Upcycling of Whey Permeate through Yeast- and Mold-Driven Fermentations under Anoxic and Oxic Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms
2.2. Experimental Design
2.3. Data Collection
3. Results
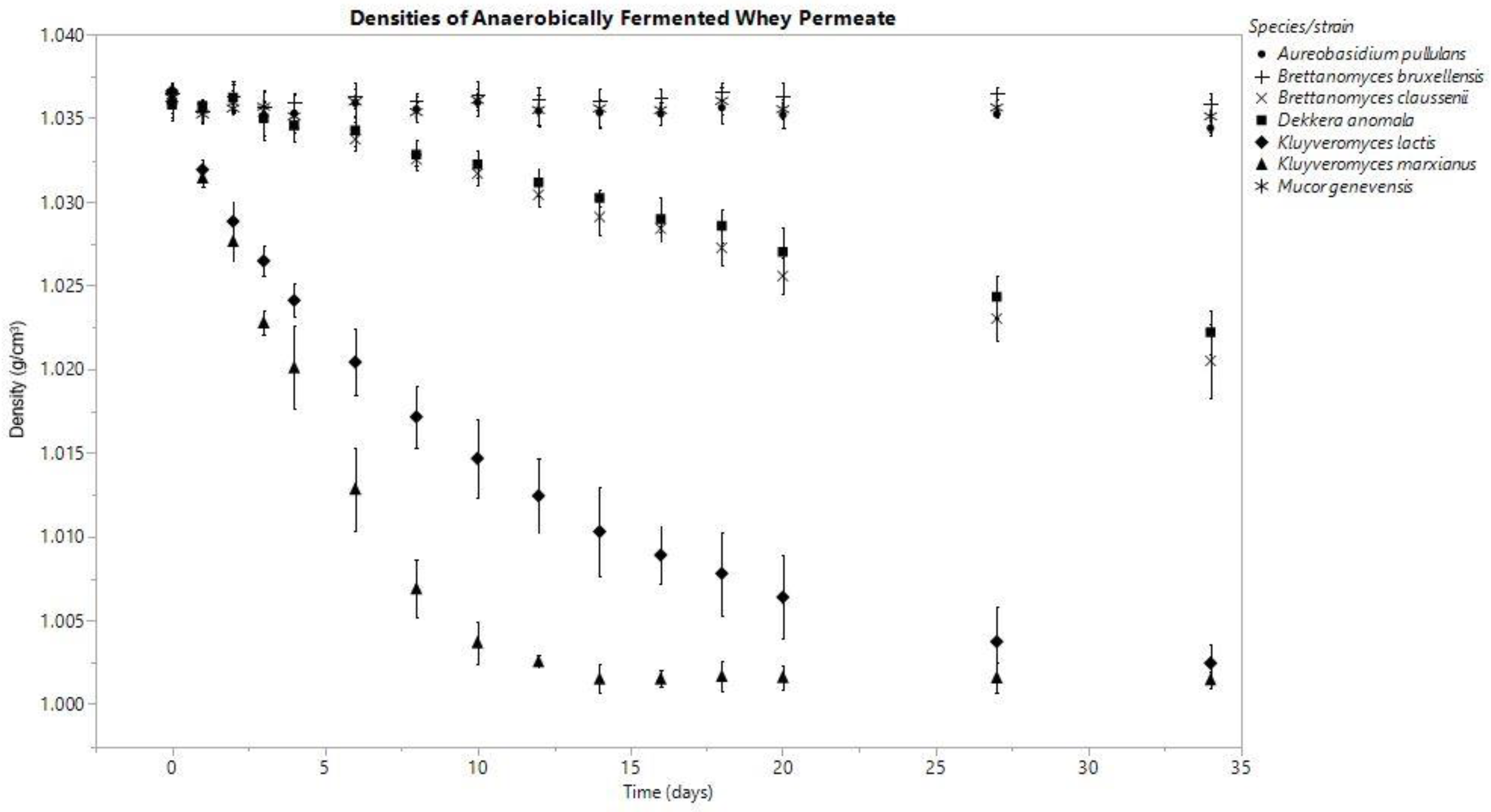
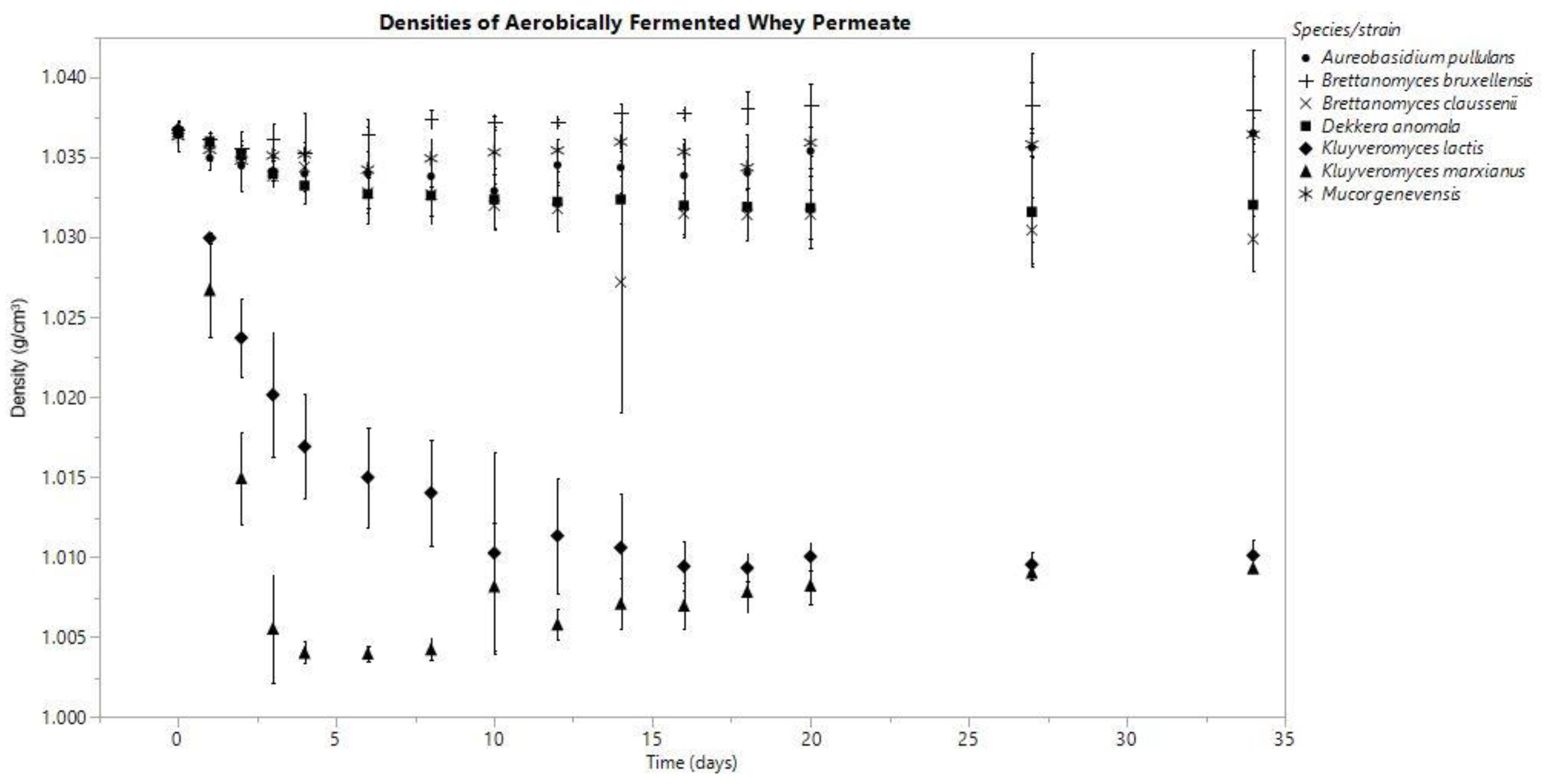
3.1. Density
3.2. Sugar Utilization
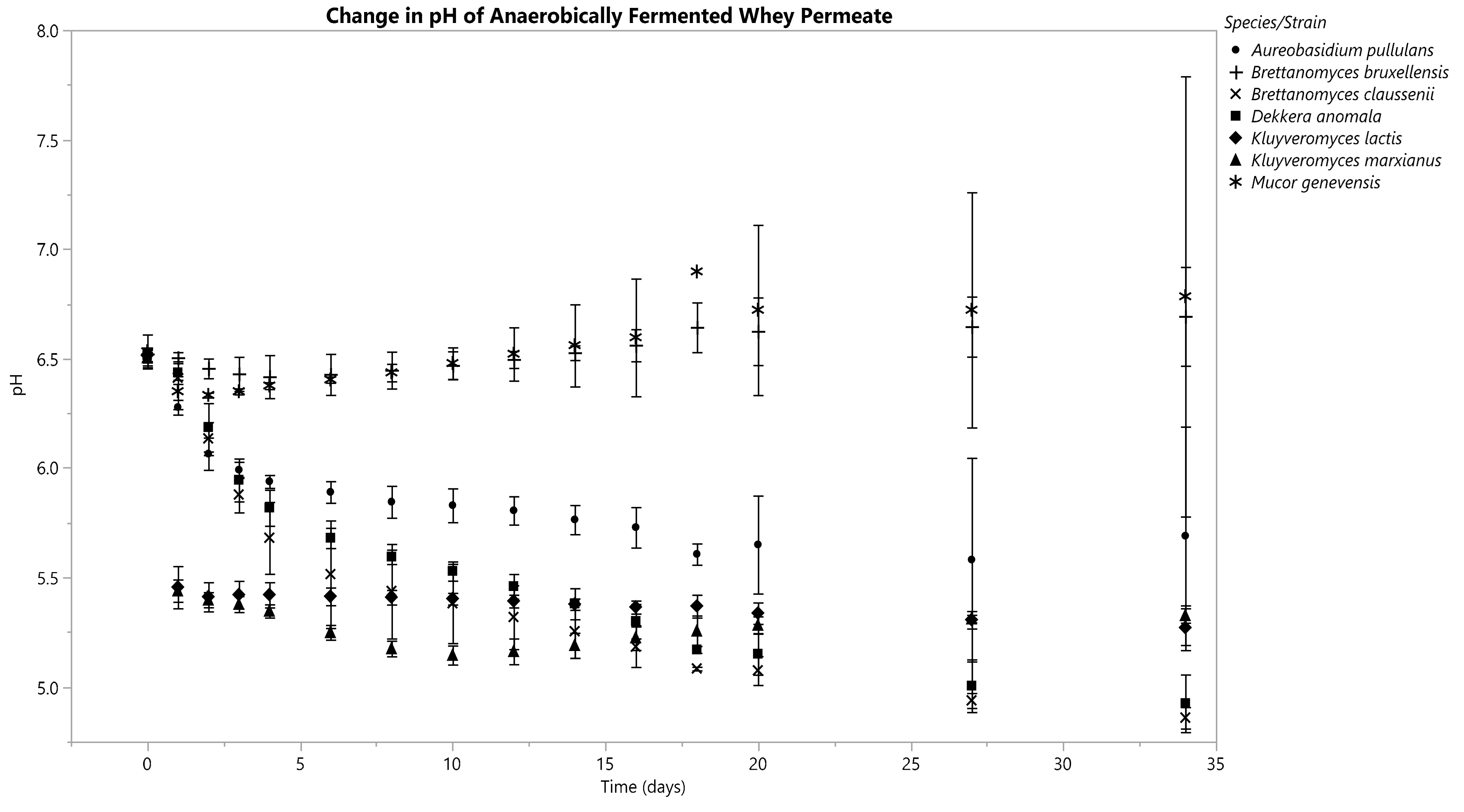
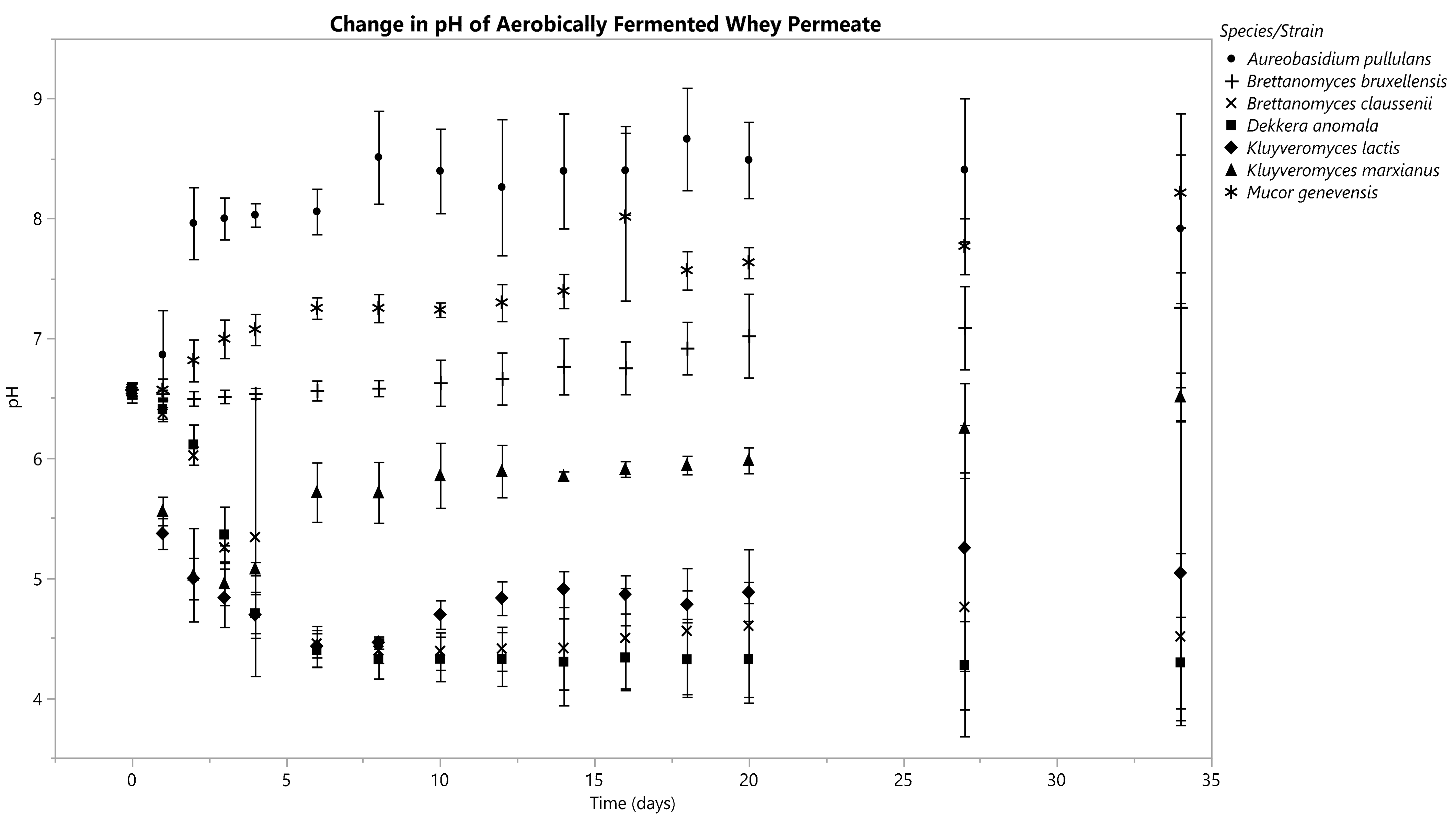
3.3. Changes in Fermentate pH
3.3.1. Anaerobic
3.3.2. Aerobic
3.4. Organic Acid Concentration (Acetic, Lactic, Tartaric, Malic)
3.4.1. Anaerobic
3.4.2. Aerobic
3.5. Ethanol Production
3.5.1. Anaerobic
3.5.2. Aerobic
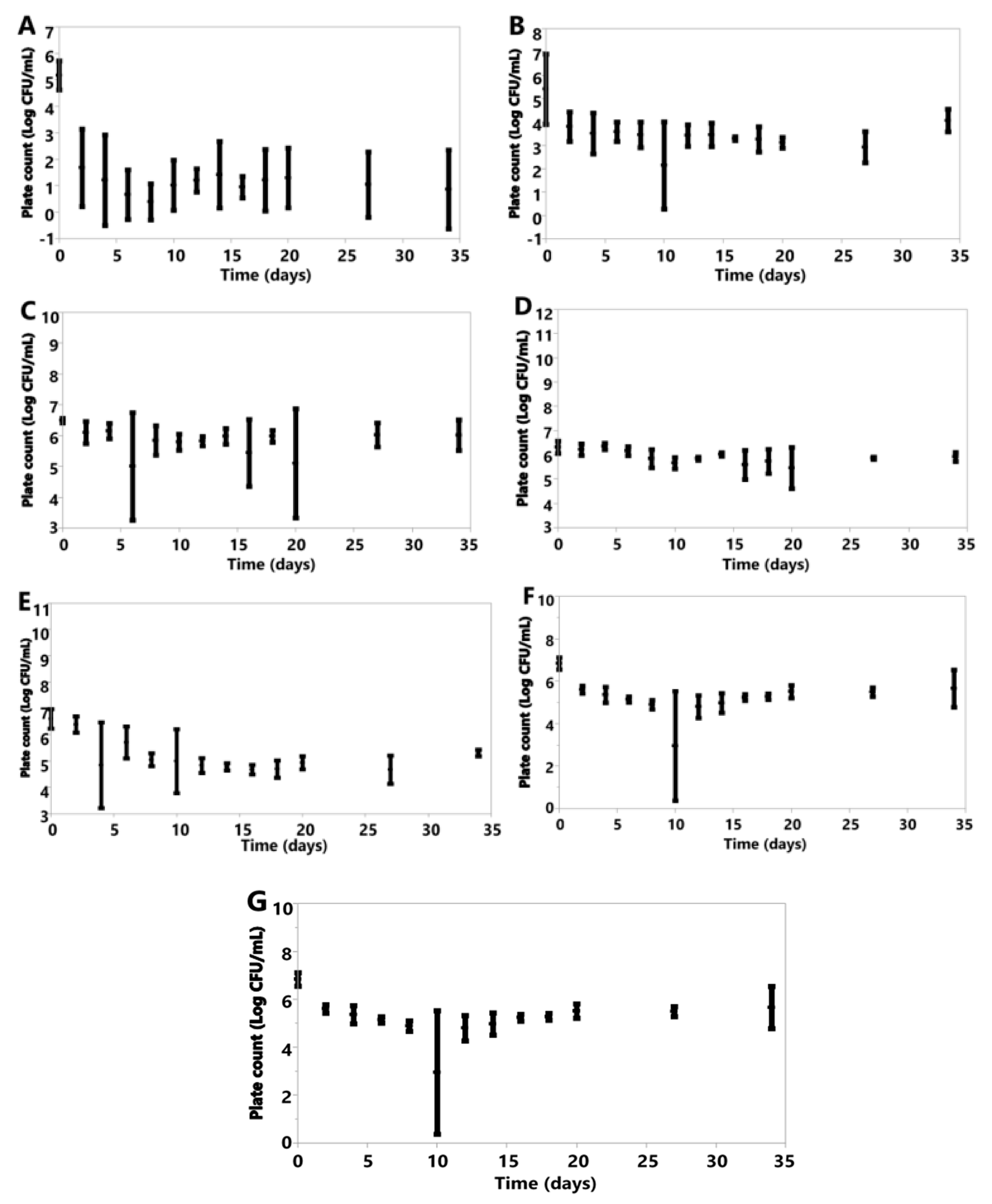
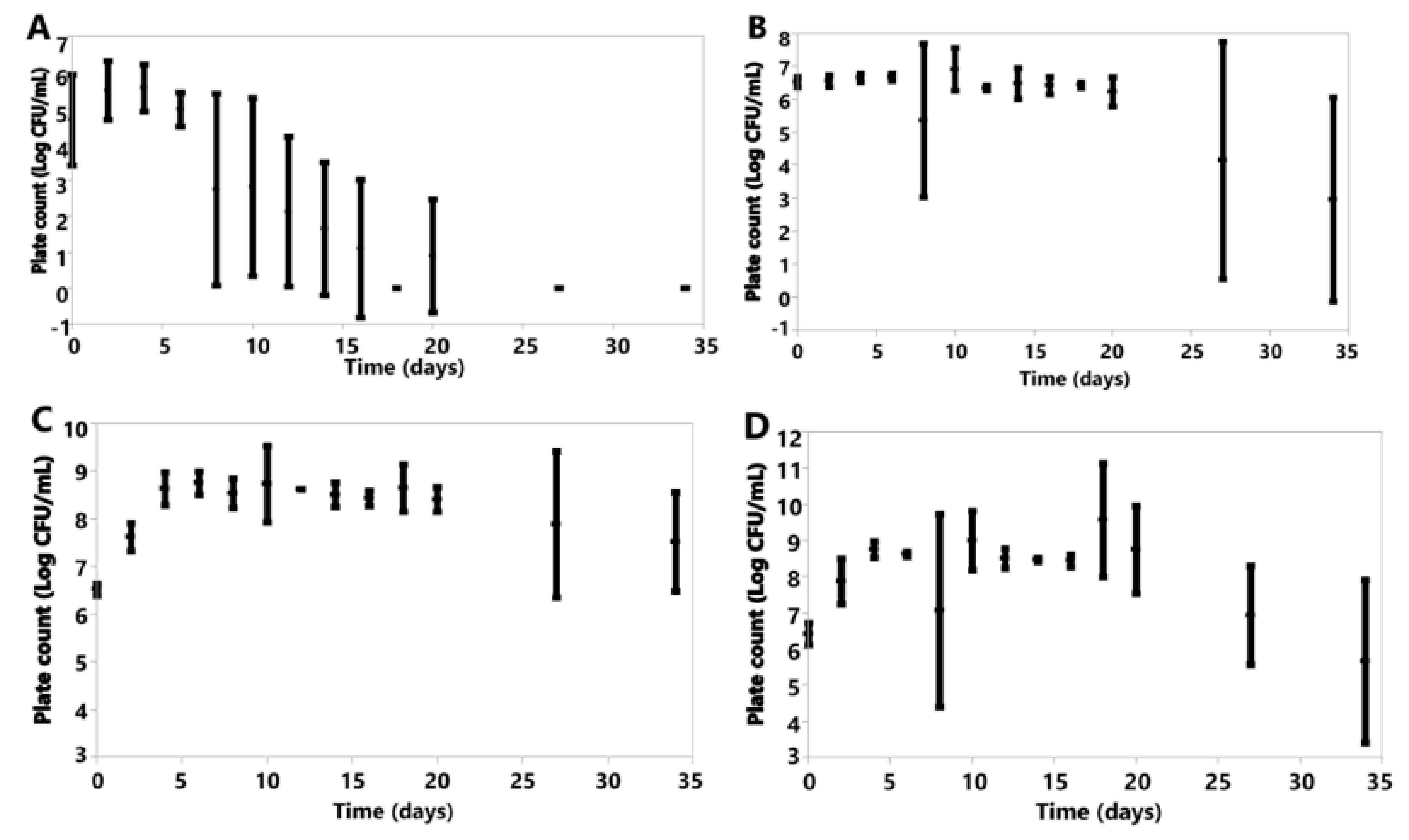
3.6. Cell Counts
3.6.1. Anaerobic
3.6.2. Aerobic
4. Discussion
4.1. Yeast Species
4.1.1. Kluyveromyces spp.
4.1.2. Brettanomyces spp. and D. anomala
4.2. Mold Species
4.2.1. M. genevensis
4.2.2. A. pullulans
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Erickson, B.E. Acid whey: Is the waste product an untapped goldmine? Chem. Eng. News Online 2017, 95, 26–30. [Google Scholar]
