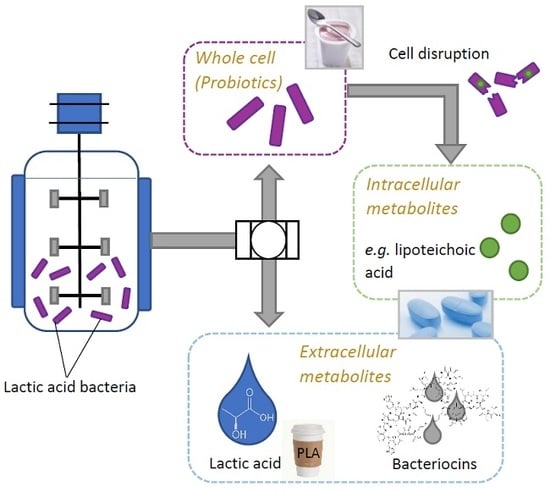
Multi-Product Lactic Acid Bacteria Fermentations: A Review
Abstract
1. Introduction
2. Potential Uses of Lactic Acid Bacteria
2.1. Products Present in Supernatant
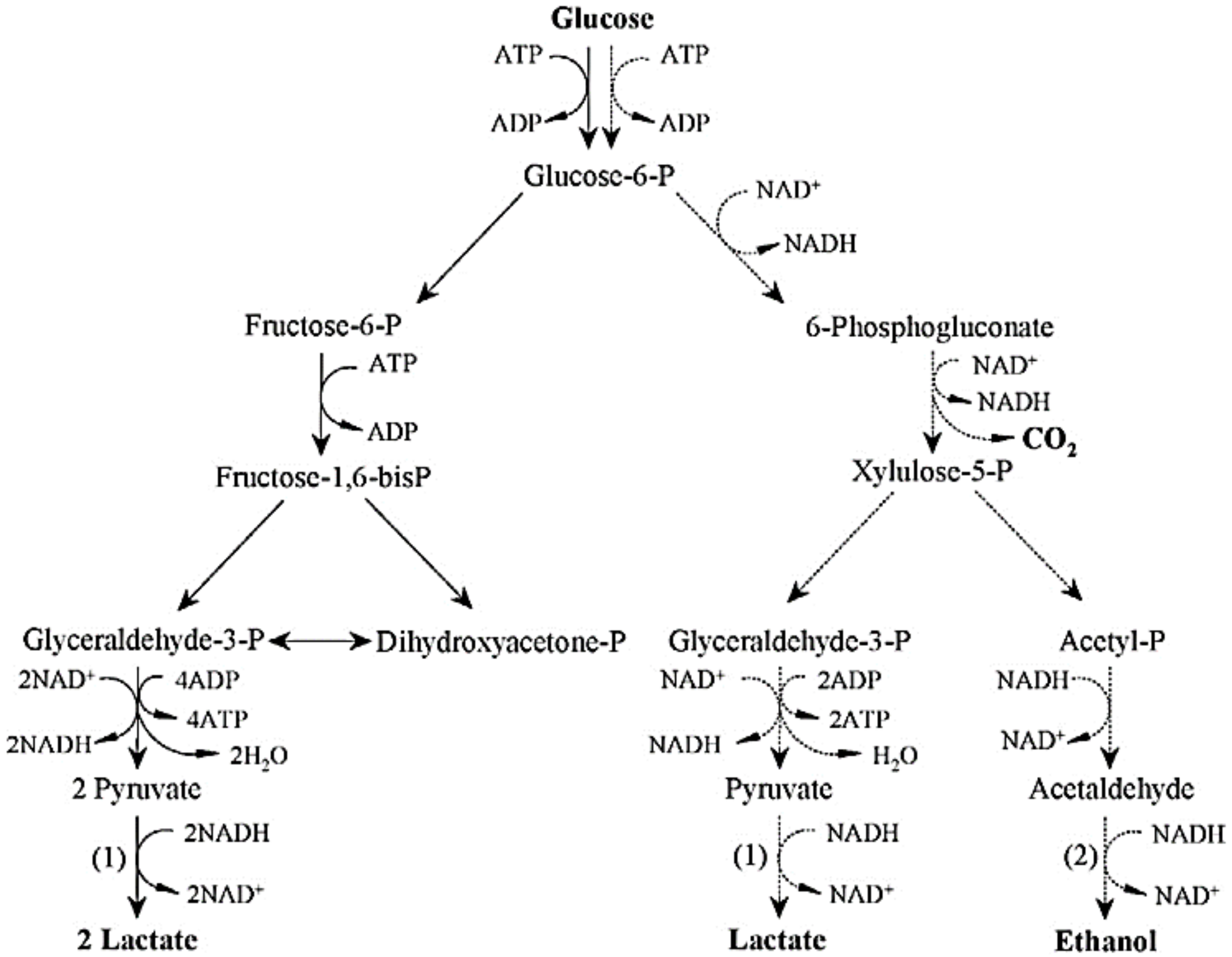
2.1.1. Lactic Acid
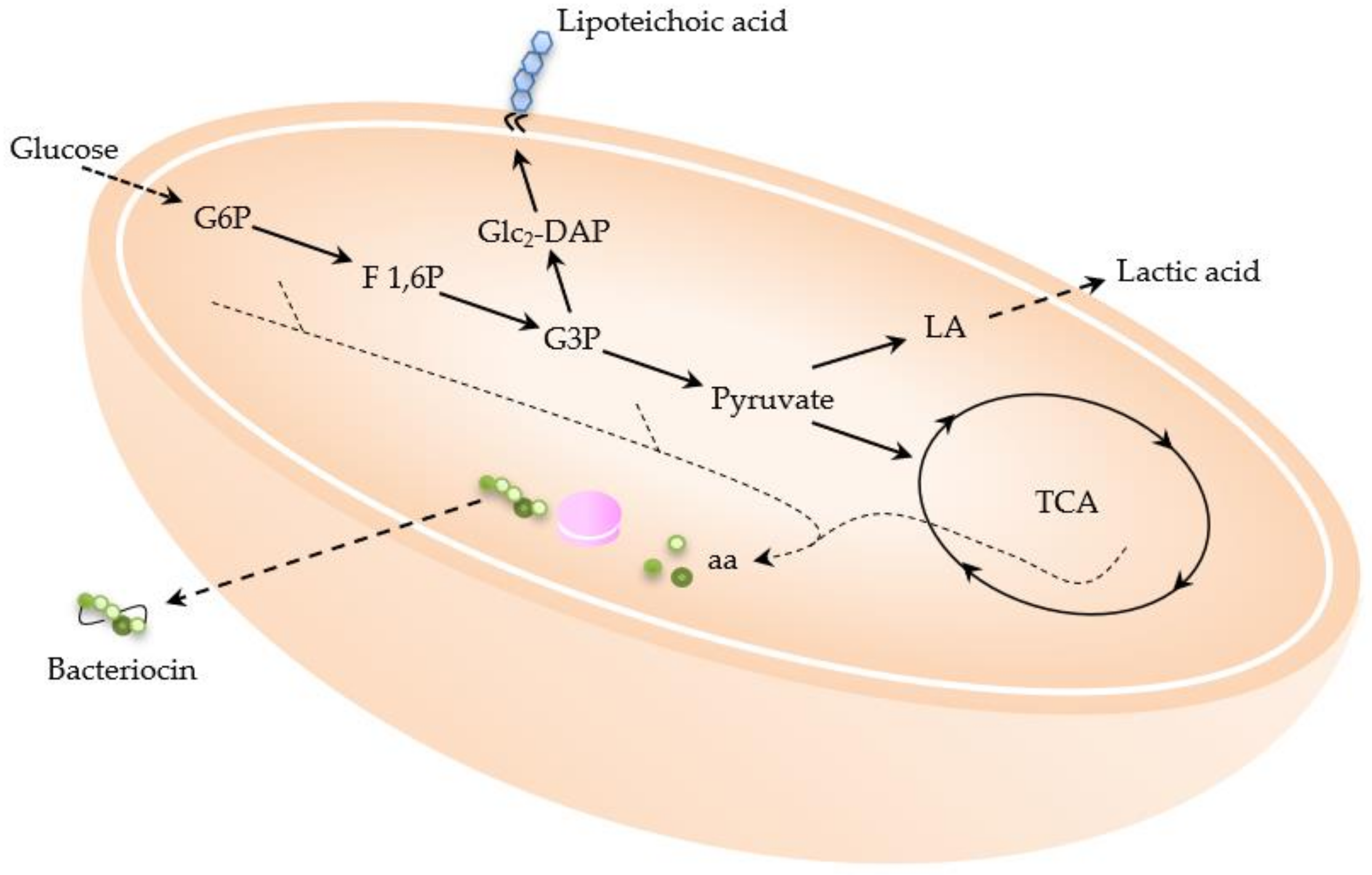
2.1.2. Bacteriocins
- Class I (lantibiotics): peptides containing lanthionine (a non-canonical amino acid). These can be elongated with a net positive charge (sub-class A, e.g., nisin), or globular with negative or no charge (sub-class B, e.g., mersacidin).
