Yeast Cellular Stress: Impacts on Bioethanol Production
Abstract
1. Introduction
2. Yeast Stress Responses
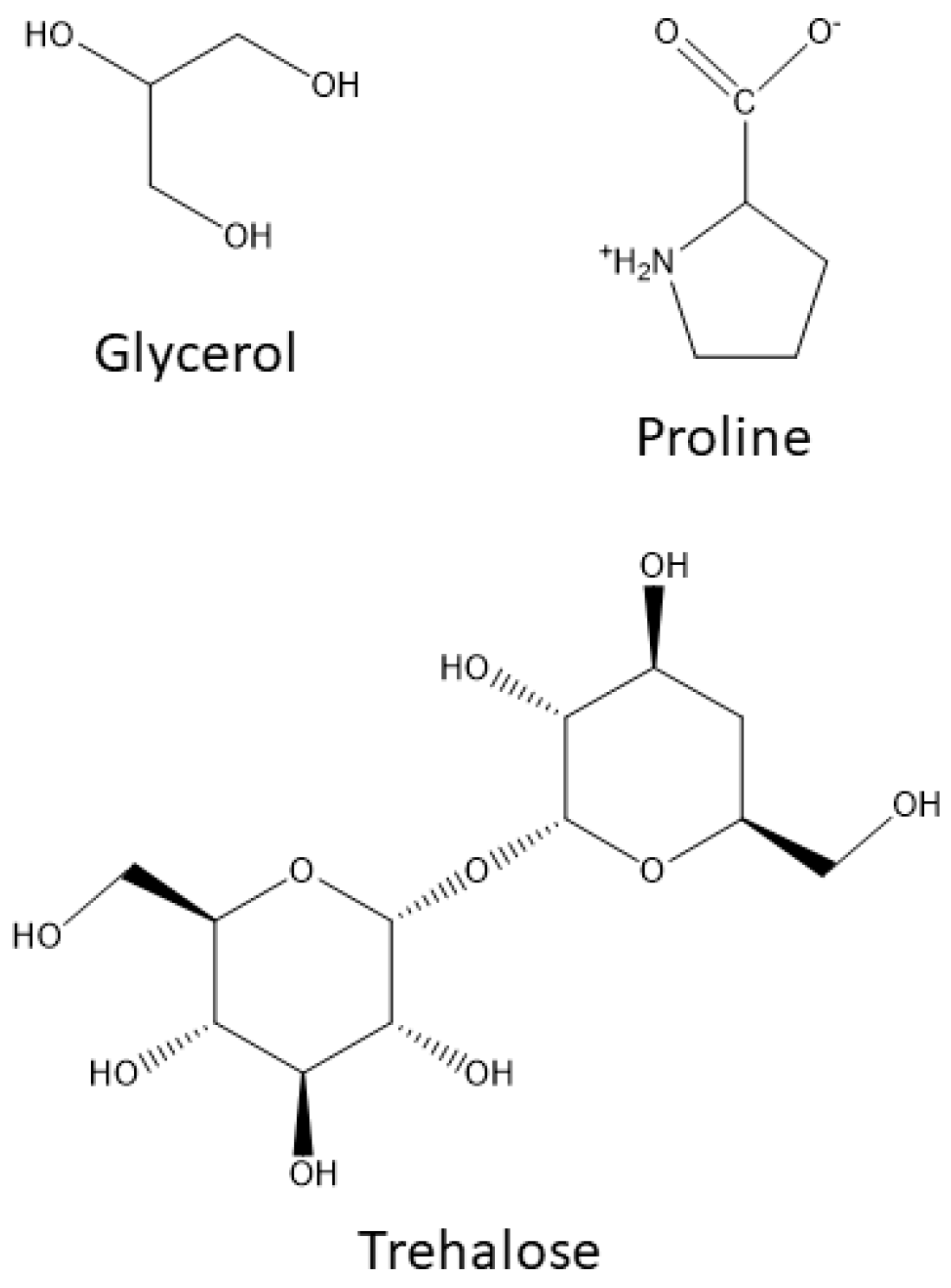
2.1. Compatible Solutes or Inert Osmolytes
2.2. The Unfolded Protein Response (UPR)
3. Yeast Stresses and Their Mitigation
3.1. Osmotic Stress
3.2. Heat Stress
3.3. Chaotrope Stress
3.4. Other Stresses
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barnett, J.A. Beginnings of microbiology and biochemistry: The contribution of yeast research. Microbiology 2003, 149, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Cray, J.A.; Bell, A.N.; Bhaganna, P.; Mswaka, A.Y.; Timson, D.J.; Hallsworth, J.E. The biology of habitat dominance; can microbes behave as weeds? Microb. Biotechnol. 2013, 6, 453–492. [Google Scholar] [CrossRef] [PubMed]
- Hagman, A.; Säll, T.; Compagno, C.; Piskur, J. Yeast “make-accumulate-consume” life strategy evolved as a multi-step process that predates the whole genome duplication. PLoS ONE 2013, 8, e68734. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.M.; Liu, P.; Fay, J.C. Evolution of ecological dominance of yeast species in high-sugar environments. Evolution 2015, 69, 2079–2093. [Google Scholar] [CrossRef] [PubMed]
- Goffeau, A.; Barrell, B.G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnston, M.; et al. Life with 6000 genes. Science 1996, 274, 546–567. [Google Scholar] [CrossRef]
- Engel, S.R.; Dietrich, F.S.; Fisk, D.G.; Binkley, G.; Balakrishnan, R.; Costanzo, M.C.; Dwight, S.S.; Hitz, B.C.; Karra, K.; Nash, R.S.; et al. The reference genome sequence of Saccharomyces cerevisiae: Then and now. G3 Genes Genomes Genet. 2014, 4, 389–398. [Google Scholar] [CrossRef]
- Cherry, J.M.; Hong, E.L.; Amundsen, C.; Balakrishnan, R.; Binkley, G.; Chan, E.T.; Christie, K.R.; Costanzo, M.C.; Dwight, S.S.; Engel, S.R.; et al. Saccharomyces Genome Database: The genomics resource of budding yeast. Nucleic Acids Res. 2012, 40, D700–D705. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, M.; Burtner, C.R.; Kennedy, B.K. Recent developments in yeast aging. PLoS Genet. 2007, 3, e84. [Google Scholar] [CrossRef] [PubMed]
- Barberis, M.; Todd, R.G.; van der Zee, L. Advances and challenges in logical modeling of cell cycle regulation: Perspective for multi-scale, integrative yeast cell models. FEMS Yeast Res. 2017, 17, fow103. [Google Scholar] [CrossRef] [PubMed]
- Petranovic, D.; Vemuri, G.N. Impact of yeast systems biology on industrial biotechnology. J. Biotechnol. 2009, 144, 204–211. [Google Scholar] [CrossRef]
- Bilsland, E.; Sparkes, A.; Williams, K.; Moss, H.J.; de Clare, M.; Pir, P.; Rowland, J.; Aubrey, W.; Pateman, R.; Young, M.; et al. Yeast-based automated high-throughput screens to identify anti-parasitic lead compounds. Open Biol. 2013, 3, 120158. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bilsland, E.; Pir, P.; Gutteridge, A.; Johns, A.; King, R.D.; Oliver, S.G. Functional expression of parasite drug targets and their human orthologs in yeast. PLoS Negl. Trop. Dis. 2011, 5, e1320. [Google Scholar] [CrossRef] [PubMed]
- Ruetenik, A.; Barrientos, A. Exploiting post-mitotic yeast cultures to model neurodegeneration. Front. Mol. Neurosci. 2018, 11, 400. [Google Scholar] [CrossRef] [PubMed]
- Lindström, M.; Liu, B. Yeast as a model to unravel mechanisms behind FUS toxicity in amyotrophic lateral sclerosis. Front. Mol. Neurosci. 2018, 11, 218. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J. Yeast systems biology: Model organism and cell factory. Biotechnol. J. 2019, 14, 1800421. [Google Scholar] [CrossRef]
- Seynnaeve, D.; Vecchio, M.D.; Fruhmann, G.; Verelst, J.; Cools, M.; Beckers, J.; Mulvihill, D.P.; Winderickx, J.; Franssens, V. Recent insights on Alzheimer’s disease originating from yeast models. Int. J. Mol. Sci. 2018, 19, 1947. [Google Scholar] [CrossRef]
- Raghavan, V.; Aquadro, C.F.; Alani, E. Baker’s Yeast Clinical Isolates Provide a Model for How Pathogenic Yeasts Adapt to Stress. Trends Genet. 2019, 35, 804–817. [Google Scholar] [CrossRef]
- Daignan-Fornier, B.; Pinson, B. Yeast to study human purine metabolism diseases. Cells 2019, 8, 67. [Google Scholar] [CrossRef]
- Cazzanelli, G.; Pereira, F.; Alves, S.; Francisco, R.; Azevedo, L.; Dias Carvalho, P.; Almeida, A.; Côrte-Real, M.; Oliveira, M.J.; Lucas, C. The yeast Saccharomyces cerevisiae as a model for understanding RAS proteins and their role in human tumorigenesis. Cells 2018, 7, 14. [Google Scholar] [CrossRef]
- Di Gregorio, S.E.; Duennwald, M.L. ALS yeast models—Past success stories and new opportunities. Front. Mol. Neurosci. 2018, 11, 394. [Google Scholar] [CrossRef]
- Macedo, N.; Brigham, C.J. From beverages to biofuels: The journeys of ethanol-producing microorganisms. Int. J. Biotechnol. Wellness Ind. 2014, 3, 79–87. [Google Scholar]
- Naghshbandi, M.P.; Tabatabaei, M.; Aghbashlo, M.; Gupta, V.K.; Sulaiman, A.; Karimi, K.; Moghimi, H.; Maleki, M. Progress toward improving ethanol production through decreased glycerol generation in Saccharomyces cerevisiae by metabolic and genetic engineering approaches. Renew. Sustain. Energy Rev. 2019, 115, 109353. [Google Scholar] [CrossRef]
- Timilsina, G.R. Biofuels in the long-run global energy supply mix for transportation. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2014, 372, 20120323. [Google Scholar] [CrossRef] [PubMed]
- Gasparatos, A.; Stromberg, P.; Takeuchi, K. Sustainability impacts of first-generation biofuels. Anim. Front. 2013, 3, 12–26. [Google Scholar] [CrossRef]
- Lee, R.A.; Lavoie, J.-M. From first-to third-generation biofuels: Challenges of producing a commodity from a biomass of increasing complexity. Anim. Front. 2013, 3, 6–11. [Google Scholar] [CrossRef]
- Das Neves, M.A.; Kimura, T.; Shimizu, N.; Nakajima, M. State of the art and future trends of bioethanol production. Dynam. Biochem. Proc. Biotechnol. Mol. Biol. 2007, 1, 1–14. [Google Scholar]