- Menchik, P.; Zuber, T.; Zuber, A.; Moraru, C.I. Short communication: Composition of coproduct streams from dairy processing: Acid whey and milk permeate. J. Dairy Sci. 2019, 102, 3978–3984. [Google Scholar] [CrossRef]
- Sanjay Gami, G.G.; Czymmek, K.; Ganoe, K.; Ketterings, Q. Acid Whey pH and Nutrient Content. Agron. Fact Sheet Ser. Fact Sheet 2016, 90, 1–2. [Google Scholar]
- State, N.Y. Part 360: Solid Waste Management Facilities General Requirements; D. o. E. Conservation: New York, NY, USA, 2013; pp. 4017–4307. [Google Scholar]
- Staff, J.U.S. Sales of Non-Alcoholic Beverages Grow More Than $2 Billion in 2017; Coca-Cola, Journey: Atlanta, GA, USA, 2018. [Google Scholar]
- Jayabalan, R.; Malbaša, R.V.; Lončar, E.S.; Vitas, J.S.; Sathishkumar, M. A Review on Kombucha Tea—Microbiology, Composition, Fermentation, Beneficial Effects, Toxicity, and Tea Fungus. Compr. Rev. Food Sci. Food Saf. 2014, 13, 538–550. [Google Scholar] [CrossRef]
- Marsh, A.J.; Hill, C.; Ross, R.P.; Cotter, P.D. Fermented beverages with health-promoting potential: Past and future perspectives. Trends Food Sci. Technol. 2014, 38, 113–124. [Google Scholar] [CrossRef]
- French, P. In Focus: Top Trends in Lower Alcohol Beer; Union Press Ltd.: London, UK, 2018. [Google Scholar]