- Class II: heat-stable peptides, not containing lanthionine. Their sub-classes depend manly on the activity (sub-class A, e.g., pediocin; sub-class B, e.g., lactococcin, plantaricin; sub-class C, e.g., acidocin).
- Class III: large, heat-labile peptides, not very well characterized. They are lytic proteins often classified as murein-hydrolases (e.g., helveticin).
2.2. Use of LAB-Biomass
2.2.1. Probiotics
2.2.2. Lipoteichoic Acid (LTA)
2.3. Other Compounds with Industrial Potential Produced by LAB
3. Perspectives of Multi-Product Processes
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Qiu, Y.; Lei, P.; Zhang, Y.; Sha, Y.; Zhan, Y.; Xu, Z.; Li, S.; Xu, H.; Ouyang, P. Recent advances in bio-based multi-products of agricultural Jerusalem artichoke resources. Biotechnol. Biofuels 2018, 11, 151. [Google Scholar] [CrossRef] [PubMed]
- Zondervan, E.; Nawaz, M.; de Haan, A.B.; Woodley, J.M.; Gani, R. Optimal design of a multi-product biorefinery system. Comput. Chem. Eng. 2011, 35, 1752–1766. [Google Scholar] [CrossRef]
- Santos, F. Sugarcane Biorefinery, Technology and Perspectives, 1st ed.; Academic Press: London, UK, 2020. [Google Scholar]
- Kamm, B.; Kamm, M. Biorefineries-Multi product processes. Adv. Biochem. Eng. Biotechnol. 2006, 105, 175–204. [Google Scholar]
- Mazzoli, R.; Bosco, F.; Mizrahi, I.; Bayer, E.A.; Pessione, E. Towards lactic acid bacteria-based biorefineries. Biotechnol. Adv. 2014, 32, 1216–1236. [Google Scholar] [CrossRef] [PubMed]
- Vinderola, G.; Ouwehand, A.C.; Salminen, S.; von Wright, A. Lactic Acid Bacteria: Microbiological and Functional Aspects, 5th ed.; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Salvetti, E.; Harris, H.M.B.; Felis, G.E.; O’Toole, P.W. Comparative genomics of the genus Lactobacillus reveals robust phylogroups that provide the basis for reclassification. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef]
- Endo, A.; Tanizawa, Y.; Arita, M. Isolation and identification of lactic acid bacteria from environmental samples. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2019; pp. 3–13. [Google Scholar]
- Sabatini, N. A Comparison of the Volatile Compounds, in Spanish-style, Greek-style and Castelvetrano-style Green Olives of the Nocellara del Belice Cultivar: Alcohols, Aldehydes, Ketones, Esters and Acids. In Olives and Olive Oil in Health and Disease Prevention; Elsevier Inc.: Burlington, VT, USA, 2010; pp. 219–231. [Google Scholar]
- Ruiz Rodríguez, L.G.; Mohamed, F.; Bleckwedel, J.; Medina, R.; De Vuyst, L.; Hebert, E.M.; Mozzi, F. Diversity and functional properties of lactic acid bacteria isolated from wild fruits and flowers present in northern Argentina. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Ghaffar, T.; Irshad, M.; Anwar, Z.; Aqil, T.; Zulifqar, Z.; Tariq, A.; Kamran, M.; Ehsan, N.; Mehmood, S. Recent trends in lactic acid biotechnology: A brief review on production to purification. J. Radiat. Res. Appl. Sci. 2014, 7, 222–229. [Google Scholar] [CrossRef]
- Champagne, C.P. The lactic acid bacteria. Int. Dairy J. 1994, 4, 665–666. [Google Scholar] [CrossRef]
- Börner, R.A.; Kandasamy, V.; Axelsen, A.M.; Nielsen, A.T.; Bosma, E.F. Genome editing of lactic acid bacteria: Opportunities for food, feed, pharma and biotech. FEMS Microbiol. Lett. 2019, 366, i30–i41. [Google Scholar] [CrossRef]
- Van Pijkeren, J.-P.; Barrangou, R. Genome Editing of Food-Grade Lactobacilli To Develop Therapeutic Probiotics. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Barrangou, R.; Horvath, P. CRISPR: New Horizons in Phage Resistance and Strain Identification. Annu. Rev. Food Sci. Technol. 2012, 3, 143–162. [Google Scholar] [CrossRef] [PubMed]
- Duar, R.M.; Lin, X.B.; Zheng, J.; Martino, M.E.; Grenier, T.; Pérez-Muñoz, M.E.; Leulier, F.; Gänzle, M.; Walter, J. Lifestyles in transition: Evolution and natural history of the genus Lactobacillus. FEMS Microbiol. Rev. 2017, 41, S27–S48. [Google Scholar] [CrossRef] [PubMed]
- Sadiq, F.A.; Yan, B.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W. Lactic Acid Bacteria as Antifungal and Anti-Mycotoxigenic Agents: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1403–1436. [Google Scholar] [CrossRef]
- Rahbar Saadat, Y.; Yari Khosroushahi, A.; Pourghassem Gargari, B. A comprehensive review of anticancer, immunomodulatory and health beneficial effects of the lactic acid bacteria exopolysaccharides. Carbohydr. Polym. 2019, 217, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Venegas-Ortega, M.G.; Flores-Gallegos, A.C.; Martínez-Hernández, J.L.; Aguilar, C.N.; Nevárez-Moorillón, G.V. Production of Bioactive Peptides from Lactic Acid Bacteria: A Sustainable Approach for Healthier Foods. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1039–1051. [Google Scholar] [CrossRef]
- Russo, P.; Spano, G.; Capozzi, V. Safety evaluation of starter cultures. In Starter Cultures in Food Production; John Wiley & Sons, Ltd.: Chichester, UK, 2017; pp. 101–128. [Google Scholar]
- Vinusha, K.S.; Deepika, K.; Johnson, T.S.; Agrawal, G.K.; Rakwal, R. Proteomic studies on lactic acid bacteria: A review. Biochem. Biophys. Rep. 2018, 14, 140–148. [Google Scholar] [CrossRef]
- Kim, A.R.; Ahn, K.B.; Yun, C.H.; Park, O.J.; Perinpanayagam, H.; Yoo, Y.J.; Kum, K.Y.; Han, S.H. Lactobacillus plantarum Lipoteichoic Acid Inhibits Oral Multispecies Biofilm. J. Endod. 2019, 45, 310–315. [Google Scholar] [CrossRef]