- Paulino de Souza, J.; Dias do Prado, C.; Eleutherio, E.C.A.; Bonatto, D.; Malavazi, I.; Ferreira da Cunha, A. Improvement of Brazilian bioethanol production - Challenges and perspectives on the identification and genetic modification of new strains of Saccharomyces cerevisiae yeasts isolated during ethanol process. Fungal Biol. 2018, 122, 583–591. [Google Scholar] [CrossRef]
- Banerjee, A. Food, feed, fuel: Transforming the competition for grains. Dev. Chang. 2011, 42, 529–557. [Google Scholar] [CrossRef]
- Meyer, P.M.; Rodrigues, P.H.; Millen, D.D. Impact of biofuel production in Brazil on the economy, agriculture, and the environment. Anim. Front. 2013, 3, 28–37. [Google Scholar] [CrossRef][Green Version]
- Ewing, M.; Msangi, S. Biofuels production in developing countries: Assessing tradeoffs in welfare and food security. Environ. Sci. Policy 2009, 12, 520–528. [Google Scholar] [CrossRef]
- Eisentraut, A. Sustainable Production of Second-Generation Biofuels; OECD/IEA: Paris, France, 2010.
- Cray, J.A.; Stevenson, A.; Ball, P.; Bankar, S.B.; Eleutherio, E.C.; Ezeji, T.C.; Singhal, R.S.; Thevelein, J.M.; Timson, D.J.; Hallsworth, J.E. Chaotropicity: A key factor in product tolerance of biofuel-producing microorganisms. Curr. Opin. Biotechnol. 2015, 33, 228–259. [Google Scholar] [CrossRef] [PubMed]
- Mohd Azhar, S.H.; Abdulla, R.; Jambo, S.A.; Marbawi, H.; Gansau, J.A.; Mohd Faik, A.A.; Rodrigues, K.F. Yeasts in sustainable bioethanol production: A review. Biochem. Biophys. Rep. 2017, 10, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Jönsson, L.J.; Martín, C. Pretreatment of lignocellulose: Formation of inhibitory by-products and strategies for minimizing their effects. Bioresour. Technol. 2016, 199, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Beckner, M.; Ivey, M.L.; Phister, T.G. Microbial contamination of fuel ethanol fermentations. Lett. Appl. Microbiol. 2011, 53, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Narendranath, N.; Hynes, S.; Thomas, K.; Ingledew, W. Effects of lactobacilli on yeast-catalyzed ethanol fermentations. Appl. Environ. Microbiol. 1997, 63, 4158–4163. [Google Scholar] [CrossRef] [PubMed]
- Gibson, B.R.; Lawrence, S.J.; Leclaire, J.P.; Powell, C.D.; Smart, K.A. Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol. Rev. 2007, 31, 535–569. [Google Scholar] [CrossRef]
- Maisonnave, P.; Sanchez, I.; Moine, V.; Dequin, S.; Galeote, V. Stuck fermentation: Development of a synthetic stuck wine and study of a restart procedure. Int. J. Food Microbiol. 2013, 163, 239–247. [Google Scholar] [CrossRef]
- Liu, Y.; Ishii, S.; Tokai, M.; Tsutsumi, H.; Ohki, O.; Akada, R.; Tanaka, K.; Tsuchiya, E.; Fukui, S.; Miyakawa, T. The Saccharomyces cerevisiae genes (CMP1 and CMP2) encoding calmodulin-binding proteins homologous to the catalytic subunit of mammalian protein phosphatase 2B. Mol. Gen. Genet. 1991, 227, 52–59. [Google Scholar] [CrossRef]
- Cyert, M.S.; Kunisawa, R.; Kaim, D.; Thorner, J. Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc. Natl. Acad. Sci. USA 1991, 88, 7376–7380. [Google Scholar] [CrossRef] [PubMed]
- Cyert, M.S.; Thorner, J. Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol. Cell. Biol. 1992, 12, 3460–3469. [Google Scholar] [CrossRef]
- Matheos, D.P.; Kingsbury, T.J.; Ahsan, U.S.; Cunningham, K.W. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 1997, 11, 3445–3458. [Google Scholar] [CrossRef] [PubMed]
- Schneiter, R.; Tatzer, V.; Gogg, G.; Leitner, E.; Kohlwein, S.D. Elo1p-dependent carboxy-terminal elongation of C14:1Delta(9) to C16:1Delta(11) fatty acids in Saccharomyces cerevisiae. J. Bacteriol. 2000, 182, 3655–3660. [Google Scholar] [CrossRef] [PubMed]
- Haro, R.; Garciadeblas, B.; Rodríguez-Navarro, A. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 1991, 291, 189–191. [Google Scholar] [CrossRef]
- Mori, K.; Kawahara, T.; Yoshida, H.; Yanagi, H.; Yura, T. Signalling from endoplasmic reticulum to nucleus: Transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells 1996, 1, 803–817. [Google Scholar] [CrossRef] [PubMed]
- Brewster, J.L.; de Valoir, T.; Dwyer, N.D.; Winter, E.; Gustin, M.C. An osmosensing signal transduction pathway in yeast. Science 1993, 259, 1760–1763. [Google Scholar] [CrossRef]
- Wiederrecht, G.; Seto, D.; Parker, C.S. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell 1988, 54, 841–853. [Google Scholar] [CrossRef]
- Nikawa, J.; Yamashita, S. IRE1 encodes a putative protein kinase containing a membrane-spanning domain and is required for inositol phototrophy in Saccharomyces cerevisiae. Mol. Microbiol. 1992, 6, 1441–1446. [Google Scholar] [CrossRef]
- Tokunaga, M.; Kawamura, A.; Kohno, K. Purification and characterization of BiP/Kar2 protein from Saccharomyces cerevisiae. J. Biol. Chem. 1992, 267, 17553–17559. [Google Scholar]
- Martinez-Pastor, M.T.; Marchler, G.; Schuller, C.; Marchler-Bauer, A.; Ruis, H.; Estruch, F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 1996, 15, 2227–2235. [Google Scholar] [CrossRef]
- Stukey, J.E.; McDonough, V.M.; Martin, C.E. The OLE1 gene of Saccharomyces cerevisiae encodes the delta 9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J. Biol. Chem. 1990, 265, 20144–20149. [Google Scholar]
- Serrano, R.; Kielland-Brandt, M.C.; Fink, G.R. Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+-ATPases. Nature 1986, 319, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Serrano, R. Characterization of the plasma membrane ATPase of Saccharomyces cerevisiae. Mol. Cell. Biochem. 1978, 22, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Brandriss, M.C. Proline biosynthesis in Saccharomyces cerevisiae: Molecular analysis of the PRO1 gene, which encodes gamma-glutamyl kinase. J. Bacteriol. 1992, 174, 4148–4156. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tomenchok, D.M.; Brandriss, M.C. Gene-enzyme relationships in the proline biosynthetic pathway of Saccharomyces cerevisiae. J. Bacteriol. 1987, 169, 5364–5372. [Google Scholar] [CrossRef] [PubMed]