- Smith, M.T. The Yeast; Elsevier Science: New York, NY, USA, 2010. [Google Scholar]
- Martin Lo, S.A.-S.; Hsu, C. Bioprocessing for value-added products from renewable resources. In Chapter 22. Bioconversion of Whey Lactose into Microbial Exopolysaccharides; Elsevier: Amsterdam, The Netherlands; Boston, MA, USA, 2007; pp. 559–583. [Google Scholar]
- Carlile, M.J.; Watkinson, S.C.; Gooday, G.W. The Fungi; Elsevier: London, UK, 2001. [Google Scholar]
- Jones, R.P.; Greenfield, P.F. Alcohol fermentation by yeasts—The effect of environmental and other variables. Process Biochem. 1981, 16, 42–49. [Google Scholar]
- FI Monila, P.S.; Jong, S.C. Validation of the species concept in the genus Dekkera by restriction analysis of genes coding for rRNA. Int. J. Syst. Bacteriol. 1993, 43, 32–35. [Google Scholar]
- Hernâni Gerós, F.C.; Leão, C. Utilization and Transport of Acetic Acid in Dekkera anomala and Their Implications on the Survival of the Yeast in Acidic Environments. J. Food Prot. 1999, 63, 96–101. [Google Scholar] [CrossRef]
- Salman Zafar, M.O. Ethanol production from crude whey by Kluyveromyces marxianus. Biochem. Eng. J. 2006, 27, 295–298. [Google Scholar] [CrossRef]