- Lebeer, S.; Claes, I.J.J.J.; Vanderleyden, J. Anti-inflammatory potential of probiotics: Lipoteichoic acid makes a difference. Trends Microbiol. 2012, 20, 5–10. [Google Scholar] [CrossRef]
- Ryu, Y.H.; Baik, J.E.; Yang, J.S.; Kang, S.S.; Im, J.; Yun, C.H.; Kim, D.W.; Lee, K.; Chung, D.K.; Ju, H.R.; et al. Differential immunostimulatory effects of Gram-positive bacteria due to their lipoteichoic acids. Int. Immunopharmacol. 2009, 9, 127–133. [Google Scholar] [CrossRef]
- Pinu, F.R.; Villas-Boas, S.G. Extracellular microbial metabolomics: The state of the art. Metabolites 2017, 7, 43. [Google Scholar] [CrossRef]
- Gou, M.; Song, W. The growing U.S. bioeconomy: Drivers, development and constraints. N. Biotechnol. 2019, 49, 48–57. [Google Scholar] [CrossRef]
- Castro-Aguirre, E.; Iñiguez-Franco, F.; Samsudin, H.; Fang, X.; Auras, R. Poly(lactic acid)—Mass production, processing, industrial applications, and end of life. Adv. Drug Deliv. Rev. 2016, 107, 333–366. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.C.; Yuan, S.F.; Wang, C.A.; Huang, Y.J.; Gou, G.L.; Hwang, W.S. Production of optically pure l-lactic acid from lignocellulosic hydrolysate by using a newly isolated and d-lactate dehydrogenase gene-deficient Lactobacillus paracasei strain. Bioresour. Technol. 2015, 198, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Villadsen, J. Innovative technology to meet the demands of the white biotechnology revolution of chemical production. Chem. Eng. Sci. 2007, 62, 6957–6968. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Z.; Qin, P.; Tan, T. Fermentative l-(+)-lactic acid production from defatted rice bran. RSC Adv. 2014, 4, 8907–8913. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.A.; Tashiro, Y.; Zendo, T.; Sonomoto, K. Improved lactic acid productivity by an open repeated batch fermentation system using Enterococcus mundtii QU 25. RSC Adv. 2013, 3, 8437–8445. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.A.; Xiao, Y.; Tashiro, Y.; Wang, Y.; Zendo, T.; Sakai, K.; Sonomoto, K. Fed-batch fermentation for enhanced lactic acid production from glucose/xylose mixture without carbon catabolite repression. J. Biosci. Bioeng. 2015, 119, 153–158. [Google Scholar] [CrossRef]
- Wang, Y.; Abdel-Rahman, M.A.; Tashiro, Y.; Xiao, Y.; Zendo, T.; Sakai, K.; Sonomotes, K. L-(+)-Lactic acid production by co-fermentation of cellobiose and xylose without carbon catabolite repression using Enterococcus mundtii QU 25. RSC Adv. 2014, 4, 22013–22021. [Google Scholar] [CrossRef]
- Wee, Y.J.; Yun, J.S.; Park, D.H.; Ryu, H.W. Biotechnological production of L(+)-lactic acid from wood hydrolyzate by batch fermentation of Enterococcus faecalis. Biotechnol. Lett. 2004, 26, 71–74. [Google Scholar] [CrossRef]
- Wee, Y.J.; Kim, J.N.; Yun, J.S.; Ryu, H.W. Utilization of sugar molasses for economical L(+)-lactic acid production by batch fermentation of Enterococcus faecalis. Enzyme. Microb. Technol. 2004, 35, 568–573. [Google Scholar] [CrossRef]
- Subramanian, M.R.; Talluri, S.; Christopher, L.P. Production of lactic acid using a new homofermentative Enterococcus faecalis isolate. Microb. Biotechnol. 2015, 8, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.F.; Hsu, T.C.; Wang, C.A.; Jang, M.F.; Kuo, Y.C.; Alper, H.S.; Guo, G.L.; Hwang, W.S. Production of optically pure l(+)-lactic acid from waste plywood chips using an isolated thermotolerant Enterococcus faecalis SI at a pilot scale. J. Ind. Microbiol. Biotechnol. 2018, 45, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Wee, Y.J.; Kim, H.O.; Yun, J.S.; Ryw, H.W. Pilot-scale lactic acid production via batch culturing of Lactobacillus sp. RKY2 using corn steep liquor as a nitrogen source. Food Technol. Biotechnol. 2006, 44, 293–298. [Google Scholar]
- Yi, X.; Zhang, P.; Sun, J.; Tu, Y.; Gao, Q.; Zhang, J.; Bao, J. Engineering wild-type robust Pediococcus acidilactici strain for high titer l- and d-lactic acid production from corn stover feedstock. J. Biotechnol. 2016, 217, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Hama, S.; Mizuno, S.; Kihara, M.; Tanaka, T.; Ogino, O.; Noda, H.; Kondo, A. Production of d-lactic acid from hardwood pulp by mechanical milling followed by simultaneous saccharification and fermentation using metabolically engineered Lactobacillus plantarum. Bioresour. Technol. 2015, 187, 167–172. [Google Scholar] [CrossRef]
- Sauer, M.; Porro, D.; Mattanovich, D.; Branduardi, P. 16 years research on lactic acid production with yeast–ready for the market? Biotechnol. Genet. Eng. Rev. 2010, 27, 229–256. [Google Scholar] [CrossRef]
- Yamada, R.; Wakita, K.; Mitsui, R.; Ogino, H. Enhanced d-lactic acid production by recombinant Saccharomyces cerevisiae following optimization of the global metabolic pathway. Biotechnol. Bioeng. 2017, 114, 2075–2084. [Google Scholar] [CrossRef]
- Baek, S.H.; Kwon, E.Y.; Bae, S.J.; Cho, B.R.; Kim, S.Y.; Hahn, J.S. Improvement of d-Lactic Acid Production in Saccharomyces cerevisiae Under Acidic Conditions by Evolutionary and Rational Metabolic Engineering. Biotechnol. J. 2017, 12. [Google Scholar] [CrossRef]
- Baek, S.H.; Kwon, E.Y.; Kim, Y.H.; Hahn, J.S. Metabolic engineering and adaptive evolution for efficient production of D-lactic acid in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2016, 100, 2737–2748. [Google Scholar] [CrossRef]