- Brandriss, M.C.; Falvey, D.A. Proline biosynthesis in Saccharomyces cerevisiae: Analysis of the PRO3 gene, which encodes delta 1-pyrroline-5-carboxylate reductase. J. Bacteriol. 1992, 174, 3782–3788. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Watanabe, Y.; Takaesu, G.; Hagiwara, M.; Irie, K.; Matsumoto, K. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 1997, 17, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Dodou, E.; Treisman, R. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 1997, 17, 1848–1859. [Google Scholar] [CrossRef] [PubMed]
- Torres, L.; Martín, H.; García-Saez, M.I.; Arroyo, J.; Molina, M.; Sánchez, M.; Nombela, C. A protein kinase gene complements the lytic phenotype of Saccharomyces cerevisiae lyt2 mutants. Mol. Microbiol. 1991, 5, 2845–2854. [Google Scholar] [CrossRef]
- Bell, W.; Klaassen, P.; Ohnacker, M.; Boller, T.; Herweijer, M.; Schoppink, P.; Van der Zee, P.; Wiemken, A. Characterization of the 56-kDa subunit of yeast trehalose-6-phosphate synthase and cloning of its gene reveal its identity with the product of CIF1, a regulator of carbon catabolite inactivation. Eur. J. Biochem. 1992, 209, 951–959. [Google Scholar] [CrossRef]
- De Virgilio, C.; Bürckert, N.; Bell, W.; Jenö, P.; Boller, T.; Wiemken, A. Disruption of TPS2, the gene encoding the 100-kDa subunit of the trehalose-6-phosphate synthase/phosphatase complex in Saccharomyces cerevisiae, causes accumulation of trehalose-6-phosphate and loss of trehalose-6-phosphate phosphatase activity. Eur. J. Biochem. 1993, 212, 315–323. [Google Scholar] [CrossRef]
- Phizicky, E.M.; Schwartz, R.C.; Abelson, J. Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J. Biol. Chem. 1986, 261, 2978–2986. [Google Scholar] [PubMed]
- Xu, Q.; Teplow, D.; Lee, T.D.; Abelson, J. Domain structure in yeast tRNA ligase. Biochemistry 1990, 29, 6132–6138. [Google Scholar] [CrossRef] [PubMed]
- Causton, H.C.; Ren, B.; Koh, S.S.; Harbison, C.T.; Kanin, E.; Jennings, E.G.; Lee, T.I.; True, H.L.; Lander, E.S.; Young, R.A. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001, 12, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Piper, P.W. The heat shock and ethanol stress responses of yeast exhibit extensive similarity and functional overlap. FEMS Microbiol. Lett. 1995, 134, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Schüller, C.; Brewster, J.; Alexander, M.; Gustin, M.; Ruis, H. The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 1994, 13, 4382–4389. [Google Scholar] [CrossRef] [PubMed]
- Moskvina, E.; Imre, E.M.; Ruis, H. Stress factors acting at the level of the plasma membrane induce transcription via the stress response element (STRE) of the yeast Saccharomyces cerevisiae. Mol. Microbiol. 1999, 32, 1263–1272. [Google Scholar] [CrossRef]
- Moskvina, E.; Schüller, C.; Maurer, C.; Mager, W.; Ruis, H. A search in the genome of Saccharomyces cerevisiae for genes regulated via stress response elements. Yeast 1998, 14, 1041–1050. [Google Scholar] [CrossRef]
- Ding, J.; Huang, X.; Zhang, L.; Zhao, N.; Yang, D.; Zhang, K. Tolerance and stress response to ethanol in the yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2009, 85, 253. [Google Scholar] [CrossRef]
- Crawford, R.A.; Pavitt, G.D. Translational regulation in response to stress in Saccharomyces cerevisiae. Yeast 2019, 36, 5–21. [Google Scholar] [CrossRef]
- Alexandre, H.; Ansanay-Galeote, V.; Dequin, S.; Blondin, B. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett. 2001, 498, 98–103. [Google Scholar] [CrossRef]
- Hallsworth, J.E. Stress-free microbes lack vitality. Fungal Biol. 2018, 122, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Aertsen, A.; Michiels, C.W. Diversify or die: Generation of diversity in response to stress. Crit. Rev. Microbiol. 2005, 31, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Estruch, F. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev. 2000, 24, 469–486. [Google Scholar] [CrossRef] [PubMed]
- Rangel, D.E.N.; Finlay, R.D.; Hallsworth, J.E.; Dadachova, E.; Gadd, G.M. Fungal strategies for dealing with environment- and agriculture-induced stresses. Fungal Biol. 2018, 122, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.; Bandara, A.; Fraser, S.; Chambers, P.; Stanley, G.A. The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. J. Appl. Microbiol. 2010, 109, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Nakamura, T.; Ando, A.; Tokuyasu, K.; Shima, J. Genome-wide screening of the genes required for tolerance to vanillin, which is a potential inhibitor of bioethanol fermentation, in Saccharomyces cerevisiae. Biotechnol. Biofuels 2008, 1, 3. [Google Scholar] [CrossRef] [PubMed]
- Swinnen, S.; Schaerlaekens, K.; Pais, T.; Claesen, J.; Hubmann, G.; Yang, Y.; Demeke, M.; Foulquié-Moreno, M.R.; Goovaerts, A.; Souvereyns, K.; et al. Identification of novel causative genes determining the complex trait of high ethanol tolerance in yeast using pooled-segregant whole-genome sequence analysis. Genome Res. 2012, 22, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wang, M.; Tan, T.; Li, J.; Yang, H.; Leach, L.; Zhang, R.; Luo, Z. Genetic dissection of ethanol tolerance in the budding yeast Saccharomyces cerevisiae. Genetics 2007, 175, 1479–1487. [Google Scholar] [CrossRef]
- Haas, R.; Horev, G.; Lipkin, E.; Kesten, I.; Portnoy, M.; Buhnik-Rosenblau, K.; Soller, M.; Kashi, Y. Mapping Ethanol Tolerance in Budding Yeast Reveals High Genetic Variation in a Wild Isolate. Front. Genet. 2019, 10, 998. [Google Scholar] [CrossRef]
