33, 301.e301–301.e308. [Google Scholar] [CrossRef]
- Painting, K.; Kirsop, B. A quick method for estimating the percentage of viable cells in a yeast population, using methylene blue staining. World J. Microbiol. Biotechnol. 1990, 6, 346–347. [Google Scholar] [CrossRef]
- Sandhu, D.K.; Waraich, M.K. Conversion of cheese whey to single-cell protein. Biotechnol. Bioeng. 1983, 25, 797–808. [Google Scholar] [CrossRef] [PubMed]
- Marcus, J.F.; DeMarsh, T.A.; Alcaine, S.D. Upcycling of whey permeate through yeast- and mold-driven fermentations under anoxic and oxic conditions. Fermentation 2021, 7, 16. [Google Scholar] [CrossRef]
- Li, B.; Wang, Z.; Li, S.; Donelan, W.; Wang, X.; Cui, T.; Tang, D. Preparation of lactose-free pasteurized milk with a recombinant thermostable β-glucosidase from Pyrococcus furiosus. BMC Biotechnol. 2013, 13, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vervoort, Y.; Herrera-Malaver, B.; Mertens, S.; Guadalupe Medina, V.; Duitama, J.; Michiels, L.; Derdelinckx, G.; Voordeckers, K.; Verstrepen, K.J. Characterization of the recombinant Brettanomyces anomalus β-glucosidase and its potential for bioflavouring. J. Appl. Microbiol. 2016, 121, 721–733. [Google Scholar] [CrossRef] [Green Version]
- Moktaduzzaman, M.; Galafassi, S.; Capusoni, C.; Vigentini, I.; Ling, Z.; Piškur, J.; Compagno, C. Galactose utilization sheds new light on sugar metabolism in the sequenced strain Dekkera bruxellensis CBS 2499. FEMS Yeast Res. 2015, 15, fou009. [Google Scholar] [CrossRef] [Green Version]
- Tiukova, I.A.; Møller-Hansen, I.; Belew, Z.M.; Darbani, B.; Boles, E.; Nour-Eldin, H.H.; Linder, T.; Nielsen, J.; Borodina, I. Identification and characterisation of two high-affinity glucose transporters from the spoilage yeast Brettanomyces bruxellensis. FEMS Microbiol. Lett. 2019, 366, fnz222. [Google Scholar] [CrossRef] [Green Version]
- Johnston, M. A model fungal gene regulatory mechanism: The GAL genes of Saccharomyces cerevisiae. Microbiol. Rev. 1987, 51, 458–476. [Google Scholar] [CrossRef]
- Sellick, C.A.; Campbell, R.N.; Reece, R.J. Chapter 3 galactose metabolism in yeast—Structure and regulation of the leloir pathway enzymes and the genes encoding them. In International Review of Cell and Molecular Biology; Jeon, K.W., Ed.; International Review of Cell and Molecular Biology; Elsevier: Amsterdam, The Netherlands, 2008; Volume 269, pp. 111–150. [Google Scholar]
- Silveira, W.B.; Passos, F.J.V.; Mantovani, H.C.; Passos, F.M.L. Ethanol production from cheese whey permeate by Kluyveromyces marxianus UFV-3: A flux analysis of oxido-reductive metabolism as a function of lactose concentration and oxygen levels. Enzyme Microb. Technol. 2005, 36, 930–936. [Google Scholar] [CrossRef]
- Ozmihci, S.; Kargi, F. Ethanol production from cheese whey powder solution in a packed column bioreactor at different hydraulic residence times. Biochem. Eng. J. 2008, 42, 180–185. [Google Scholar] [CrossRef]
- Diniz, R.H.S.; Rodrigues, M.Q.R.B.; Fietto, L.G.; Passos, F.M.L.; Silveira, W.B. Optimizing and validating the production of ethanol from cheese whey permeate by Kluyveromyces marxianus UFV-3. Biocatal. Agric. Biotechnol. 2014, 3, 111–117. [Google Scholar] [CrossRef]
- Speers, R.A.; Rogers, P.; Smith, B. Non-linear modelling of industrial brewing fermentations. J. Inst. Brew. 2003, 109, 229–235. [Google Scholar] [CrossRef]
- Coote, N.; Kirsop, B.H. Factors responsible for the decrease in pH during beer fermentations. J. Inst. Brew. 1976, 82, 149–153. [Google Scholar] [CrossRef]
- Cássio, F.; Leão, C.; van Uden, N. Transport of lactate and other short-chain monocarboxylates in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1987, 53, 509–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werner-Washburne, M.; Braun, E.; Johnston, G.C.; Singer, R.A. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol. Rev. 1993, 57, 383–401. [Google Scholar] [CrossRef]