- López-Gómez, J.P.; Alexandri, M.; Schneider, R.; Venus, J. A review on the current developments in continuous lactic acid fermentations and case studies utilising inexpensive raw materials. Process Biochem. 2019, 79, 1–10. [Google Scholar] [CrossRef]
- Lee, R.K.; Ryu, H.W.; Oh, H.; Kim, M.; Wee, Y.J. Cell-recycle continuous fermentation of Enterococcus faecalis RKY1 for economical production of lactic acid by reduction of yeast extract supplementation. J. Microbiol. Biotechnol. 2014, 24, 661–666. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, M.A.; Tashiro, Y.; Zendo, T.; Sakai, K.; Sonomoto, K. Highly efficient L-lactic acid production from xylose in cell recycle continuous fermentation using Enterococcus mundtii QU 25. RSC Adv. 2016, 6, 17659–17668. [Google Scholar] [CrossRef]
- Kwon, S.; Yoo, I.K.; Lee, W.G.; Chang, H.N.; Chang, Y.K. High-rate continuous production of lactic acid by Lactobacillus rhamnosus in a two-stage membrane cell-recycle bioreactor. Biotechnol. Bioeng. 2001, 73, 25–34. [Google Scholar] [CrossRef]
- Wee, Y.J.; Ryu, H.W. Lactic acid production by Lactobacillus sp. RKY2 in a cell-recycle continuous fermentation using lignocellulosic hydrolyzates as inexpensive raw materials. Bioresour. Technol. 2009, 100, 4262–4270. [Google Scholar] [CrossRef] [PubMed]
- Melzoch, K.; Votruba, J.; Hábová, V.; Rychtera, M. Lactic acid production in a cell retention continuous culture using lignocellulosic hydrolysate as a substrate. J. Biotechnol. 1997, 56, 25–31. [Google Scholar] [CrossRef]
- Zhang, J.; Hao, N.; Chen, G.Q. Effect of expressing polyhydroxybutyrate synthesis genes (phbCAB) in Streptococcus zooepidemicus on production of lactic acid and hyaluronic acid. Appl. Microbiol. Biotechnol. 2006, 71, 222–227. [Google Scholar] [CrossRef]
- Demichelis, F.; Pleissner, D.; Fiore, S.; Mariano, S.; Navarro, I.M.; Schneider, R.; Venus, J. Investigation of food waste valorization through sequential lactic acid fermentative production and anaerobic digestion of fermentation residues. Bioresour. Technol. 2017, 241, 508–516. [Google Scholar] [CrossRef]
- Schiraldi, C.; Adduci, V.; Valli, V.; Maresca, C.; Giuliano, M.; Lamberti, M.; Carteni, M.; de Rosa, M. High cell density cultivation of probiotics and lactic acid production. Biotechnol. Bioeng. 2003, 82, 213–222. [Google Scholar] [CrossRef]
- Djukić-Vuković, A.P.; Mojović, L.V.; Vukašinović-Sekulić, M.S.; Nikolić, S.B.; Pejin, J.D. Integrated production of lactic acid and biomass on distillery stillage. Bioprocess Biosyst. Eng. 2013, 36, 1157–1164. [Google Scholar] [CrossRef]
- Djukić-Vuković, A.P.; Mojović, L.V.; Semenčenko, V.V.; Radosavljević, M.M.; Pejin, J.D.; Kocić-Tanackov, S.D. Effective valorisation of distillery stillage by integrated production of lactic acid and high quality feed. Food Res. Int. 2015, 73, 75–80. [Google Scholar] [CrossRef]
- Djukić-Vuković, A.P.; Jokić, B.M.; Kocić-Tanackov, S.D.; Pejin, J.; Mojović, L.V. Mg-modified zeolite as a carrier for Lactobacillus rhamnosus in L(+) lactic acid production on distillery wastewater. J. Taiwan Inst. Chem. Eng. 2016, 59, 262–266. [Google Scholar] [CrossRef]
- Cui, F.; Wan, C.; Li, Y.; Liu, Z.; Rajashkara, G. Co-production of Lactic Acid and Lactobacillus rhamnosus Cells from Whey Permeate with Nutrient Supplements. Food Bioprocess Technol. 2012, 5, 1278–1286. [Google Scholar] [CrossRef]
- De Vuyst, L.; Leroy, F. Bacteriocins from lactic acid bacteria: Production, purification, and food applications. J. Mol. Microbiol. Biotechnol. 2007, 13, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Hoover, D. Bacteriocins and their Food Applications. Compr. Rev. Food Sci. Food Saf. 2003, 2, 82–100. [Google Scholar] [CrossRef]
- Cheigh, C.I.; Choi, H.J.; Park, H.; Kim, S.B.; Kook, M.C.; Kim, T.S.; Hwang, J.K.; Pyun, Y.R. Influence of growth conditions on the production of a nisin-like bacteriocin by Lactococcus lactis subsp. lactis A164 isolated from kimchi. J. Biotechnol. 2002, 95, 225–235. [Google Scholar] [CrossRef]
- Neysens, P.; Messens, W.; De Vuyst, L. Effect of sodium chloride on growth and bacteriocin production by Lactobacillus amylovorus DCE 471. Int. J. Food Microbiol. 2003, 88, 29–39. [Google Scholar] [CrossRef]
- Callewaert, R.; De Vuyst, L. Bacteriocin production with Lactobacillus amylovorus DCE 471 is improved and stabilized by fed-batch fermentation. Appl. Environ. Microbiol. 2000, 66, 606–613. [Google Scholar] [CrossRef]
- Parente, E.; Brienza, C.; Ricciardi, A.; Addario, G. Growth and bacteriocin production by Enterococcus faecium DPC1146 in batch and continuous culture. J. Ind. Microbiol. Biotechnol. 1997, 18, 62–67. [Google Scholar] [CrossRef]
- Avonts, L.; Van Uytven, E.; De Vuyst, L. Cell growth and bacteriocin production of probiotic Lactobacillus strains in different media. Int. Dairy J. 2004, 14, 947–955. [Google Scholar] [CrossRef]
- Todorov, S.D.; van Reenan, C.A.; Dicks, L.M.T. Optimization of bacteriocin production by Lactobacillus plantarum ST13BR, a strain isolated from barley beer. J. Gen. Appl. Microbiol. 2004, 50, 149–157. [Google Scholar] [CrossRef]
- Todorov, S.D.; Dicks, L.M.T. Effect of medium components on bacteriocin production by Lactobacillus pentosus ST151BR, a strain isolated from beer produced by the fermentation of maize, barley and soy flour. World J. Microbiol. Biotechnol. 2004, 20, 643–650. [Google Scholar] [CrossRef]