- Liu, R.; Liang, L.; Choudhury, A.; Garst, A.D.; Eckert, C.A.; Oh, E.J.; Winkler, J.; Gill, R.T. Multiplex navigation of global regulatory networks (MINR) in yeast for improved ethanol tolerance and production. Metab. Eng. 2019, 51, 50–58. [Google Scholar] [CrossRef]
- Kasavi, C.; Eraslan, S.; Arga, K.Y.; Oner, E.T.; Kirdar, B. A system based network approach to ethanol tolerance in Saccharomyces cerevisiae. BMC Syst. Biol. 2014, 8, 90. [Google Scholar] [CrossRef] [PubMed]
- Ismail, A.; Ali, A. Selection of high ethanol-yielding Saccharomyces. Folia Microbiol. 1971, 16, 350–354. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.; Oliver, S. Isolation of ethanol-tolerant mutants of yeast by continuous selection. Eur. J. Appl. Microbiol. Biotechnol. 1982, 16, 119–122. [Google Scholar] [CrossRef]
- Jiménez, J.; Benítez, T. Selection of ethanol-tolerant yeast hybrids in pH-regulated continuous culture. Appl. Environ. Microbiol. 1988, 54, 917–922. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, A.; Sindhu, R.; Arun, K.; Pandey, A.; Binod, P.; Gnansounou, E. Advances in Biofuel Production by Strain Development in Yeast from Lignocellulosic Biomass. Bioprocess. Biomol. Prod. 2019, 289–302. [Google Scholar]
- Miyagawa, K.-I.; Ishiwata-Kimata, Y.; Kohno, K.; Kimata, Y. Ethanol stress impairs protein folding in the endoplasmic reticulum and activates Ire1 in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 2014, 78, 1389–1391. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Tapia, E.; Nana, R.K.; Querol, A.; Pérez-Torrado, R. Ethanol cellular defense induce unfolded protein response in yeast. Front. Microbiol. 2016, 7, 189. [Google Scholar] [CrossRef]
- Alexandre, H.; Rousseaux, I.; Charpentier, C. Relationship between ethanol tolerance, lipid composition and plasma membrane fluidity in Saccharomyces cerevisiae and Kloeckera apiculata. FEMS Microbiol. Lett. 1994, 124, 17–22. [Google Scholar] [CrossRef]
- Vanegas, J.M.; Contreras, M.F.; Faller, R.; Longo, M.L. Role of unsaturated lipid and ergosterol in ethanol tolerance of model yeast biomembranes. Biophys. J. 2012, 102, 507–516. [Google Scholar] [CrossRef]
- Shi, D.-J.; Wang, C.-L.; Wang, K.-M. Genome shuffling to improve thermotolerance, ethanol tolerance and ethanol productivity of Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 2009, 36, 139–147. [Google Scholar] [CrossRef]
- Alper, H.; Moxley, J.; Nevoigt, E.; Fink, G.R.; Stephanopoulos, G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 2006, 314, 1565–1568. [Google Scholar] [CrossRef] [PubMed]
- Snoek, T.; Nicolino, M.P.; Van den Bremt, S.; Mertens, S.; Saels, V.; Verplaetse, A.; Steensels, J.; Verstrepen, K.J. Large-scale robot-assisted genome shuffling yields industrial Saccharomyces cerevisiae yeasts with increased ethanol tolerance. Biotechnol. Biofuels 2015, 8, 32. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Chen, D.; Su, Q.; Yuan, X.; Liu, K.; Huang, L.; Fang, J.; Chen, J.; He, W.; Chen, Y. Improved ethanol tolerance and production of Saccharomyces cerevisiae by global transcription machinery engineering via directed evolution of the SPT8 gene. Food Biotechnol. 2019, 33, 155–173. [Google Scholar] [CrossRef]
- Jetti, K.D.; GNS, R.R.; Garlapati, D.; Nammi, S.K. Improved ethanol productivity and ethanol tolerance through genome shuffling of Saccharomyces cerevisiae and Pichia stipitis. Int. Microbiol. 2019, 22, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Mitsui, R.; Yamada, R.; Ogino, H. Improved stress tolerance of Saccharomyces cerevisiae by CRISPR-Cas-mediated genome evolution. Appl. Biochem. Biotechnol. 2019, 189, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Yuan, X.; Liang, L.; Fang, J.; Chen, Y.; He, W.; Xue, T. Using CRISPR/Cas9 for multiplex genome engineering to optimize the ethanol metabolic pathway in Saccharomyces cerevisiae. Biochem. Eng. J. 2019, 145, 120–126. [Google Scholar] [CrossRef]
- Brown, A.D. Compatible solutes and extreme water stress in eukaryotic micro-organisms. Adv. Microb. Physiol. 1978, 17, 181–242. [Google Scholar]
- Hallsworth, J.E.; Prior, B.A.; Nomura, Y.; Iwahara, M.; Timmis, K.N. Compatible solutes protect against chaotrope (ethanol)-induced, nonosmotic water stress. Appl. Environ. Microbiol. 2003, 69, 7032–7034. [Google Scholar] [CrossRef]
- Cray, J.A.; Russell, J.T.; Timson, D.J.; Singhal, R.S.; Hallsworth, J.E. A universal measure of chaotropicity and kosmotropicity. Environ. Microbiol. 2013, 15, 287–296. [Google Scholar] [CrossRef]
- Timson, D.J. The roles and applications of chaotropes and kosmotropes in industrial fermentation processes. World J. Microbiol. Biotechnol. 2020, 36, 89. [Google Scholar] [CrossRef]
- McCammick, E.M.; Gomase, V.S.; McGenity, T.J.; Timson, D.J.; Hallsworth, J.E. Water-Hydrophobic Compound Interactions with the Microbial Cell. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1451–1466. [Google Scholar] [CrossRef]
- Singer, M.A.; Lindquist, S. Multiple Effects of Trehalose on Protein Folding In Vitro and In Vivo. Mol. Cell 1998, 1, 639–648. [Google Scholar] [CrossRef]
- Lever, M.; Blunt, J.; Maclagan, R. Some ways of looking at compensatory kosmotropes and different water environments. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2001, 130, 471–486. [Google Scholar] [CrossRef]
- Moelbert, S.; Normand, B.; De Los Rios, P. Kosmotropes and chaotropes: Modelling preferential exclusion, binding and aggregate stability. Biophys. Chem. 2004, 112, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002, 66, 300–372. [Google Scholar] [CrossRef] [PubMed]
- Nevoigt, E.; Stahl, U. Osmoregulation and glycerol metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 1997, 21, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Mager, W.H.; de Boer, A.H.; Siderius, M.H.; Voss, H.-P. Cellular responses to oxidative and osmotic stress. Cell Stress Chaperones 2000, 5, 73. [Google Scholar] [CrossRef]
