Abstract
Pectinases are the group of enzymes that catalyze the degradation of pectic substances. It has wide applications in food industries for the production and clarification of wines and juices. The aim of this study was to isolate, screen and characterize pectinase from fungi isolated from various soil samples and evaluate its application in juice clarification. Fungal strains were isolated and screened primarily using 1% citruspectin incorporated potato dextrose agar (PDA) and secondarily using pectinase screening agar medium (PSAM) for pectinolytic organisms. The enzyme was produced by submerged state fermentation and assayed using the dinitro salicylic acid (DNS) method. From 20 different soil samples, 55 fungal isolates were screened primarily and, among them, only 14 isolates were subjected for secondary screening. Out of 14, only four strains showed the highest pectinolytic activity. Among four strains, Aspergillus spp. Gm showed the highest enzyme production at a 48-h incubation period, 1% substrate concentration, and 30 °C temperature. The thermal stability assessment resulted that the activity of pectinase enzyme declines by 50% within 10 min of heating at 60 °C. The optimum temperature, pH, and substrate concentration for the activity of enzyme was 30 °C (75.4 U/mL), 5.8 (72.3 U/mL), and 0.5% (112.0 U/mL), respectively. Furthermore, the yield of the orange juice, the total soluble solid (TSS), and clarity (% transmittance) was increased as the concentration of the pectinase increased, indicating its potential use in juice processing. Overall, the strain Aspergillus spp. Gm was identified as a potent strain for pectinase production in commercial scale.
1. Introduction
Pectin is an important component of the middle lamella and primary cell wall of higher plants. Pectins are high molecular weight acidic heteropolysaccharide primarily made up of α (1−4) linked d-galacturonic acid residues [1].Three major pectic polysaccharides groups are recognized, all containing d-galacturonic acid to a greater or a lesser extent. They are homogalacturonan (HG), rhamnogalacturonan I (RGI), and rhamnogalacturonan II (RGII) [2,3,4].
Pectinases are a group of enzymes that degrade pectic substance and are classified according to their mechanism of action. For example, methylesterases remove methoxy groups from highly or partially esterified galacturonan. Polygalacturonases catalyze the hydrolysis of the glycosidic bonds in a random fashion (endopolygalacturonase) or from the nonreducing end of homogalacturonan releasing galacturonic or digalacturunic acid residues (exopoly-glacturonases) [5,6]. Pectinolytic enzymes, or pectinases, are also classified according to their mode of action and their substrate: polygalacturonases, which are sub classified as endo-polygalacturonases (E.C. 3.2.1.15) and exo-polygalacturonases (E.C. 3.2.1.67); lyases, which are sub classified into pectatelyases (E.C. 4.2.2.9 and EC. 4.2.2.2) or pectin lyases (E.C. 4.2.2.10); and pectin methylesterases (E.C. 3.1.1.11). It is recommended to use a combination of different kinds of pectinases, along with other enzymes such as cellulases and hemicelullases, as multiple enzymes can degrade different parts of the polymer, resulting in the maximal degradation of the pectin in various raw materials such as in citrus juice processing [7,8].Studies have reported that pectinase of microbial origin accounts for 25% of global food and industrial enzymes sale and their market is increasing continuously [9]. Additionally, enzymes comprise a well-established global market projected to reach USD 6.3 billion in 2021 [7]. Microorganisms including fungi are promising sources of enzymes. Fungi produces numerous extracellular enzymes that possess a special effect in the decomposition of organic matter. These include pectinolytic enzymes which are excreted to break down the middle lamella in plants so that it can insert fungal hyphae and extract nutrients from the plant [10,11]. In addition to fungi, pectinolytic enzymes are naturally produced by many other organisms like bacteria, insects, nematodes, and protozoans [12]. For the commercial production of pectinases, Aspergillus spp., Erwinia spp., Bacillus spp., and Penicillium spp. have been extensively used [9,13].
Pectinases have crucial roles in food industries. These enzymes are useful for fruit juice extraction and wine clarification; tea, cocoa, and coffee concentration and fermentation; vegetable oil extraction; preparation of jam and jellies; and pickling [14,15]. Furthermore, these enzymes are used in paper and pulp industries, bleaching of paper, bio-scouring of cotton, retting and degumming of plant fibers, oil extraction, wastewater treatment, poultry feed additives, protoplast fusion technology, and bioenergy production [10,15].