- Karimi, G.E.; Taherzadeh, M.J. Ethanol production from dilute-acid pretreated rice straw by simultaneous saccharification and fermentation with Mucor indicus, Rhizopus oryzae, and Saccharomyces cerevisiae. Enzyme Microb. Technol. 2006, 40, 138–144. [Google Scholar] [CrossRef]
- Roukas, T. Pullulan production from deproteinized whey by Aureobasidium pullulans. J. Ind. Microbiol. Biotechnol. 1999, 22, 617–621. [Google Scholar] [CrossRef] [PubMed]
- D’Amore, T.; Russel, I.; Stewart, G.G. Sugar Utilization by yeast during fermentation. J. Ind. Microbiol. 1988, 4, 315–324. [Google Scholar] [CrossRef]
- Simova, E.; Beshkova, D.; Angelov, A.; Hristozova, T.; Frengova, G.; Spasov, Z. Lactic acid bacteria and yeasts in kefir grains and kefir made from them. J. Ind. Microbiol. Biotechnol. 2002, 28, 1–6. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, V.S.; Sutton, C.; Bencivengo, M.; Sullivan, B.; Holsinger, V.H. Influence of lactose hydrolysis and solids concentration on alcohol production by yeast in acid whey ultrafiltrate. Biotechnol. Bioeng. 1977, 19, 1689–1702. [Google Scholar] [CrossRef]