- Takagi, H. Proline as a stress protectant in yeast: Physiological functions, metabolic regulations, and biotechnological applications. Appl. Microbiol. Biotechnol. 2008, 81, 211–223. [Google Scholar] [CrossRef]
- Francois, J.; Parrou, J.L. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2001, 25, 125–145. [Google Scholar] [CrossRef]
- Winderickx, J.; de Winde, J.H.; Crauwels, M.; Hino, A.; Hohmann, S.; Van Dijck, P.; Thevelein, J.M. Regulation of genes encoding subunits of the trehalose synthase complex in Saccharomyces cerevisiae: Novel variations of STRE-mediated transcription control? Mol. Gen. Genet. 1996, 252, 470–482. [Google Scholar] [CrossRef]
- Gorner, W.; Durchschlag, E.; Martinez-Pastor, M.T.; Estruch, F.; Ammerer, G.; Hamilton, B.; Ruis, H.; Schuller, C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes. Dev. 1998, 12, 586–597. [Google Scholar] [CrossRef]
- Conlin, L.K.; Nelson, H.C.M. The Natural Osmolyte Trehalose Is a Positive Regulator of the Heat-Induced Activity of Yeast Heat Shock Transcription Factor. Mol. Cell. Biol. 2007, 27, 1505–1515. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ng, B.S.; Thibault, G. Endoplasmic reticulum stress response in yeast and humans. Biosci. Rep. 2014, 34. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.E. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005, 69, 262–291. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, T.N.; Sidrauski, C.; Dörfler, S.; Walter, P. Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J. 1999, 18, 3119–3132. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.S.; Shamu, C.E.; Walter, P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 1993, 73, 1197–1206. [Google Scholar] [CrossRef]
- Kimata, Y.; Kimata, Y.I.; Shimizu, Y.; Abe, H.; Farcasanu, I.C.; Takeuchi, M.; Rose, M.D.; Kohno, K. Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol. Biol. Cell 2003, 14, 2559–2569. [Google Scholar] [CrossRef]
- Okamura, K.; Kimata, Y.; Higashio, H.; Tsuru, A.; Kohno, K. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem. Biophys. Res. Commun. 2000, 279, 445–450. [Google Scholar] [CrossRef]
- Sidrauski, C.; Walter, P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 1997, 90, 1031–1039. [Google Scholar] [CrossRef]
- Gardner, B.M.; Walter, P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science 2011, 333, 1891–1894. [Google Scholar] [CrossRef]
- Ho, N.; Yap, W.S.; Xu, J.; Wu, H.; Koh, J.H.; Goh, W.W.B.; George, B.; Chong, S.C.; Taubert, S.; Thibault, G. Stress sensor Ire1 deploys a divergent transcriptional program in response to lipid bilayer stress. J. Cell Biol. 2020, 219. [Google Scholar] [CrossRef]
- Sidrauski, C.; Cox, J.S.; Walter, P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 1996, 87, 405–413. [Google Scholar] [CrossRef]
- Mori, K.; Ogawa, N.; Kawahara, T.; Yanagi, H.; Yura, T. Palindrome with spacer of one nucleotide is characteristic of the cis-acting unfolded protein response element in Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 9912–9920. [Google Scholar] [CrossRef] [PubMed]
- Welihinda, A.A.; Tirasophon, W.; Green, S.R.; Kaufman, R.J. Gene induction in response to unfolded protein in the endoplasmic reticulum is mediated through Ire1p kinase interaction with a transcriptional coactivator complex containing Ada5p. Proc. Natl. Acad. Sci. USA 1997, 94, 4289–4294. [Google Scholar] [CrossRef] [PubMed]
- Moir, R.D.; Gross, D.A.; Silver, D.L.; Willis, I.M. SCS3 and YFT2 link transcription of phospholipid biosynthetic genes to ER stress and the UPR. PLoS Genet. 2012, 8, e1002890. [Google Scholar] [CrossRef] [PubMed]
- Travers, K.J.; Patil, C.K.; Wodicka, L.; Lockhart, D.J.; Weissman, J.S.; Walter, P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 2000, 101, 249–258. [Google Scholar] [CrossRef]
- Thibault, G.; Ismail, N.; Ng, D.T. The unfolded protein response supports cellular robustness as a broad-spectrum compensatory pathway. Proc. Natl. Acad. Sci. USA 2011, 108, 20597–20602. [Google Scholar] [CrossRef]
- Fun, X.H.; Thibault, G. Lipid bilayer stress and proteotoxic stress-induced unfolded protein response deploy divergent transcriptional and non-transcriptional programmes. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2020, 1865. [Google Scholar] [CrossRef]
- Krysan, D.J. The cell wall and endoplasmic reticulum stress responses are coordinately regulated in Saccharomyces cerevisiae. Commun. Integr. Biol. 2009, 2, 233–235. [Google Scholar] [CrossRef]
- Chowdhury, S.; Smith, K.W.; Gustin, M.C. Osmotic stress and the yeast cytoskeleton: Phenotype-specific suppression of an actin mutation. J. Cell Biol. 1992, 118, 561–571. [Google Scholar] [CrossRef]
- Mas, G.; De Nadal, E.; Dechant, R.; De La Concepción, M.L.R.; Logie, C.; Jimeno-González, S.; Chávez, S.; Ammerer, G.; Posas, F. Recruitment of a chromatin remodelling complex by the Hog1 MAP kinase to stress genes. EMBO J. 2009, 28, 326–336. [Google Scholar] [CrossRef]
- Silva, R.D.; Sotoca, R.; Johansson, B.; Ludovico, P.; Sansonetty, F.; Silva, M.T.; Peinado, J.M.; Côrte-Real, M. Hyperosmotic stress induces metacaspase-and mitochondria-dependent apoptosis in Saccharomyces cerevisiae. Mol. Microbiol. 2005, 58, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Peña, J.M.; García, R.; Nombela, C.; Arroyo, J. The high-osmolarity glycerol (HOG) and cell wall integrity (CWI) signalling pathways interplay: A yeast dialogue between MAPK routes. Yeast 2010, 27, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Jendretzki, A.; Wittland, J.; Wilk, S.; Straede, A.; Heinisch, J.J. How do I begin? Sensing extracellular stress to maintain yeast cell wall integrity. Eur. J. Cell Biol. 2011, 90, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Sanz, A.B.; García, R.; Rodríguez-Peña, J.M.; Arroyo, J. The CWI pathway: Regulation of the transcriptional adaptive response to cell wall stress in yeast. J. Fungi 2018, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef]