Enzyme breakdown of the biomolecules depends upon the type of microorganisms, fermentation condition such as pH, incubation time or cultivation time, carbon and nitrogen source, types and concentration of substrate, temperature, agitation, and use of different enzyme preparations [16,17]. The application of new enzymes with desirable biochemical and physicochemical characteristics and low-cost production in commercial processes has always been regarded as essential research. Keeping all the advantages into consideration, the objectives of this study were (1) to isolate and screen pectinase-producing fungi from the soil samples, (2) to optimize different parameters for maximum enzyme production and evaluate the enzyme activity with various parameters, and (3) to evaluate its potentiality in juice clarification.
2. Materials and Methods
2.1. Soil Sampling
Soil samples were collected from various regions of Nepal, geographically located at Gulmi (GPS location: 28°03′60.00″ N 83°14′60.00″ E), Manang (GPS location: 28°33′7″ N84°14′27″ E), and Kathmandu (GPS location: 27°42’6.08″N 85°19′14.16″E). These sampling sites varied in altitude ranging from 1700 to 5416 m and were rich in spoiled fruits, agro-industrial wastes, fruit pulp, composts, decaying leaves, and organic fertilizers. Altogether, 20 soil samples were collected from the depth of 20 cm into sterile zip-lock bags and transported to the laboratory for analysis.
2.2. Isolation, Screening, and Identification of Pectinolytic Fungi
Potato dextrose agar (PDA, Himedia) incorporated with 1% citrus pectin was used for the isolation and screening of pectinolytic fungi from the soil samples. For isolation, a 5gm soil sample was transferred to 45 mL of distilled water (10−1) serially diluted up to 10−6. From dilutions, 0.1 mL was inoculated by spread plate method in the prepared agar plates and incubated at 25 °C for 3–5 days. Different colonies were selected and subcultured onto the two different PDA plates containing pectin for duplication and incubated at 25°C for 3–5 days.Fungal colonies with distinct morphology were selected and subcultured repeatedly to obtain pure culture by point inoculation. For primary screening, 1% (w/v) cetyltrimethylammonium bromide (CTAB) was flooded on one agar plate containing selected fungal colonies and incubated at 25 °C for 1 h. The zone of hydrolysis around the colonies indicated the pectinolytic activity of fungi and the colonies were preserved for further study.The primary screened fungal isolates were inoculated on pectinase screening agar medium (PSAM) and incubated at 25 °C for 42 h. The PSAM contains (in g L−1): (NH4)2HPO4, 3.0; KH2PO4, 2.0; K2HPO4, 3.0; MgSO4, 0.1, pectin, 10.0; and agar, 25.0. The pH of the media was adjusted to 4.5. After incubation, these plates were flooded with CTAB for secondary screening [4,12]. Furthermore, fungal strains were identified by using lactophenol cotton blue stain method to observe their morphology, hyphal characteristics, presence or absence of asexual spores, the arrangement of conidia, and the reproductive structuresunder the bright field microscope under 40X magnification [18,19].
2.3. Enzyme Production
2.3.1. Submerged Fermentation for Enzyme Production
Submerged fermentation (SmF) was carried out for the enzyme production. In a conical flask, 50 mL of Hankin’s broth was prepared, sterilized and cooled as described previously [20]. Then, 1 mL pre-fermenter inoculum was inoculated. The flasks were incubated in a shaking incubator at 150 rpm and 30 °C for 5–7 days [21]. The pectinase activity in the fermented broth was monitored by using the dinitro salicylic acid (DNS) assay method as described previously [22].Two milliliters of the fermented broth was pipette out into a sterile tube and centrifuged at 8000 rpm for 20 min. The supernatant obtained was used as the crude enzyme and used for analysis. About 1 mL of the crude enzyme and 1ml of 3% pectin were mixed in a sterile tube and incubated at 50°C for 15 min. After incubation, 1 mL of the DNS (Dinitrosalicylic acid) reagent was added to stop the hydrolysis reaction. The mixture was then shaken to mix the content and then placed in a boiling water bath for 30 min for color development. The absorbance was then read at 540 nm spectrophotometrically running the enzyme and substrate blanks in parallel. The blank containing 1 mL of 0.5% pectin, 1 mL of sodium acetate buffer (0.1 M, pH 4.2) and 2 mL of DNS reagent was used as a control. One unit of enzyme activity was defined as the amount of enzyme which liberated 1 μmol of galacturonic acid per hour under standard assay conditions.