- Malheiros, P.S.; Sant’Anna, V.; Todorov, S.D.; Franco, B.D.G.M. Optimization of growth and bacteriocin production by Lactobacillus sakei subsp. Sakei 2a. Braz. J. Microbiol. 2015, 46, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Ogunbanwo, S.T.; Sanni, A.I.; Onilude, A.A. Influence of cultural conditions on the production of bacteriocin by Lactobacillus brevis OG1. Afr. J. Biotechnol. 2003, 2, 182–192. [Google Scholar]
- Lejeune, R.; Callewaert, R.; Crabbé, K.; De Vuyst, L. Modelling the growth and bacteriocin production by Lactobacillus amylovorus DCE 471 in batch cultivation. J. Appl. Microbiol. 1998, 84, 159–168. [Google Scholar] [CrossRef]
- Todorov, S.; Gotcheva, B.; Dousset, X.; Onno, B.; Ivanova, I. Influence of growth medium on bacteriocin production in Lactobacillus plantarum st31. Biotechnol. Biotechnol. Equip. 2000, 14, 50–55. [Google Scholar] [CrossRef]
- Zamfir, M.; Callewaert, R.; Cornea, P.C.; De Vuyst, L. Production kinetics of acidophilin 801, a bacteriocin produced by Lactobacillus acidophilus IBB 801. FEMS Microbiol. Lett. 2000, 190, 305–308. [Google Scholar] [CrossRef][Green Version]
- Messens, W.; Neysens, P.; Vansieleghem, W.; Vanderhoeven, J.; De Vuyst, L. Modeling growth and bacteriocin production by Lactobacillus amylovorus DCE 471 in response to temperature and pH values used for sourdough fermentations. Appl. Environ. Microbiol. 2002, 68, 1431–1435. [Google Scholar] [CrossRef]
- Li, C.; Bai, J.; Cai, Z.; Ouyang, F. Optimization of a cultural medium for bacteriocin production by Lactococcus lactis using response surface methodology. J. Biotechnol. 2002, 93, 27–34. [Google Scholar] [CrossRef]
- Da Luz, J.A.; Hans, E.; Zeng, A.-P. Automated fast filtration and on-filter quenching improve the intracellular metabolite analysis of microorganisms. Eng. Life Sci. 2014, 14, 135–142. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document: The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Kumar, H.; Salminen, S.; Verhagen, H.; Rowland, I.; Heimbach, J.; Bañares, S.; Young, T.; Nomoto, K.; Lalonde, M. Novel probiotics and prebiotics: Road to the market. Curr. Opin. Biotechnol. 2015, 32, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Aleixandre-Tudó, J.L.; Castelló-Cogollos, L.; Aleixandre, J.L.; Aleixandre-Benavent, R. Tendencies and Challenges in Worldwide Scientific Research on Probiotics. Probiotics Antimicrob. Proteins 2019. [Google Scholar] [CrossRef] [PubMed]
- Ouwehand, A.C.; Salminen, S.; Isolauri, E. Probiotics: An overview of beneficial effects. Antonie van Leeuwenhoek. Int. J. Gen. Mol. Microbiol. 2002, 82, 279–289. [Google Scholar] [CrossRef]
- Leahy, S.C.; Higgins, D.G.; Fitzgerald, G.F.; Van Sinderen, D. Getting better with bifidobacteria. J. Appl. Microbiol. 2005, 98, 1303–1315. [Google Scholar] [CrossRef]
- Pérez-Ramos, A.; Mohedano, M.L.; López, P.; Spano, G.; Fiocco, D.; Russo, D.; Capozzi, V. In situ β-glucan fortification of cereal-based matrices by Pediococcus parvulus 2.6, Technological aspects and prebiotic potential. Int. J. Mol. Sci. 2017, 18, 1588. [Google Scholar] [CrossRef]
- Deo, D.; Davray, D.; Kulkarni, R.A. Diverse repertoire of exopolysaccharide biosynthesis gene clusters in Lactobacillus revealed by comparative analysis in 106 sequenced genomes. Microorganisms 2019, 7, 444. [Google Scholar] [CrossRef]
- Lolou, V.; Panayiotidis, M.I. Functional role of probiotics and prebiotics on skin health and disease. Fermentation 2019, 5, 41. [Google Scholar] [CrossRef]
- Mattila-Sandholm, T.; Myllärinen, P.; Crittenden, R.; Mogensen, G.; Fondén, R.; Saarela, M. Technological challenges for future Probiotic foods. Int. Dairy J. 2002, 12, 173–182. [Google Scholar] [CrossRef]
- Plessas, S.; Bosnea, L.; Alexopoulus, A.; Bezirtzoglou, E. Potenctial effects of probiotics in cheese and yogurt production: A review. Eng. Life Sci. 2012, 12, 433–440. [Google Scholar] [CrossRef]
- Rivière, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and butyrate-producing colon bacteria: Importance and strategies for their stimulation in the human gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef]
- Fiocco, D.; Longo, A.; Arena, M.P.; Russo, P.; Spano, G.; Capozzi, V. How probiotics face food stress: They get by with a little help. Crit. Rev. Food Sci. Nutr. 2019, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Todorov, S.D.; LeBlanc, J.G.; Franco, B.D.G.M. Evaluation of the probiotic potential and effect of encapsulation on survival for Lactobacillus plantarum ST16Pa isolated from papaya. World J. Microbiol. Biotechnol. 2012, 28, 973–984. [Google Scholar] [CrossRef] [PubMed]
- Puebla-Barragan, S.; Reid, G. Forty-five-year evolution of probiotic therapy. Microb. Cell 2019, 6, 184–196. [Google Scholar] [CrossRef] [PubMed]
- Emerenini, E.; Afolabi, O.R.; Okolie, P.I.; Akintokun, A.K. Isolation and Molecular Characterization of Lactic Acid Bacteria Isolated from Fresh Fruits and Vegetables Using Nested PCR Analysis. Br. Microbiol. Res. J. 2013, 3, 368–377. [Google Scholar] [CrossRef]
- Moraes, P.M.; Martins Perin, L.; Silva Júnior, A.; Nero, L.A. Comparison of phenotypic and molecular tests to identify lactic acid bacteria. Braz. J. Microbiol. 2013, 44, 109–112. [Google Scholar] [CrossRef]
- Castillo, N.A.; Perdigán, G.; De Moreno De Leblanc, A. Oral administration of a probiotic Lactobacillus modulates cytokine production and TLR expression improving the immune response against Salmonella enterica serovar Typhimurium infection in mice. BMC Microbiol. 2011, 11. [Google Scholar] [CrossRef]