- Baetz, K.; Moffat, J.; Haynes, J.; Chang, M.; Andrews, B. Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4. Mol. Cell. Biol. 2001, 21, 6515–6528. [Google Scholar] [CrossRef]
- Kim, K.-Y.; Truman, A.W.; Levin, D.E. Yeast Mpk1 mitogen-activated protein kinase activates transcription through Swi4/Swi6 by a noncatalytic mechanism that requires upstream signal. Mol. Cell. Biol. 2008, 28, 2579–2589. [Google Scholar] [CrossRef]
- Kim, K.-Y.; Truman, A.W.; Caesar, S.; Schlenstedt, G.; Levin, D.E. Yeast Mpk1 cell wall integrity mitogen-activated protein kinase regulates nucleocytoplasmic shuttling of the Swi6 transcriptional regulator. Mol. Biol. Cell 2010, 21, 1609–1619. [Google Scholar] [CrossRef]
- Jung, U.S.; Levin, D.E. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 1999, 34, 1049–1057. [Google Scholar] [CrossRef]
- Lagorce, A.; Hauser, N.C.; Labourdette, D.; Rodriguez, C.; Martin-Yken, H.; Arroyo, J.; Hoheisel, J.D.; François, J. Genome-wide analysis of the response to cell wall mutations in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2003, 278, 20345–20357. [Google Scholar] [CrossRef]
- O’Rourke, S.M.; Herskowitz, I.; O’Shea, E.K. Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 2002, 18, 405–412. [Google Scholar] [CrossRef]
- Posas, F.; Chambers, J.R.; Heyman, J.A.; Hoeffler, J.P.; de Nadal, E.; Ariño, J.n. The transcriptional response of yeast to saline stress. J. Biol. Chem. 2000, 275, 17249–17255. [Google Scholar] [CrossRef] [PubMed]
- Rep, M.; Krantz, M.; Thevelein, J.M.; Hohmann, S. The transcriptional response of Saccharomyces cerevisiae to osmotic shock Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J. Biol. Chem. 2000, 275, 8290–8300. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.R.; Tyers, M.; Perret, M.; Craig, B.M.; Fang, K.S.; Gustin, M.C. Regulation of cell cycle progression by Swe1p and Hog1p following hypertonic stress. Mol. Biol. Cell 2001, 12, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Beese, S.E.; Negishi, T.; Levin, D.E. Identification of positive regulators of the yeast fps1 glycerol channel. PLoS Genet 2009, 5, e1000738. [Google Scholar] [CrossRef]
- Lee, J.; Reiter, W.; Dohnal, I.; Gregori, C.; Beese-Sims, S.; Kuchler, K.; Ammerer, G.; Levin, D.E. MAPK Hog1 closes the S. cerevisiae glycerol channel Fps1 by phosphorylating and displacing its positive regulators. Genes Dev. 2013, 27, 2590–2601. [Google Scholar] [CrossRef] [PubMed]
- Auesukaree, C. Molecular mechanisms of the yeast adaptive response and tolerance to stresses encountered during ethanol fermentation. J. Biosci. Bioeng. 2017, 124, 133–142. [Google Scholar] [CrossRef]
- Fleet, G.M. Yeasts-growth during fermentation. In Wine Microbiology & Biotechnology; Harword Academic Publishers: Amsterdam, The Netherlands, 1993; pp. 27–54. [Google Scholar]
- Van Uden, N. Effect of alcohols on the temperature relations of growth and death in yeasts. In Alcohol Toxicity in Yeast and Bacteria; CRC Press: Boca Raton, FL, USA, 1989; pp. 77–88. [Google Scholar]
- Verghese, J.; Abrams, J.; Wang, Y.; Morano, K.A. Biology of the heat shock response and protein chaperones: Budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol. Mol. Biol. Rev. 2012, 76, 115–158. [Google Scholar] [CrossRef]
- Swan, T.M.; Watson, K. Stress tolerance in a yeast lipid mutant: Membrane lipids influence tolerance to heat and ethanol independently of heat shock proteins and trehalose. Can. J. Microbiol. 1999, 45, 472–479. [Google Scholar] [CrossRef]
- Swan, T.M.; Watson, K. Stress tolerance in a yeast sterol auxotroph: Role of ergosterol, heat shock proteins and trehalose. FEMS Microbiol. Lett. 1998, 169, 191–197. [Google Scholar] [CrossRef]
- Endress, E.; Bayerl, S.; Prechtel, K.; Maier, C.; Merkel, R.; Bayerl, T.M. The effect of cholesterol, lanosterol, and ergosterol on lecithin bilayer mechanical properties at molecular and microscopic dimensions: A solid-state NMR and micropipet study. Langmuir 2002, 18, 3293–3299. [Google Scholar] [CrossRef]
- Hsueh, Y.-W.; Gilbert, K.; Trandum, C.; Zuckermann, M.; Thewalt, J. The Effect of Ergosterol on Dipalmitoylphosphatidylcholine Bilayers: A Deuterium NMR and Calorimetric Study. Biophys. J. 2005, 88, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Bittman, R.; Duportail, G.; Heissler, D.; Vilcheze, C.; London, E. Effect of the structure of natural sterols and sphingolipids on the formation of ordered sphingolipid/sterol domains (rafts) Comparison of cholesterol to plant, fungal, and disease-associated sterols and comparison of sphingomyelin, cerebrosides, and ceramide. J. Biol. Chem. 2001, 276, 33540–33546. [Google Scholar] [PubMed]
- Caspeta, L.; Chen, Y.; Ghiaci, P.; Feizi, A.; Buskov, S.; Hallstrom, B.M.; Petranovic, D.; Nielsen, J. Biofuels. Altered sterol composition renders yeast thermotolerant. Science 2014, 346, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Fattah, W.; Fadil, M.; Nigam, P.; Banat, I. Isolation of thermotolerant ethanologenic yeasts and use of selected strains in industrial scale fermentation in an Egyptian distillery. Biotechnol. Bioeng. 2000, 68, 531–535. [Google Scholar] [CrossRef]
- Rajoka, M.; Ferhan, M.; Khalid, A. Kinetics and thermodynamics of ethanol production by a thermotolerant mutant of Saccharomyces cerevisiae in a microprocessor-controlled bioreactor. Lett. Appl. Microbiol. 2005, 40, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Banat, I.; Marchant, R. Characterization and potential industrial applications of five novel, thermotolerant, fermentative, yeast strains. World J. Microbiol. Biotechnol. 1995, 11, 304–306. [Google Scholar] [CrossRef]