2.3.2. Effect of Various Factors on Enzyme Production
To study the effect of incubation period, submerged state fermentation was carried out and DNS assay was performed every 24 h until 6 days. The effect of substrate concentration on pectinase production was evaluated using Hankin’s broth medium containing different concentrations of pectin (0.5, 1 and 2%) and 1 mL fungal inoculum. The enzyme production was assessed by DNS method after incubating the culture flask at 30 °C for 48 h. To study the effect of temperature on pectinase production, Hankin’s broth containing 1% pectin was prepared. Then, 1 mL of the inoculum was added and incubated separately at 25, 30, and 40 °C for 48 h. After incubation, the enzyme assay was performed using the DNS method.
2.3.3. Enzyme Extraction and Partial Purification
The fermentation broth was filtered through Whatman No.1 filter paper and centrifuged (10,000 rpm, 30 min, at 4 °C) for 2–3 times to remove the spores and mycelia of the organism. The filtration process was carried out in a cold condition in ice water to prevent enzyme deactivation. The supernatant represented the soluble crude extract. The crude enzyme was then mixed with three volumes of ice-cold acetone and was allowed to stand for 15 min. The entire content was centrifuged at 4000 rpm at 4 °C for 20 min. The enzyme precipitate was dissolved in a sodium acetate buffer (0.1M, pH 4.2) and was stored at 4 °C for further use [23].
2.4. Characterization of Partially Purified Pectinase Enzyme
2.4.1. Thermal Stability
To assess the thermal stability, the partially purified enzyme extract was heated at 60 °C until 1 h had passed. At 10 min intervals, 1 mL aliquots were withdrawn and mixed with 1 mL of 0.3% (w/v) pectin in test tubes and the tubes were incubated at 30 °C for 10 min. The enzyme activity was measured as described by the DNS assay method [24,25].
2.4.2. Effect of Temperature on Enzyme Activity
For temperature assessment, 1 mL of enzyme suspended on sodium acetate buffer (0.1 M, pH 4.2) was added to 1 mL of 0.3% (w/v) pectin solution in sterile tubes. The tubes were incubated from 25 to 55 °C for 15 min. After incubation, 2 mL of the DNS reagent was added and incubated in a boiling water bath for 15 min. The enzyme activity was monitored by the DNS assay method.
2.4.3. Effect of pH on Enzyme Activity
The effect of pH on enzyme activity was tested using sodium acetate buffer (0.1 M; pH range, 3.2–5.8), sodium phosphate buffer (0.1 M; pH range, 5.9–7.1) and Tris-HCl buffer (0.1 M; pH range, 7.2–9.0). Tubes containing 0.5 mL respective buffers were mixed with 0.5 mL of the enzyme. Then, 1 mL of 0.3% (w/v) pectin solution was added and all the tubes were incubated at 30 °C for 10 min. Afterward, 2 mL of the DNS reagent was added and incubated in boiling water for 15 min. The enzyme activity was measured by DNS assay method.
2.4.4. Effect of Substrate Concentration on Enzyme Activity
Pectin substrates of different concentrations (0.5%, 1.0%, 1.5% and 2.0%) were prepared in different tubes to evaluate the effect of substrate concentration on enzyme activity. One mL of enzyme suspended in acetate buffer (0.1M, pH 4.2) was mixed with 1 mL of the respective substrate concentrations. Determination of the pectinase activity was done by the DNS assay method.
2.5. Application of Pectinase Enzyme
Mature ripened orange fruits (Citrus sinensis) were commercially purchased from a local grocery store and stored in food processing laboratory.Prior to juice extraction, the fruits were sorted, washed and peeled. The juice was extracted using sterile small-scale juice extractor available in the laboratory. The extracted juice was pasteurized at 85 °C for 3 min to inactivate the natural fruit enzymes and cooled down to 4 °C. Then, different conical flasks containing 20 mL of orange juice were added with varying concentrations of the enzyme (0%, 0.25%, 0.5%, 0.75% and 1%). All the flasks were incubated at 30 °C for 1 h. After incubation, the samples were then heated at 85 °C for 3 min to inactivate the enzyme.The juice treated with different enzyme concentrations was filtered using Whatman No. 1 filter paper. The volume of fruit juice obtained was measured using a 50 mL measuring cylinder. The total soluble solids (TSS) was determined using a Brix refractometer (model no. CTL-REFM-BR32, LW Scientific) and expressed as degree Brix (°Brix). For clarity, the juice was shaken and 10 mL of juice was centrifuged at 5000 rpm for 10 min. The supernatant portion of the juice was used to determine percent transmittance and absorbance at 540 nm by spectrophotometer (SL−150, Elico) [26].