- García-Ruiz, A.; González de Llano, D.; Esteban-Fernández, A.; Requena, T.; Bartolomé, B.; Moreno-Arribas, M.V. Assessment of probiotic properties in lactic acid bacteria isolated from wine. Food Microbiol. 2014, 44, 220–225. [Google Scholar] [CrossRef]
- Dlamini, Z.C.; Langa, R.L.S.; Aiyegoro, O.A.; Okoh, A.I. Safety Evaluation and Colonisation Abilities of Four Lactic Acid Bacteria as Future Probiotics. Probiotics Antimicrob. Proteins 2019, 11, 397–402. [Google Scholar] [CrossRef]
- Mathara, J.M.; Schillinger, U.; Kutima, P.M.; Mbugua, S.K.; Guigas, C.; Franz, C.; Holzapfel, W.H. Functional properties of Lactobacillus plantarum strains isolated from Maasai traditional fermented milk products in Kenya. Curr. Microbiol. 2008, 56, 315–321. [Google Scholar] [CrossRef]
- Evivie, S.E.; Huo, G.C.; Igene, J.O.; Bian, X. Some current applications, limitations and future perspectives of lactic acid bacteria as probiotics. Food Nutr. Res. 2017, 61, 1318034. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.K.; Salminen, S. The coming of age of probiotics. Trends Food Sci. Technol. 1995, 6, 241–245. [Google Scholar] [CrossRef]
- Klewicki, R.; Klewicka, E. Antagonistic activity of lactic acid bacteria as probiotics against selected bacteria of the Enterobaceriacae family in the presence of polyols and their galactosyl derivatives. Biotechnol. Lett. 2004, 26, 317–320. [Google Scholar] [CrossRef] [PubMed]
- Gad, M.; Ravn, P.; Søborg, D.A.; Lund-Jensen, K.; Ouwehand, A.C.; Jensen, S.S. Regulation of the IL-10/IL-12 axis in human dendritic cells with probiotic bacteria. FEMS Immunol. Med. Microbiol. 2011, 63, 93–107. [Google Scholar] [CrossRef]
- Haghighi, H.R.; Gong, J.; Gyles, C.L.; Hayes, M.A.; Sanei, B.; Parvizi, P.; Gisavi, H.; Chambers, J.R.; Sharif, S. Modulation of antibody-mediated immune response by probiotics in chickens. Clin. Diagn. Lab. Immunol. 2005, 12, 1387–1392. [Google Scholar] [CrossRef]
- Frank, D.N.; St Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef]
- Özdemir, Ö. Various effects of different probiotic strains in allergic disorders: An update from laboratory and clinical data. Clin. Exp. Immunol. 2010, 160, 295–304. [Google Scholar] [CrossRef]
- Malanovic, N.; Lohner, K. Gram-positive bacterial cell envelopes: The impact on the activity of antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2016, 1858, 936–946. [Google Scholar] [CrossRef]
- Kusumoto, S.; Fukase, K.; Fujimoto, Y. Synthesis of Lipopolysaccharide, Peptidoglycan, and Lipoteichoic Acid Fragments. In Comprehensive Glycoscience: From Chemistry to Systems Biology; Elsevier Inc.: Burlington, VT, USA, 2007; pp. 685–711. [Google Scholar]
- Reichmann, N.T.; Gründling, A. Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol. Lett. 2011, 319, 97–105. [Google Scholar] [CrossRef]
- Brown, S.; Santa Maria, J.P.; Walker, S. Wall Teichoic Acids of Gram-Positive Bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef]
- Villéger, R.; Saad, N.; Grenier, K.; Falourd, X.; Foucat, L.; Urdaci, M.C.; Bressollier, P.; Ouk, T.S. Characterization of lipoteichoic acid structures from three probiotic Bacillus strains: Involvement of d-alanine in their biological activity. Antonie Van Leeuwenhoek 2014, 106, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Den Camp, H.J.M.O.; Veerkamp, J.H.; Oosterhof, A.; van Halbeek, H. Structure of the lipoteichoic acids from Bifidobacterium bifidum spp. pennsylvanicum. Biochim. Biophys. Acta BBA Lipids Lipid Metab. 1984, 795, 301–313. [Google Scholar] [CrossRef]
- Ginsburg, I. Role of lipoteichoic acid in infection and inflammation. Lancet Infect. Dis. 2002, 2, 171–179. [Google Scholar] [CrossRef]
- Weill, F.S.; Cela, E.M.; Paz, M.L.; Ferrari, A.; Leoni, J.; González-Maglio, D.H. Lipoteichoic acid from Lactobacillus rhamnosus GG as an oral photoprotective agent against UV-induced carcinogenesis. Br. J. Nutr. 2013, 109, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Brauweiler, A.M.; Goleva, E.; Leung, D.Y.M. Staphylococcus aureus Lipoteichoic Acid Damages the Skin Barrier through an IL-1–Mediated Pathway. J. Investig. Dermatol. 2019, 139, 1753–1761. [Google Scholar] [CrossRef] [PubMed]
- Coley, J.; Duckworth, M.; Baddiley, J. Extraction and purification of lipoteichoic acids from gram-positive bacteria. Carbohydr. Res. 1975, 40, 41–52. [Google Scholar] [CrossRef]
- Morath, S.; Geyer, A.; Hartung, T. Structure-function relationship of cytokine induction by lipoteichoic acid from Staphylococcus aureus. J. Exp. Med. 2001, 193, 393–397. [Google Scholar] [CrossRef]
- Mays, Z.J.; Nair, N.U. Synthetic biology in probiotic lactic acid bacteria: At the frontier of living therapeutics. Curr. Opin. Biotechnol. 2018, 53, 224–231. [Google Scholar] [CrossRef]
- Felis, G.E.; Salvetti, E.; Torriani, S. Systematics of Lactic Acid Bacteria: Current Status. In Biotechnology of Lactic Acid Bacteria: Novel Applications, 2nd ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2015; pp. 25–31. [Google Scholar]
- Ogawa, J.; Kishino, S.; Ando, A.; Sugimoto, S.; Mihara, K.; Shimizu, S. Production of conjugated fatty acids by lactic acid bacteria. J. Biosci. Bioeng. 2005, 100, 355–364. [Google Scholar] [CrossRef]