- Piper, P.W.; Talreja, K.; Panaretou, B.; Moradas-Ferreira, P.; Byrne, K.; Praekelt, U.M.; Meacock, P.; Récnacq, M.; Boucherie, H. Induction of major heat-shock proteins of Saccharomyces cerevisiae, including plasma membrane Hsp30, by ethanol levels above a critical threshold. Microbiology 1994, 140, 3031–3038. [Google Scholar] [CrossRef]
- Benjaphokee, S.; Hasegawa, D.; Yokota, D.; Asvarak, T.; Auesukaree, C.; Sugiyama, M.; Kaneko, Y.; Boonchird, C.; Harashima, S. Highly efficient bioethanol production by a Saccharomyces cerevisiae strain with multiple stress tolerance to high temperature, acid and ethanol. New Biotechnol. 2012, 29, 379–386. [Google Scholar] [CrossRef]
- Birch, R.M.; Walker, G.M. Influence of magnesium ions on heat shock and ethanol stress responses of Saccharomyces cerevisiae. Enzyme Microb. Technol. 2000, 26, 678–687. [Google Scholar] [CrossRef]
- Ball, P.; Hallsworth, J.E. Water structure and chaotropicity: Their uses, abuses and biological implications. Phys. Chem. Chem. Phys. 2015, 17, 8297–8305. [Google Scholar] [CrossRef] [PubMed]
- Abu-Hamdiyyah, M. The Effect of Urea on the Structure of Water and Hydrophobic Bonding1. J. Phys. Chem. 1965, 69, 2720–2725. [Google Scholar] [CrossRef]
- Sahle, C.J.; Schroer, M.A.; Juurinen, I.; Niskanen, J. Influence of TMAO and urea on the structure of water studied by inelastic X-ray scattering. Phys. Chem. Chem. Phys. 2016, 18, 16518–16526. [Google Scholar] [CrossRef] [PubMed]
- Timson, D.J. Four Challenges for Better Biocatalysts. Fermentation 2019, 5, 39. [Google Scholar] [CrossRef]
- Geiler-Samerotte, K.A.; Dion, M.F.; Budnik, B.A.; Wang, S.M.; Hartl, D.L.; Drummond, D.A. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Telini, B.P.; Menoncin, M.; Bonatto, D. Does Inter-Organellar Proteostasis Impact Yeast Quality and Performance During Beer Fermentation? Front. Genet. 2020, 11, 2. [Google Scholar] [CrossRef]
- Tran, D.M.; Ishiwata-Kimata, Y.; Mai, T.C.; Kubo, M.; Kimata, Y. The unfolded protein response alongside the diauxic shift of yeast cells and its involvement in mitochondria enlargement. Sci. Rep. 2019, 9, 12780. [Google Scholar] [CrossRef] [PubMed]
- Hallsworth, J.E. Ethanol-induced water stress in yeast. J. Ferment. Bioeng. 1998, 85, 125–137. [Google Scholar] [CrossRef]
- De Klerk, C.; Fosso-Kankeu, E.; Du Plessis, L.; Marx, S. Assessment of the viability of Saccharomyces cerevisiae in response to synergetic inhibition during bioethanol production. Curr. Sci. 2018, 115. [Google Scholar] [CrossRef]
- Fosso-Kankeu, E.; Marx, S.; Meyer, A. Simulated inhibitory effects of typical byproducts of biomass pretreatment process on the viability of Saccharomyces cerevisiae and bioethanol production yield. Afr. J. Biotechnol. 2015, 14, 2383–2394. [Google Scholar]
- Eardley, J.; Dedi, C.; Dymond, M.; Hallsworth, J.E.; Timson, D.J. Evidence for chaotropicity/kosmotropicity offset in a yeast growth model. Biotechnol. Lett. 2019, 41, 1309–1318. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, T.; Timasheff, S.N. The stabilization of proteins by osmolytes. Biophys. J. 1985, 47, 411–414. [Google Scholar] [CrossRef]
- Gekko, K.; Timasheff, S.N. Thermodynamic and kinetic examination of protein stabilization by glycerol. Biochemistry 1981, 20, 4677–4686. [Google Scholar] [CrossRef] [PubMed]
- Gekko, K. Calorimetric Study on Thermal Denaturation of Lysozyme in Polyol-Water Mixtures. J. Biochem. 1982, 91, 1197–1204. [Google Scholar] [CrossRef] [PubMed]
- Timson, D.J.; Eardley, J. Destressing yeast for higher biofuel yields: Can Excess Chaotropicity Be Mitigated? Appl. Biochem. Biotechnol. 2020, 192, 1368–1375. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, D.J. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 1998, 14, 1511–1527. [Google Scholar] [CrossRef]
- Garre, E.; Raginel, F.; Palacios, A.; Julien, A.; Matallana, E. Oxidative stress responses and lipid peroxidation damage are induced during dehydration in the production of dry active wine yeasts. Int. J. Food Microbiol. 2010, 136, 295–303. [Google Scholar] [CrossRef]
- Ikner, A.; Shiozaki, K. Yeast signaling pathways in the oxidative stress response. Mutat. Res./Fundam. Mol. Mech. Mutagenesis 2005, 569, 13–27. [Google Scholar] [CrossRef]
- Moradas-Ferreira, P.; Costa, V.; Piper, P.; Mager, W. The molecular defences against reactive oxygen species in yeast. Mol. Microbiol. 1996, 19, 651–658. [Google Scholar] [CrossRef]
- Girard, P.; Boiteux, S. Repair of oxidized DNA bases in the yeast Saccharomyces cerevisiae. Biochimie 1997, 79, 559–566. [Google Scholar] [CrossRef]
- Pan, X.; Ye, P.; Yuan, D.S.; Wang, X.; Bader, J.S.; Boeke, J.D. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 2006, 124, 1069–1081. [Google Scholar] [CrossRef] [PubMed]
- Herrero, E.; Ros, J.; Bellí, G.; Cabiscol, E. Redox control and oxidative stress in yeast cells. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2008, 1780, 1217–1235. [Google Scholar] [CrossRef] [PubMed]
- Ariño, J. Integrative responses to high pH stress in S. cerevisiae. Omics 2010, 14, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Serra-Cardona, A.; Canadell, D.; Ariño, J. Coordinate responses to alkaline pH stress in budding yeast. Microb. Cell 2015, 2, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Viladevall, L.; Serrano, R.; Ruiz, A.; Domenech, G.; Giraldo, J.; Barcelo, A.; Arino, J. Characterization of the calcium-mediated response to alkaline stress in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 43614–43624. [Google Scholar] [CrossRef]
- Stathopoulos, A.M.; Cyert, M.S. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes. Dev. 1997, 11, 3432–3444. [Google Scholar] [CrossRef]
- Casado, C.; González, A.; Platara, M.; Ruiz, A.; Ariño, J. The role of the protein kinase A pathway in the response to alkaline pH stress in yeast. Biochem. J. 2011, 438, 523–533. [Google Scholar] [CrossRef]