- Brown, S.W.; Oliver, S.G.; Harrison, D.E.F.; Righelato, R.C. Ethanol inhibition of yeast growth and fermentation: Differences in the magnitude and complexity of the effect. Appl. Microbiol. Biotechnol. 1981, 11, 151–155. [Google Scholar] [CrossRef]
- Maudy, T.; Smith, A.M.v.G. Dekkera anomala sp. nov., the teleomorph of Brettanomyces anomalus, recovered from spoiled soft drinks. Antonievan Leeuwenhoek 1984, 50, 143–148. [Google Scholar]
- Jan Steensels, L.D.; Malcorps, P.; Derdlinckx, G.; Verachtert, H.; Verstrepen, K.J. Brettanomyces yeasts—From spoilage organisms to valuable contributors to industrial fermentations. Int. J. Food Microbiol. 2015, 206, 24–38. [Google Scholar] [CrossRef]
- Jackson, R.S. Wine Science: Principles and Applications F. S. A. Technology. Science Direct; Academic Press: Cambridge, MA, USA, 2008. [Google Scholar]
- Orlowski, M. Mucor Dimorphism. Microbiol. Rev. 1991, 55, 234–258. [Google Scholar] [CrossRef]
- Walsh, M.; Bombyk, R.A.; Wagh, A.; Bingham, A.; Berreau, L.M. Synthesis of lactose monolaurate as influenced by various lipases and solvents. J. Mol. Catal. B Enzym. 2009, 60, 171–177. [Google Scholar] [CrossRef]