- Lee, J.; Lee, M.H.; Cho, E.J.; Lee, S. High-yield methods for purification of α-linolenic acid from Perilla frutescens var. japonica oil. Appl. Biol. Chem. 2016, 59, 89–94. [Google Scholar] [CrossRef]
- Ayorinde, F.O.; Osman, G.; Shepard, R.L.; Powers, F.T. Synthesis of azelaic acid and suberic acid from Vernonia galamensis oil. J. Am. Oil Chem. Soc. 1988, 65, 1774–1777. [Google Scholar] [CrossRef]
- Choi, K.; Jeon, B.S.; Kim, B.C.; Oh, M.K.; Um, Y.; Sang, B.I. In situ biphasic extractive fermentation for hexanoic acid production from sucrose by Megasphaera elsdenii NCIMB 702410. Appl. Biochem. Biotechnol. 2013, 171, 1094–1107. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, V.; Liu, J.; Dantoft, S.H.; Solem, C.; Jensen, P.R. Synthesis of (3R)-acetoin and 2,3-butanediol isomers by metabolically engineered Lactococcus lactis. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Suomalainen, T.; Mäyrä-Mäkinen, A.; Huttunen, E. Antimicrobial activity of 2-Pyrrolidone-5-Carboxylic acid produced by lactic acid bacteria. J. Food Prot. 1997, 60, 786–790. [Google Scholar] [CrossRef]
- Le Lay, C.; Coton, E.; Le Blay, G.; Chobert, J.M.; Haertlé, T.; Choiset, Y.; Van Long, N.N.; Meslet-Cladière, L.; Mounier, J. Identification and quantification of antifungal compounds produced by lactic acid bacteria and propionibacteria. Int. J. Food Microbiol. 2016, 239, 79–85. [Google Scholar] [CrossRef]
- Parada, J.L.; Caron, C.R.; Medeiros, A.B.P.; Soccol, C.R. Bacteriocins from lactic acid bacteria: Purification, properties and use as biopreservatives. Braz. Arch. Biol. Technol. 2007, 50, 521–542. [Google Scholar] [CrossRef]
- Ebner, P.; Reichert, S.; Luqman, A.; Krismer, B.; Popella, P.; Götz, F. Lantibiotic production is a burden for the producing staphylococci. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Corsetti, A.; Gobbetti, M.; Rossi, J.; Damiani, P. Antimould activity of sourdough lactic acid bacteria: Identification of a mixture of organic acids produced by Lactobacillus sanfrancisco CB1. Appl. Microbiol. Biotechnol. 1998, 50, 253–256. [Google Scholar] [CrossRef]
- Kwak, M.K.; Liu, R.; Kwon, J.O.; Kim, M.K.; Kim, A.H.; Kang, S.O. Cyclic dipeptides from lactic acid bacteria inhibit proliferation of the influenza a virus. J. Microbiol. 2013, 51, 836–843. [Google Scholar] [CrossRef]
- Ryan, L.A.M.; Fabio, D.B.; Arendt, E.K.; Koehler, P. Detection and quantitation of 2,5-diketopiperazines in wheat sourdough and bread. J. Agric. Food Chem. 2009, 57, 9563–9568. [Google Scholar] [CrossRef]
- Raveschot, C.; Cudennec, B.; Coutte, F.; Flahaut, C.; Fremont, M.; Drider, D.; Dhulster, P. Production of bioactive peptides by Lactobacillus species: From gene to application. Front. Microbiol. 2018, 9, 2354. [Google Scholar] [CrossRef]
- Oberman, H.; Piątkiewicz, A.; Libudzisz, Z. Production of diacetyl and acetoin by lactic acid bacteria. Food Nahrung. 1982, 26, 615–623. [Google Scholar] [CrossRef]
- Krampitz, L.O. Preparation and determination of acetoin, diacetyl, and acetolactate. Methods Enzymol. 1957, 3, 277–283. [Google Scholar] [CrossRef]
- Liu, Q.; Huang, X.; Yang, D.; Si, T.; Pan, S.; Yang, F. Yield improvement of exopolysaccharides by screening of the Lactobacillus acidophilus ATCC and optimization of the fermentation and extraction conditions. EXCLI J. 2016, 15, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Niku-Paavola, M.-L.; Laitila, A.; Mattila-Sandholm, T.; Haikara, A. New types of antimicrobial compounds produced by Lactobacillus plantarum. J. Appl. Microbiol. 1999, 86, 29–35. [Google Scholar] [CrossRef]
- Lavermicocca, P.; Valerio, F.; Evidente, A.; Lazzaroni, S.; Corsetti, A.; Gobbetti, M. Purification and characterization of novel antifungal compounds from the sourdough Lactobacillus plantarum strain 21B. Appl. Environ. Microbiol. 2000, 66, 4084–4090. [Google Scholar] [CrossRef]
- Vollenweider, S.; Grassi, G.; König, I.; Puhan, Z. Purification and structural characterization of 3-hydroxypropionaldehyde and its derivatives. J. Agric. Food Chem. 2003, 51, 3287–3293. [Google Scholar] [CrossRef]
- Patra, F.; Tomar, S.K.; Arora, S. Technological and functional applications of low-calorie sweeteners from lactic acid bacteria. J. Food Sci. 2009, 74, R16–R23. [Google Scholar] [CrossRef]
- Leblanc, J.G.; Laiño, J.E.; del Valle, M.J.; Vannini, V.; van Sinderen, D.; Taranto, M.P.; de Valdez, G.F.; de Giori, G.S.; Sesma, F. B-Group vitamin production by lactic acid bacteria - current knowledge and potential applications. J. Appl. Microbiol. 2011, 111, 1297–1309. [Google Scholar] [CrossRef]
- Ge, Y.-Y.; Zhang, J.-R.; Corke, H.; Gan, R.-Y. Screening and Spontaneous Mutation of Pickle-Derived Lactobacillus plantarum with Overproduction of Riboflavin, Related Mechanism, and Food Application. Foods 2020, 9, 88. [Google Scholar] [CrossRef]
- Bertsch, A.; Roy, D.; LaPointe, G. Enhanced exopolysaccharide production by Lactobacillus rhamnosus in co-culture with Saccharomyces cerevisiae. Appl. Sci. 2019, 9. [Google Scholar] [CrossRef]
- Ziadi, M.; Bouzaiene, T.; M’Hir, S.; Zaafouri, K.; Mokhtar, F.; Hamdi, M.; Boisset-Helbert, C. Evaluation of the efficiency of ethanol precipitation and ultrafiltration on the purification and characteristics of exopolysaccharides produced by three lactic acid bacteria. Biomed. Res. Int. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Tagliazucchi, D.; Martini, S.; Solieri, L. Bioprospecting for Bioactive Peptide Production by Lactic Acid Bacteria Isolated from Fermented Dairy Food. Fermentation 2019, 5, 96. [Google Scholar] [CrossRef]