2.6. Statistical Analysis
All the data were statistically analyzed using Office Excel 2019 and OriginPro 8.5 software. The statistically significant (p-value < 0.05) of various factors on enzyme activity was determined using one-way ANOVA.
3. Results and Discussion
3.1. Isolation, Screening, and Identification of the Fungal Isolates
A total of 55 fungal strains were isolated and screened primarily. Based on the zone of hydrolysis, only 14 strains were subjected for secondary screening. During secondary screening, 4 strains designated as Gm, Lco, C, and T showed the highest pectinolytic activity with the zone of hydrolysis of 35, 32, 25, and 15 mm in diameter, respectively. These four strains were morphologically and culturally identified as Aspergillus spp. Gm, Penicillium spp Lco, Fusarium spp. C, and Aspergillus spp. T. Based on primary and secondary screening, Aspergillus spp. Gm was found to be a potent strain and hence was selected for further study. The Photograph 1A shows the zone of hydrolysis observed during primary and secondary screening for enzyme production. Similarly, Figure 1B,C show the colony morphology and lactophenol cotton blue staining results of the isolate, respectively. The strains of Aspergillus spp. are promising sources of pectinase enzyme. Several previous studies have demonstrated the significance of Aspergillus spp. in the commercial production of pectinase enzyme [12,25].
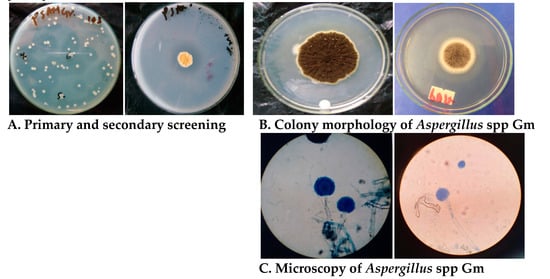
Figure 1.
(A)Primary and secondary screening for Aspergillus spp. Gm.(B)Colony morphology of Aspergillus spp. Gm. (C) Lactophenol cotton staining of Aspergillus spp. Gm.
3.2. Optimization of the Fermentation Conditions for Maximum Enzyme Production
The strain Aspergillus spp. Gm was used to study the optimum conditions for maximum enzyme production. This strain showed the highest (106.7 U/mL) pectinase production at 48 h of incubation period. After 48 h, the enzyme production was found to be decreased (Figure 2A). Various other studies have reported the highest enzyme activity of Aspergillus niger at 48–72 h, after which the enzyme production was found to be declined [27,28]. The cause of decrease in enzyme production after certain time interval during incubation might be due to the exhaustion of essential supplements and/or accumulation of toxic metabolites in the culture medium [29]. The shorter fermentation period of 48 h could be advantageous for production of pectinase at industrial scale.
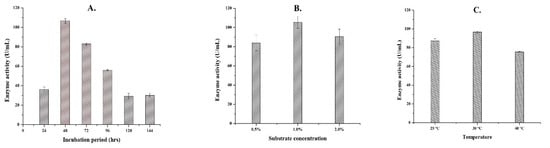
Figure 2.
Optimization of the fermentation conditions for maximum enzyme production from strain Aspergillus spp. Gm. (A), effect of incubation period; (B), effect of substrate concentration; and (C), effect of temperature on enzyme production.
With 1% substrate concentration, strain Gm produces maximum pectinase of 105.2 U/mL. The enzyme production was found to be decreased when the substrate was used more than 1% concentration (Figure 2B). A higher substrate concentration increases the viscosity of culture media and creates nutrient rich environment. Higher nutrients and substrate in the fermentation media inhibit microbial growth lowering enzyme production [30,31]. Previous researches have also stated that lower substrate concentration is effective for enzyme production [32,33]. The requirement of low substrate concentration might be cost effective for large scale production of pectinase enzyme.
At 30 °C, activity of strain Gm was reported to be the highest and produces maximum (96.7 U/mL) pectinase. At a higher temperature, enzyme production declined (Figure 2C). Enzymes are usually denatured at higher temperature resulting decreased activity [34]. Previous studies have reported that the maximum yield of pectinase enzyme from the members of genus Aspergillus spp. occur at the temperature range of 30–40 °C [35,36]. Among all the parameters analyzed, only temperature was found to be statistically significant (p < 0.05). From these optimization data, this study found that 48 h of incubation period, 1% substrate concentration, and 30 °C temperature could be the optimum cultural conditions for strain Gm to produce maximum pectinase enzyme.