- Saha, B.C. Production, purification and properties of endoglucanase from a newly isolated strain of Mucor circinelloides. Process Biochem. 2004, 39, 1871–1876. [Google Scholar] [CrossRef]
- Bartnicki-Garcia, S. Control of Dimorphism in Mucor by Hexoses: Inhibition of Hyphal Morphogenesis. J. Bacteriol. 1968, 96, 1586–1594. [Google Scholar] [CrossRef] [PubMed]
- Clark-Walker, G.D. Relationship between Dimorphology and Respiration in Mucor genevensis Studied with Chloramphenicol. J. Bacteriol. 1973, 116, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Anh LeDuy, J.-J.Y.; Chagraoui, A. Enhanced Production of Pullulan from Lactose Adaptation and by Mixed Culture Techniques. Biotechnol. Lett. 1983, 5, 49–54. [Google Scholar]
- Denis Rho, A.M.; Luong, J.H.T.; LeDuy, A. Oxygen requirement in pullulan fermentation. Appl. Microbiol. Biotechnol. 1988, 28, 361–366. [Google Scholar]
- Kuan-Chen Cheng, A.D.; Catchmark, J.M. Pullulan: Biosynthesis, production and applications. Appl. Microbiol. Biotechnol. 2011, 92, 29–44. [Google Scholar] [CrossRef]
- Bureau, T.A.T. “Kombucha”. Does TTB Regulate Kombucha Retrieved 5/8/2019. 2019. Available online: https://ttb.gov/kombucha/index.shtml (accessed on 2 January 2021).