- Ariyanti, D.; Hadiyanto, H. Ethanol production from whey by Kluyveromyces marxianus in batch fermentation system: Kinetics parameters estimation. Bull. Chem. React. Eng. Catal. 2013, 7, 179. [Google Scholar] [CrossRef]
- Zafar, S.; Owais, M. Ethanol production from crude whey by Kluyveromyces marxianus. Biochem. Eng. J. 2006, 27, 295–298. [Google Scholar] [CrossRef]
- Custer, M. Onderzoekingen over Het Gistgeslacht. Ph.D. Thesis, TU Delft, Delft, The Netherlands, 3 May 1940. [Google Scholar]
- Fonseca, A.; Spencer-Martins, I.; Van Uden, N. Transport of lactic acid in Kluyveromyces marxianus: Evidence for a monocarboxylate uniport. Yeast 1991, 7, 775–780. [Google Scholar] [CrossRef]
| Supplemented Medium | ||||||
|---|---|---|---|---|---|---|
| Lactose * | Glucose + Galactose | Glucose | Galactose | |||
| Species | Timepoint | Lactose (g/L) | Glucose (g/L) | Galactose (g/L) | Glucose (g/L) | Galactose (g/L) |
| B. claussenii | Day 0 | 32.86 ± 1.95 | 19.07 ± 0.15 | 18.84 ± 0.49 | 37.12 ± 0.51 | 35.03 ± 0.34 |
| Day 18 | ND | ND | 15.28 ± 0.56 | ND | 0.01 ± 0.02 | |
| K. marxianus | Day 0 | 33.17 ± 1.55 | 18.95 ± 0.12 | 18.39 ± 0.29 | 38 ± 1.05 | 34.95 ± 0.49 |
| Day 7 | ND | ND | ND | ND | ND | |
| S. cerevisiae | Day 0 | 32.87 ± 2.05 | 18.64 ± 0.17 | 18.45 ± 0.32 | 37.42 ± 0.12 | 35.58 ± 0.63 |
| Day 7 | 34.16 ± 0.61 | ND | ND | ND | ND | |
| Species | Timepoint | Supplemented Medium | |||
|---|---|---|---|---|---|
| Lactose | Glucose + Galactose | Glucose | Galactose | ||
| B. claussenii | Day 18 | 1.78 ± 0.07 a | 0.77 ± 0.10 b | 1.86 ± 0.09 a | 1.66 ± 0.10 a |
| K. marxianus | Day 7 | 2.05 ± 0.03 a,b | 1.99 ± 0.01 b | 1.99 ± 0.03 b | 2.11 ± 0.04 a |
| S. cerevisiae | Day 7 | ND | 2.00 ± 0.09 a | 2.02 ± 0.06 a | 1.94 ± 0.03 a |
| Species | Timepoint | Lactose | Glucose + Galactose | Glucose | Galactose |
|---|---|---|---|---|---|
| Lactic Acid (g/L) | |||||
| B. claussenii | Day 0 | 0.101 ± 0.002 | 0.102 ± 0.008 | 0.104 ± 0.004 | 0.104 ± 0.003 |
| Day 18 | ND | ND | ND | ND | |
| K. marxianus | Day 0 | 0.1 ± 0.004 | 0.102 ± 0.006 | 0.116 ± 0.016 | 0.102 ± 0.007 |
| Day 7 | ND | 0.048 ± 0.041 | ND | ND | |
| S. cerevisiae | Day 0 | 0.101 ± 0.002 | 0.104 ± 0.003 | 0.104 ± 0.002 | 0.105 ± 0.001 |
| Day 7 | ND | 0.099 ± 0.011 a,b | 0.132 ± 0.018 a | 0.096 ± 0.014 b | |
| Acetic Acid (g/L) | |||||
| B. claussenii | Day 0 | ND | ND | 0.013 ± 0.023 | ND |
| Day 18 | 2.728 ± 0.425 a | 2.305 ± 0.77 a | 1.66 ± 0.339 a | 3.096 ± 0.696 a | |
| K. marxianus | Day 0 | ND | ND | 0.014 ± 0.024 | ND |
| Day 7 | 0.184 ± 0.033 a | 0.326 ± 0.082 a | 0.311 ± 0.042 a | 0.299 ± 0.08 a | |
| S. cerevisiae | Day 0 | ND | ND | ND | ND |
| Day 7 | ND | 0.513 ± 0.382 a | 0.348 ± 0.139 a | 0.547 ± 0.309 a | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rivera Flores, V.K.; DeMarsh, T.A.; Gibney, P.A.; Alcaine, S.D. Fermentation of Dairy-Relevant Sugars by Saccharomyces, Kluyveromyces, and Brettanomyces: An Exploratory Study with Implications for the Utilization of Acid Whey, Part I. Fermentation 2021, 7, 266. https://doi.org/10.3390/fermentation7040266
Rivera Flores VK, DeMarsh TA, Gibney PA, Alcaine SD. Fermentation of Dairy-Relevant Sugars by Saccharomyces, Kluyveromyces, and Brettanomyces: An Exploratory Study with Implications for the Utilization of Acid Whey, Part I. Fermentation. 2021; 7(4):266. https://doi.org/10.3390/fermentation7040266
Chicago/Turabian StyleRivera Flores, Viviana K., Timothy A. DeMarsh, Patrick A. Gibney, and Samuel D. Alcaine. 2021. "Fermentation of Dairy-Relevant Sugars by Saccharomyces, Kluyveromyces, and Brettanomyces: An Exploratory Study with Implications for the Utilization of Acid Whey, Part I" Fermentation 7, no. 4: 266. https://doi.org/10.3390/fermentation7040266
APA StyleRivera Flores, V. K., DeMarsh, T. A., Gibney, P. A., & Alcaine, S. D. (2021). Fermentation of Dairy-Relevant Sugars by Saccharomyces, Kluyveromyces, and Brettanomyces: An Exploratory Study with Implications for the Utilization of Acid Whey, Part I. Fermentation, 7(4), 266. https://doi.org/10.3390/fermentation7040266