3.3. Characteristics of Partially Purified Pectinase
The optimal working ranges of partially purified pectinase enzyme produced by strain Gm were determined based on the assessment of thermal stability, temperature, pH, and substrate concentration. The evaluation of thermal stability indicated that the activity of pectinase reduced approximately by 50% within 10 min of heating at 60 °C. Further heat treatment until 40 min resulted complete loss in the activity of pectinase enzyme (Figure 3A). Thermal stability and activity of pectinases have crucial role in biotechnological processes and food industries. Studies have shown that pectinolytic enzymes can be stable and active at wide range of temperature (30–80 °C) [25,36,37]. The optimum working temperature of pectinase enzyme extracted from strain Gm was found to be 30 °C. At this temperature, pectinase enzyme showed the highest activity of 75.4 U/mL (Figure 3B). There was a significant difference in the activity of pectinase enzyme with various thermal treatment and temperature (p < 0.05). This feature of pectinase enzyme possesses great industrial value as it makes the enzyme less susceptible to thermal inactivation. High temperature-tolerant pectinase enzymes have been previously extracted from other various members of Aspergillus [29,36,38].
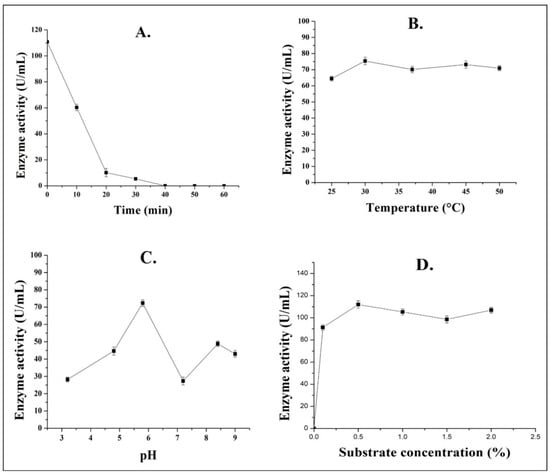
Figure 3.
Effect of various factors on enzyme activity of partially purified pectinase enzyme from strain Aspergillus spp. Gm. (A), thermal stability at 60 °C; (B), temperature; (C), pH; and (D), substrate concentration.
The enzyme produced from the strain Gm was found to be active over a wide range of pH (3.2–9.0). However, the optimum enzyme activity (72.3 U/mL) was observed at pH 5.8, indicating that the pectinase enzyme showed higher activity in theslightly acidic range (Figure 3C). The enzyme activity was significantly different with various pH (p < 0.05). Most of the studies have reported that pectinolytic enzymes have shown to exhibit higherenzymatic activity at pH range of 4.0–7.0 [25,39,40]. Although the enzyme showed highest activity at pH 5.8, a significant peak at pH 8.0 was observed. This may be due to enzyme’s activity and stability. Fungal pectinases are found to be stable from acidic to alkaline pH ranges (4.0–8.0) but their activity and stability may have different peaks at different pH ranges [41]. In a similar study, they observed the stability and optimum activity of fungal pectinases at different ranges, i.e., stability pH 4.0, whereas the optimum activity was found at pH 10.5–11 [42].
Acidic pectinase is useful in preparation of plant protoplast, to perform partial saccharification of sugars, to study phytopathogens invasion in plant, and in other various plant biotechnology applications [15]. However, the activity of pectinase enzyme was observed highest (112.0 U/mL) at 0.5% of substrate concentration. Numerically, the activity of pectinase varies with various substrate concentration (Figure 3D). However, statistically, there was no significant difference in the activity of enzyme with various substrate concentration (p < 0.05). Studies have reported that low substrate concentrations are useful to achieve higher enzyme activity [33,43]. High substrate (pectin) concentrations decrease the availability of enzymes and lower the amount of free water in the system. Free water availability is an important factor for maximum enzymatic activity [44]. Therefore, low substrate concentration (0.5%), as observed in this study, could be useful to achieve highest activity of pectinase enzyme.
3.4. Application of Pectinase Enzyme in Juice Clarification
The pectinase enzyme extracted from strain Gm was applied for juice clarification. The juice clarification experiments observed that as the concentration of enzyme increased, the yield of the juice, TSS, absorbance value, and % transmittance also increased. At 1% enzyme concentration, the yield of the orange juice, TSS, absorbance value, and % transmittance was found to be the highest, indicating its potential application in juice processing industries (Table 1). There are several studies which have reported that rise in enzyme concentration markedly improved the juice clarification [45,46].

Table 1.