- Costello, H. Global Kombucha Market 2018 Size, Demand, Types, Consumption, Key Players, Production, Top Regions, Competitive and Comparative Analysis, Growth & Forecast By 2023; Reuters Online: New York, NY, USA, 2018. [Google Scholar]
- Lawton, M.R. Leveraging endogenous barley enzymes to turn lactose-containing dairy by-products into fermentable adjuncts for Saccharomyces cerevisiae-based ethanol fermentations. J. Dairy Sci. 2019, 102, 2044–2050. [Google Scholar] [CrossRef]
- Shehadeh, A.; Kechagia, D.; Evangelou, A.; Tataridis, P.; Shehadeh, F. Effect of ethanol, glycerol, glucose and tartaric acid on the viscosity of model aqueous solutions and wine samples. Food Chem. 2019, 300, 125191. [Google Scholar] [CrossRef]
- Spiller, G.A.; Story, J.A.; Furumoto, E.J.; Chezem, J.C.; Spiller, M. Effect of tartaric acid and dietary fibre from sun-dried raisins on colonic function and on bile acid and volatile fatty acid excretion in healthy adults. Br. J. Nutr. 2003, 90, 803–807. [Google Scholar] [CrossRef]

| Lactose (g/100 g) | Glucose (g/100 g) | Galactose (g/100 g) | Total Sugar (g/100 g) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample Day | Species | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev |
| 0 | Uninoculated | 7.2 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 7.2 | 0.1 |
| 27 | A. pullulans | 6.6 | 0.2 | 0.0 | 0.0 | 0.2 | 0.1 | 6.9 | 0.3 |
| 27 | B. bruxellensis | 7.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7.4 | 0.0 |
| 27 | B. claussenii | 3.9 | 0.6 | 0.0 | 0.0 | 0.4 | 0.3 | 4.2 | 0.4 |
| 27 | D. anomala | 4.0 | 0.6 | 0.0 | 0.0 | 0.5 | 0.3 | 4.4 | 0.3 |
| 27 | K. lactis | 0.2 | 0.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.3 |
| 27 | K. marxianus | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 27 | M. genevensis | 7.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7.2 | 0.0 |
| Lactose | Glucose | Galactose | Total Sugar | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample Day | Species | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev |
| 0 | Uninoculated | 7.2 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 7.2 | 0.1 |
| 27 | A. pullulans | 7.5 | 0.9 | 0.0 | 0.0 | 0.1 | 0.1 | 7.6 | 0.8 |
| 27 | B. bruxellensis | 6.5 | 2.1 | 0.4 | 0.6 | 0.4 | 0.5 | 7.3 | 0.9 |
| 27 | B. claussenii | 5.4 | 0.6 | 0.0 | 0.1 | 0.0 | 0.1 | 5.5 | 0.7 |
| 27 | D. anomala | 5.5 | 0.8 | 0.0 | 0.0 | 0.0 | 0.0 | 5.5 | 0.8 |
| 27 | K. lactis | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 27 | K. marxianus | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 27 | M. genevensis | 7.4 | 0.3 | 0.0 | 0.0 | 0.0 | 0.0 | 7.4 | 0.3 |
| A. pullulans | B. bruxellensis | B. claussenii | D. anomala | K. lactis | K. marxianus | M. genevensis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (days) | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev |
| 0 | 0.09 | 0.16 | 0.24 | 0.06 | 0.52 | 0.46 | 0.21 | 0.02 | 0.28 | 0.15 | 0.66 | 0.67 | 0.28 | 0.08 |
| 20 | 0.56 | 0.21 | 0.07 | 0.13 | 0.66 | 0.46 | 0.54 | 0.07 | 0.60 | 0.22 | 1.17 | 0.21 | 0.56 | 0.20 |
| 34 | 0.45 | 0.23 | 0.08 | 0.14 | 0.90 | 0.33 | 0.73 | 0.29 | 0.79 | 0.09 | 1.39 | 0.23 | 0.66 | 0.55 |
| A. pullulans | B. bruxellensis | B. claussenii | D. anomala | K. lactis | K. marxianus | M. genevensis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (days) | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev |
| 0 | 0.85 | 0.06 | 0.78 | 0.08 | 0.78 | 0.09 | 0.81 | 0.03 | 0.80 | 0.04 | 0.79 | 0.06 | 0.75 | 0.06 |
| 20 | 1.27 | 0.30 | 0.70 | 0.24 | 1.37 | 0.29 | 1.21 | 0.24 | 0.84 | 0.07 | 0.81 | 0.03 | 0.00 | 0.00 |
| 34 | 1.09 | 0.34 | 0.57 | 0.36 | 1.33 | 0.19 | 1.25 | 0.13 | 0.85 | 0.13 | 0.47 | 0.09 | 0.00 | 0.00 |
| A. pullulans | B. bruxellensis | B. claussenii | D. anomala | K. lactis | K. marxianus | M. genevensis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (days) | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev |
| 0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 20 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.63 | 0.09 | 0.16 | 0.15 | 0.00 | 0.00 |
| 34 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.44 | 0.07 | 0.00 | 0.00 | 0.00 | 0.00 |
| A. pullulans | B. bruxellensis | B. claussenii | D. anomala | K. lactis | K. marxianus | M. genevensis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (days) | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev |
| 0 | 0.25 | 0.22 | 0.40 | 0.09 | 0.69 | 0.62 | 0.86 | 0.42 | 0.76 | 0.29 | 0.52 | 0.32 | 0.50 | 0.30 |
| 20 | 0.29 | 0.03 | 0.00 | 0.00 | 5.88 | 3.97 | 8.49 | 3.71 | 0.75 | 0.41 | 0.48 | 0.35 | 0.26 | 0.02 |
| 34 | 0.25 | 0.08 | 0.03 | 0.06 | 7.75 | 5.35 | 9.18 | 4.13 | 2.68 | 2.22 | 0.34 | 0.20 | 0.40 | 0.19 |
| A. pullulans | B. bruxellensis | B. claussenii | D. anomala | K. lactis | K. marxianus | M. genevensis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (days) | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev |
| 0 | 0.83 | 0.05 | 0.84 | 0.01 | 0.85 | 0.04 | 0.87 | 0.03 | 0.86 | 0.02 | 0.86 | 0.01 | 0.86 | 0.01 |
| 20 | 0.26 | 0.24 | 0.66 | 0.35 | 0.40 | 0.10 | 0.34 | 0.06 | 0.00 | 0.00 | 0.13 | 0.22 | 0.67 | 0.09 |
| 34 | 0.26 | 0.26 | 0.29 | 0.51 | 0.31 | 0.28 | 0.19 | 0.17 | 0.00 | 0.00 | 0.06 | 0.10 | 0.37 | 0.35 |
| A. pullulans | B. bruxellensis | B. claussenii | D. anomala | K. lactis | K. marxianus | M. genevensis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (days) | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev |
| 0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 20 | 0.23 | 0.20 | 0.04 | 0.06 | 0.31 | 0.38 | 0.44 | 0.56 | 0.00 | 0.00 | 0.04 | 0.06 | 0.00 | 0.00 |
| 34 | 0.39 | 0.35 | 0.16 | 0.27 | 0.65 | 0.67 | 0.83 | 0.76 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| A. pullulans | B. bruxellensis | B. claussenii | D. anomala | K. lactis | K. marxianus | M. genevensis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (days) | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev |
| 0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 20 | 0.00 | 0.00 | 0.00 | 0.00 | 1.27 | 0.25 | 1.04 | 0.16 | 3.72 | 0.62 | 4.52 | 0.02 | 0.00 | 0.00 |
| 34 | 0.00 | 0.00 | 0.00 | 0.00 | 1.81 | 0.44 | 1.63 | 0.23 | 4.33 | 0.10 | 4.47 | 0.02 | 0.00 | 0.00 |
| A. pullulans | B. bruxellensis | B. claussenii | D. anomala | K. lactis | K. marxianus | M. genevensis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (days) | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev |
| 0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 20 | 0.00 | 0.00 | 0.00 | 0.00 | 0.13 | 0.23 | 0.12 | 0.12 | 0.53 | 0.40 | 0.68 | 0.58 | 0.00 | 0.00 |
| 34 | 0.00 | 0.00 | 0.00 | 0.00 | 0.18 | 0.31 | 0.20 | 0.18 | 0.04 | 0.08 | 0.00 | 0.00 | 0.00 | 0.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcus, J.F.; DeMarsh, T.A.; Alcaine, S.D. Upcycling of Whey Permeate through Yeast- and Mold-Driven Fermentations under Anoxic and Oxic Conditions. Fermentation 2021, 7, 16. https://doi.org/10.3390/fermentation7010016
Marcus JF, DeMarsh TA, Alcaine SD. Upcycling of Whey Permeate through Yeast- and Mold-Driven Fermentations under Anoxic and Oxic Conditions. Fermentation. 2021; 7(1):16. https://doi.org/10.3390/fermentation7010016
Chicago/Turabian StyleMarcus, Justin Fisk, Timothy A. DeMarsh, and Samuel David Alcaine. 2021. "Upcycling of Whey Permeate through Yeast- and Mold-Driven Fermentations under Anoxic and Oxic Conditions" Fermentation 7, no. 1: 16. https://doi.org/10.3390/fermentation7010016
APA StyleMarcus, J. F., DeMarsh, T. A., & Alcaine, S. D. (2021). Upcycling of Whey Permeate through Yeast- and Mold-Driven Fermentations under Anoxic and Oxic Conditions. Fermentation, 7(1), 16. https://doi.org/10.3390/fermentation7010016