- Canadell, D.; García-Martínez, J.; Alepuz, P.; Pérez-Ortín, J.E.; Ariño, J. Impact of high pH stress on yeast gene expression: A comprehensive analysis of mRNA turnover during stress responses. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2015, 1849, 653–664. [Google Scholar] [CrossRef]
- Ariño, J.; Ramos, J.; Sychrova, H. Monovalent cation transporters at the plasma membrane in yeasts. Yeast 2019, 36, 177–193. [Google Scholar] [CrossRef]
- Platara, M.; Ruiz, A.; Serrano, R.; Palomino, A.; Moreno, F.; Ariño, J. The Transcriptional Response of the Yeast Na+-ATPase ENA1 Gene to Alkaline Stress Involves Three Main Signaling Pathways. J. Biol. Chem. 2006, 281, 36632–36642. [Google Scholar] [CrossRef]
- Benito, B.; Quintero, F.J.; Rodríguez-Navarro, A. Overexpression of the sodium ATPase of Saccharomyces cerevisiae: Conditions for phosphorylation from ATP and Pi. Biochim. Biophys. Acta (BBA)-Biomembr. 1997, 1328, 214–225. [Google Scholar] [CrossRef]
- Giannattasio, S.; Guaragnella, N.; Corte-Real, M.; Passarella, S.; Marra, E. Acid stress adaptation protects Saccharomyces cerevisiae from acetic acid-induced programmed cell death. Gene 2005, 354, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Tenreiro, S.; Nunes, P.c.A.; Viegas, C.A.; Neves, M.S.; Teixeira, M.C.; Cabral, M.G.; Sá-Correia, I. AQR1 gene (ORF YNL065w) encodes a plasma membrane transporter of the major facilitator superfamily that confers resistance to short-chain monocarboxylic acids and quinidine in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2002, 292, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Tenreiro, S.; Rosa, P.C.; Viegas, C.A.; Sá-Correia, I. Expression of the AZR1 gene (ORF YGR224w), encoding a plasma membrane transporter of the major facilitator superfamily, is required for adaptation to acetic acid and resistance to azoles in Saccharomyces cerevisiae. Yeast 2000, 16, 1469–1481. [Google Scholar] [CrossRef]
- Carmelo, V.; Santos, H.; Sá-Correia, I. Effect of extracellular acidification on the activity of plasma membrane ATPase and on the cytosolic and vacuolar pH of Saccharomyces cerevisiae. Biochim. Biophys. Acta (BBA)-Biomembr. 1997, 1325, 63–70. [Google Scholar] [CrossRef][Green Version]

| Protein | Systematic Gene Name | Function | Role in Stress | References |
|---|---|---|---|---|
| Cmp2p | YML057W | Calcineurin catalytic subunit | Involved in sensing alkaline stress | [39,40] |
| Cna1p | YLR433C | Calcineurin catalytic subunit | Involved in sensing alkaline stress | [39,40] |
| Cnb1p | YKL190W | Calcineurin regulatory subunit | Involved in sensing alkaline stress | [41] |
| Crz1p | YNL027W | Transcription factor | Involved in responding to alkaline stress | [42] |
| Elo1p | YJL196C | Medium-chain fatty acyl elongase | Enables production of longer fatty acids in heat and chaotrope stress | [43] |
| Ena1p | YDR040C | Sodium ion pump | Involved in responding to alkaline stress | [44] |
| Hac1p | YFL031W | Transcription factor | Activates genes involved in the unfolded protein response | [45] |
| Hog1p | YLR113W | MAP kinase | Terminal kinase of the HOG pathway. Activates genes in response to osmotic and other stresses | [46] |
| Hsf1p | YGL073W | Transcription factor | Activates genes involved in the heat shock response | [47] |
| Ire1p | YHR079C | Protein kinase and nuclease | Cleaves HAC1 RNA making it translationally competent | [48] |
| Kar2p | YJL034W | Chaperone | Detects and responds to the presence of unfolded proteins in the endoplasmic reticulum | [49] |
| Msn2p | YMR037C | Transcription factor | Activates STRE responsive genes | [50] |
| Msn4p | YKL062W | Transcription factor | Activates STRE responsive genes | [50] |
| Ole1p | YGL055W | Δ9 fatty acid desaturase | Enables production of saturated fatty acids in heat and chaotrope stress | [51] |
| Pma1p | YGL008C | Hydrogen ion pump | Pumps protons into the vacuole and extracellular medium in acid stress | [52,53] |
| Pro1p | YDR300C | γ-glutamyl kinase | Enables synthesis of proline which protects against heat and chaotrope stress | [54] |
| Pro2p | YOR323C | γ-glutamyl phosphate reductase | Enables synthesis of proline which protects against heat and chaotrope stress | [55] |
| Pro3p | YER023W | Δ1-pyrroline-5-carboxylate reductase | Enables synthesis of proline which protects against heat and chaotrope stress | [55,56] |
| Rlm1p | YPL089C | Transcription factor | Activated by Slt2p. Controls expression of genes involved in cell wall maintenance and strengthening | [57,58] |
| Slt2p | YHR030C | MAP kinase | Terminal kinase of the CWI pathway. Activates genes in response to cell wall stress | [59] |
| Tps1p | YBR126C | Trehalose-6-phosphate synthase | Enables synthesis of trehalose in heat and chaotrope stress | [60] |
| Tps2p | YDR074W | Trehalose-6-phosphate phosphatase | Enables synthesis of trehalose in heat and chaotrope stress | [61] |
| Trl1p | YJL087C | RNA ligase | Ligates HAC1 following cleavage by Ire1p | [62,63] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eardley, J.; Timson, D.J. Yeast Cellular Stress: Impacts on Bioethanol Production. Fermentation 2020, 6, 109. https://doi.org/10.3390/fermentation6040109
Eardley J, Timson DJ. Yeast Cellular Stress: Impacts on Bioethanol Production. Fermentation. 2020; 6(4):109. https://doi.org/10.3390/fermentation6040109
Chicago/Turabian StyleEardley, Joshua, and David J. Timson. 2020. "Yeast Cellular Stress: Impacts on Bioethanol Production" Fermentation 6, no. 4: 109. https://doi.org/10.3390/fermentation6040109
APA StyleEardley, J., & Timson, D. J. (2020). Yeast Cellular Stress: Impacts on Bioethanol Production. Fermentation, 6(4), 109. https://doi.org/10.3390/fermentation6040109