Effect of different pectinase concentrations on yield, total soluble solids, and transmittance.
The major impediments to exploit the commercial potential of pectinases are the yield, stability and the cost of enzyme production. In order to obtain high and commercially viable yields of pectinase enzymes, it is essential to optimize the fermentation conditions used for fungal growth and enzyme production. Optimal parameters of the pectinases enzyme biosynthesis from microbial origin varied greatly with the variation of the strains, environmental parameters, and nutritional conditions. Furthermore, the economic feasibility of the microbial enzymes production and its application generally depends on the cost of its production processes [43].
4. Conclusions
In conclusion, Aspergillus spp. Gm was identified as the promising strain for the production of pectinase enzyme via submerged fermentation. The maximum pectinase production was obtained under optimal cultivation conditions at 48 h of incubation period, 1% substrate concentration, and 30 °C temperature. The pectinase enzyme produced from strain Aspergillus spp. Gm showed high thermal stability and reported maximum enzymatic activity at 30 °C temperature, pH 5.8, and 0.5% substrate concentration. In addition, the produced pectinase enzyme in this study showed proven ability in juice clarification, indicating a potential use in food industries. However, further studies must be performed to identify the strain in genetic levels and to assure its commercial application in large-scale food formulation and processing which will be the topic of interest for our research group.
Author Contributions
S.K. (Sudeep KC), D.R.J., and B.L.: Conceived and designed the work, performed the experimental work, and collection of the samples. S.K. (Santosh Khanal), T.R.B., B.R.P. and R.D.: Provide analytical tools and contribute in data analysis. D.K.C., and J.U.: data curation, manuscript preparation and manuscript editing. N.K., and V.R.: Contribute in the overall conduct, data analysis, result presentation and submission. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
We acknowledge for all the helping hands who helped to conduct this study.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Kavuthodi, B.; Sebastian, D. Review on bacterial production of alkaline pectinase with special emphasis on Bacillus species. Biosci. Biotechnol. Res. Commun. 2018, 11, 18–30. [Google Scholar] [CrossRef]
- Alkorta, I.; Garbisu, C.; Llama, M.J.; Serra, J.L. Industrial applications of pectic enzymes: A review. Process Biochem. 1998, 33, 21–28. [Google Scholar] [CrossRef]
- Javed, R.; Nawaz, A.; Munir, M.; Hanif, M.; Mukhtar, H.; Haq, I.U.; Abdullah, R. Extraction, purification and industrial applications of pectinase: A review. J. Biotechnol. Biores. 2018, 1, 1–6. [Google Scholar]
- Carrasco, M.; Rozas, J.M.; Alcaíno, J.; Cifuentes, V.; Baeza, M. Pectinase secreted by psychrotolerant fungi: Identification, molecular characterization and heterologous expression of a cold-active polygalacturonase from Tetracladium sp. Microb. Cell Fact. 2019, 18, 45. [Google Scholar] [CrossRef]
- Cornuault, V.; Posé, S.; Knox, J.P. Disentangling pectic homogalacturonan and rhamnogalacturonan-I polysaccharides: Evidence for sub-populations in fruit parenchyma systems. Food Chem. 2018, 246, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Pedrolli, D.B.; Monteiro, A.C.; Gomes, E.; Carmona, E.C. Pectin and pectinases: Production, characterization and industrial application of microbial pectinolytic enzymes. Open Biotechnol. J. 2009, 3, 9–18. [Google Scholar] [CrossRef]
- Oumer, O.J.; Abate, D. Screening and molecular identification of pectinase producing microbes from coffee pulp. Biomed Res. Int. 2018, 2018, 2961767. [Google Scholar] [CrossRef]
- Garg, G.; Singh, A.; Kaur, A.; Singh, R.; Kaur, K.; Mahajan, R. Microbial pectinases: An ecofriendly tool of nature for industries. Biotech 2016, 6, 1–13. [Google Scholar] [CrossRef]
- Oumer, O.J. Pectinase: Substrate, production and their biotechnological applications. Int. J. Environ. Agric. Biotechnol. 2017, 2, 1007–1014. [Google Scholar] [CrossRef]
- Gummadi, S.N.; Panda, T. Purification and biochemical properties of microbial pectinases—A review. Process Biochem. 2003, 38, 987–996. [Google Scholar] [CrossRef]
- Hoondal, G.; Tiwari, R.; Tewari, R.; Dahiya, N.; Beg, Q. Microbial alkaline pectinases and their industrial applications: A review. Appl. Microbiol. Biotechnol. 2002, 59, 409–418. [Google Scholar] [PubMed]
- Khairnar, Y.; Krishna, V.K.; Boraste, A.; Gupta, N.; Trivedi, S.; Patil, P.; Gupta, G.; Gupta, M.; Jhadav, A.; Mujapara, A.; et al. Study of pectinase production in submerged fermentation using different strains of Aspergillus Niger. Int. J. Microbiol. Res. 2009, 1, 13–17. [Google Scholar] [CrossRef]
- Dhital, R.; Panta, O.P.; Karki, T.B. Optimization of cultural conditions for the production of pectinase from selected fungal strain. J. Food Sci. Technol. Nepal 2014, 8, 65–70. [Google Scholar] [CrossRef]
- Barman, S.; Sit, N.; Badwaik, L.S.; Deka, S.C. Pectinase production by Aspergillus niger using banana (Musa balbisiana) peel as substrate and its effect on clarification of banana juice. J. Food Sci. Technol. 2015, 52, 3579–3589. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kubra, K.T.; Ali, S.; Walait, M.; Sundus, H. Potential applications of pectinases in food, agricultural and environmental sectors. J. Pharm. Chem. Biol. Sci. 2018, 6, 23–34. [Google Scholar]
- Koirala, N.; Pandey, R.P.; Parajuli, P.; Jung, H.J.; Sohng, J.K. Methylation and subsequent glycosylation of 7,8-dihydroxyflavone. J. Biotech. 2014, 184, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Patidar, M.K.; Nighojkar, S.; Kumar, A.; Nighojkar, A. Pectinolytic enzymes-solid state fermentation, assay methods and applications in fruit juice industries: A review. 3 Biotech 2018, 8, 199. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, S.Y.; Choi, H.S. Molecular and morphological identification of fungal species isolated from rice meju. Food Sci. Biotechnol. 2013, 22, 721–728. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, C.; Chen, H.; Zhang, J.; Chen, W. Isolation and identification of endophytic fungi from Actinidia macrosperma and investigation of their bioactivities. Evid. -Based Complement. Altern. Med. 2012, 2012, 382742. [Google Scholar] [CrossRef][Green Version]
- Hankin, L.; Anagnostakis, S.L. The use of solid media for detection of enzyme production by fungi. Mycologia 1975, 67, 597–607. [Google Scholar] [CrossRef]
- Kashyap, D.R.; Chandra, S.; Kaul, A.; Tewari, R. Production, purification and characterization of pectinase from a Bacillus sp. DT7. World J. Microbiol. Biotechnol. 2000, 16, 277–282. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Rajendran, R.; Karthik, S.; Radhai, R.; Rajapriya, P.; Balakumar, C. Production and optimization of fungal pectinase from Fusarium sp. Int. J. Curr. Res. 2011, 3, 254–258. [Google Scholar]
- Arotupin, D.J.; Akinyosoye, F.A.; Onifade, A.K. Purification and characterization of pectinmethylesterase from Aspergillus repens isolated from cultivated soil. Afr. J. Biotechnol. 2008, 7, 1991–1998. [Google Scholar] [CrossRef]
- Maciel, M.; Herculano, P.; Porto, T.; Teixeira, M.; Moreira, K.; De Souza-Motta, C. Production and partial characterization of pectinases from forage palm by Aspergillus niger URM4645. Afr. J. Biotechnol. 2011, 10, 2469–2475. [Google Scholar]
- Rai, P.; Majumdar, G.C.; DasGupta, S.; De, S. Optimizing pectinase usage in pretreatment of mosambi juice for clarification by response surface methodology. J. Food Eng. 2004, 64, 397–403. [Google Scholar] [CrossRef]
- Abdullah, R.; Farooq, I.; Kaleem, A.; Iqtedar, M.; Iftikhar, T. Pectinase production from Aspergillus niger IBT-7 using solid state fermentation. Bangladesh J. Bot. 2018, 47, 473–478. [Google Scholar] [CrossRef]
- Rashmi, R.; Murthy, K.; Sneha, G.; Shabana, S.; Syama, A.; Radhika, V. Partial purification and biochemical characterization of extracellular pectinase from Aspergillus niger isolated from groundnut seeds. J. Appl. Biosci. 2008, 9, 378–384. [Google Scholar]
- Phutela, U.; Dhuna, V.; Sandhu, S.; Chadha, B.S. Pectinase and polygalacturonase production by a thermophilic Aspergillus fumigatus isolated from decomposting orange peels. Braz. J. Microbiol. 2005, 36, 63–69. [Google Scholar] [CrossRef]
- Biz, A.; Farias, F.C.; Motter, F.A.; De Paula, D.H.; Richard, P.; Krieger, N.; Mitchell, D.A. Pectinase activity determination: An early deceleration in the release of reducing sugars throws a spanner in the works. PLoS ONE 2014, 9, e109529. [Google Scholar] [CrossRef]
- Patil, N.P.; Chaudhari, B.L. Production and purification of pectinase by soil isolate Penicillium spp and search for better agro-residue for its SSF. Recent Res. Sci. Technol. 2010, 2, 36–42. [Google Scholar]
- Palaniyappan, M.; Vijayagopal, V.; Viswanathan, R.; Viruthagiri, T. Screening of natural substrates and optimization of operating variables on the production of pectinase by submerged fermentation using Aspergillus niger MTCC 281. Afr. J. Biotechnol. 2009, 8, 682–686. [Google Scholar]
- Akhter, N.; Morshed, M.A.; Uddin, A.; Begum, F.; Sultan, T.; Azad, A.K. Production of pectinase by Aspergillus niger cultured in solid state media. Int. J. Biosci. 2011, 1, 33–42. [Google Scholar]
- Al-Najada, A.R.; Al-Hindi, R.R.; Mohamed, S.A. Characterization of polygalacturonases from fruit spoilage Fusarium oxysporum and Aspergillus tubingensis. Afr. J. Biotechnol. 2012, 11, 8527–8536. [Google Scholar] [CrossRef]
- Singh, S.; Mandal, S.K. Optimization of processing parameters for production of pectinolytic enzymes from fermented pineapple residue of mixed Aspergillus species. Jordan J. Biol. Sci. 2012, 5, 307–314. [Google Scholar]
- Khatri, B.P.; Bhattarai, T.; Shrestha, S.; Maharjan, J. Alkaline thermostable pectinase enzyme from Aspergillus niger strain MCAS2 isolated from Manaslu Conservation Area, Gorkha, Nepal. Springerplus 2015, 4, 488. [Google Scholar] [CrossRef]
- Deshmukh, N.; Talkal, R.; Jha, K.; Singh, P.G.; Prajapati, D.C. Production, purification, characterization and comparison of polygalacturonase from various strains of Aspergillus. Int. J. Sci. Technol. Res. 2012, 1, 85–91. [Google Scholar]
- Dogan, N.; Tari, C. Characterization of three-phase partitioned exo-polygalacturonase from Aspergillus sojae with unique properties. Biochem. Eng. J. 2008, 39, 43–50. [Google Scholar] [CrossRef]
- Martins, E.S.; Silva, D.; Da Silva, R.; Gomes, E. Solid state production of thermostable pectinases from thermophilic Thermoascus aurantiacus. Process Biochem. 2002, 37, 949–954. [Google Scholar] [CrossRef]
- Mohamed, S.A.; Al-Malki, A.L.; Kumosani, T.A. Characterization of a polygalacturonase from Trichoderma harzianum grown on citrus peel with application for apple Juice. Aust. J. Basic Appl. Sci. 2009, 3, 2770–2777. [Google Scholar]
- Jayani, R.S.; Saxena, S.; Gupta, R. Microbial pectinolytic enzymes: A review. Process Biochem. 2005, 40, 2931–2944. [Google Scholar] [CrossRef]
- Ferreira, V.; da Silva, R.; Silva, D.; Gomes, E. Production of pectate lyase by Penicillium viridicatum RFC3 in solid-state and submerged fermentation. Int. J. Microbiol. 2010, 1–8. [Google Scholar] [CrossRef]
- Sandri, I.; Silveira, M. Production and application of pectinases from Aspergillus niger obtained in solid state cultivation. Beverages 2018, 4, 48. [Google Scholar] [CrossRef]
- Demir, H.; Tari, C. Valorization of wheat bran for the production of polygalacturonase in SSF of Aspergillus sojae. Ind. Crops Prod. 2014, 54, 302–309. [Google Scholar] [CrossRef]
- Kareem, S.; Adebowale, A. Clarification of orange juice by crude fungal pectinase from citrus peel. Niger. Food J. 2007, 25, 130–137. [Google Scholar] [CrossRef][Green Version]
- Joshi, V.; Parmar, M.; Rana, N. Purification and characterization of pectinase produced from apple pomace and evaluation of its efficacy in fruit juice extraction and clarification. Indian J. Nat. Prod. Resour. 2011, 2, 189–197. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).