- Claus, H.; Mojsov, K. Enzymes for wine fermentation: Current and perspective applications. Fermentation 2018, 4, 52. [Google Scholar] [CrossRef]
- Arancon, R.A.D.; Lin, C.S.K.; Chan, K.M.; Kwan, T.H.; Luque, R. Advances on waste valorization: New horizons for a more sustainable society. Energy Sci. Eng. 2013, 1, 53–71. [Google Scholar] [CrossRef]
- Bustamante, D.; Tortajada, M.; Ramón, D.; Rojas, A. Production of D-Lactic Acid by the Fermentation of Orange Peel Waste Hydrolysate by Lactic Acid Bacteria. Fermentation 2019, 6, 1. [Google Scholar] [CrossRef]
- Olszewska-Widdrat, A.; Alexandri, M.; López-Gómez, J.P.; Schneider, R.; Mandl, M.; Venus, J. Production and purification of L-lactic acid in lab and pilot scales using sweet sorghum juice. Fermentation 2019, 5. [Google Scholar] [CrossRef]
- Ghimire, A.; Trably, E.; Frunzo, L.; Pirozzi, F.; Lens, P.N.L.; Esposito, G.; Cazier, E.A.; Escudié, R. Effect of total solids content on biohydrogen production and lactic acid accumulation during dark fermentation of organic waste biomass. Bioresour. Technol. 2018, 248, 180–186. [Google Scholar] [CrossRef]
- Nguyen, Q.A.; Cho, E.; Trinh, L.T.P.; Jeong, J.S.; Bae, H.J. Development of an integrated process to produce D-mannose and bioethanol from coffee residue waste. Bioresour. Technol. 2017, 244, 1039–1048. [Google Scholar] [CrossRef]
- López-Gómez, J.P.; Alexandri, M.; Schneider, R.; Latorre-Sánchez, M.; Lozano, C.C.; Venus, J. Organic fraction of municipal solid waste for the production of L-lactic acid with high optical purity. J. Clean. Prod. 2019, 247, 119165. [Google Scholar] [CrossRef]
- Berbegal, C.; Fragasso, M.; Russo, P.; Bimbo, F.; Grieco, F.; Spano, G.; Capozzi, V. Climate changes and food quality: The potential of microbial activities as mitigating strategies in the wine sector. Fermentation 2019, 5, 85. [Google Scholar] [CrossRef]
- Sijtsema, S.J.; Snoek, H.M.; van Haaster-de Winter, M.A.; Dagevos, H. Let’s Talk about Circular Economy: A Qualitative Exploration of Consumer Perceptions. Sustainability 2019, 12, 286. [Google Scholar] [CrossRef]
- De Vero, L.; Boniotti, M.B.; Budroni, M.; Buzzini, P.; Cassanelli, S.; Comunian, R.; Gullo, M.; Logrieco, A.F.; Mannazzu, I.; Musumeci, R.; et al. Preservation, characterization and exploitation of microbial biodiversity: The perspective of the italian network of culture collections. Microorganisms 2019, 7, 685. [Google Scholar] [CrossRef] [PubMed]
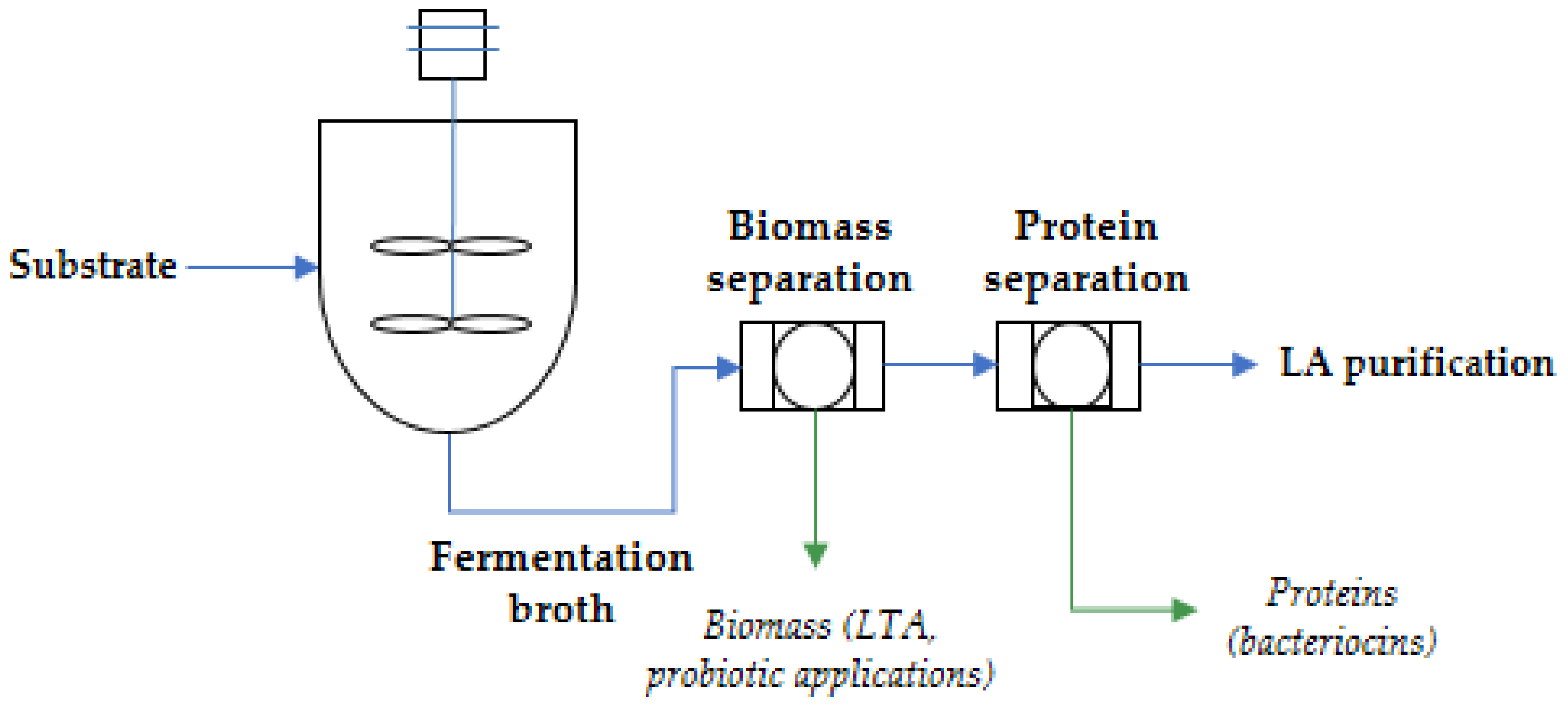
| Microorganism | Optimized Bacteriocin Level (AU∙mL−1 × 103) | Media | Relevant Media Component b | pH c | T (°C) | Other Considerations d | Reference |
|---|---|---|---|---|---|---|---|
| L. lactis subsp. lactis | 131 | M17 broth | Lactose (3.0%) | 6.0 | 30 | Early stationary phase | [60] |
| L. amylovorus | 25.6 | Sourdough simulation medium | NaCl (10 g∙L−1) | 5.4 | 37 | NR | [61] |
| L. amylovorus | 25.6 | Modified MRS | Glucose (11 g∙L−1), nitrogen mixture (25 g∙L−1) | 5.0 | 37 | Continuous fed-batch fermentation | [62] |
| L. lactis sbsp. lactis | 15.4 | Optimized medium | Various ingredients | 5.5 | 30 | Max activity after 7 h | [63] |
| L. acidophilus | 12.8 | MRS broth | Glucose (20 g∙L−1) | 6.5 | 37 | 100 rpm | [64] |
| L. plantarum | 12.8 | MRS broth | Tryptone (10 g∙L−1), meat extract (5 g∙L−1), yeast extract (5 g∙L−1), maltose (20 g∙L−1), mannose (20 g∙L−1) | 5.5–6.5 | 30 | Max activity after 14 h | [64,65] |
| L. pentosus | 6.4 | MRS | Tryptone (12.5 g∙L−1), meat extract (7.5 g∙L−1), maltose (20 g∙L−1), glucose (10–20 g∙L−1), no glycerol | n.r. | 30 | NR | [66] |
| L. sakei sbsp. sakei | 10.9 | MRS broth | Glucose (5.5 g∙L−1) and Tween-80 (10.5 µL∙mL−1) | 6.0 | 30 | Media supplemented with Tween-20, sodium citrate, KCl, and cysteine | [67] |
| Ln. mesenteroides | 10.2 | MRS | Glucose (4.5%), peptone (8%), yeast extract (1.5%) | 5.5 | 25 | NR | [67] |
| L. plantarum | 9.0 | MRS | KH2PO4 (2, 5, and 10 g∙L−1) | n.r. | 30 | [64,65] | |
| L. brevis | 6.4 | MRS | Yeast extract (2–3%), NaCl (1–2%), glucose (1%), Tween-80 (0.5%) | n.c. | 30 | Final pH between 3.86 and 4.04 | [68] |
| L. amylovorus | 6.4 | MRS | Glucose (20 g∙L−1) | n.r. | 37 | NR | [69] |
| L. plantarum | 6.4 | MRS | Meat extract (1.5%), yeast extract (1%), biotin (0.01 mg∙L−1) | 6.5 | 30 | NR | [70] |
| L. plantarum | 6.4 | MRS | Meat and yeast extract (20 g∙L−1), maltose (3.0%), no glycerol | 6.5 | 30 | NR | [64,65] |
| L. acidophilus | 6.4 | MRS | Glucose (20 g∙L−1) | 6.0 | 37 | 100 rpm | [71] |
| L. amylovorus | 5.5 | Sourdough simulation medium | Yeast extract (12 g∙L−1), tryptone (10 g∙L−1) | 5.4 | 37 | NR | [72] |
| L. lactis | 2.1 | Cultural medium | Soybean peptone (4.49 g∙L−1), KH2PO4 (28.42 g∙L−1) | 6.8 | 30 | Shaking at 180 rpm | [73] |
| Metabolite | Titer | Microorganism | Location within The Fermentation | Biological Activity | Downstream | Reference |
|---|---|---|---|---|---|---|
| 2,3 butanediol | 32 g/L | Lactococcus lactis | Supernatant | Bulk chemical in plastic industry | Distillation, stream stripping, pervaporation | [120] |
| 2-pyrrolidone-5-carboxylic acid (Pyroglutamic acid) | - | Lactobacillus spp. Pediococcus spp. | Supernatant | Antimicrobial | Ethanol precipitation, gel filtration, and anion exchange | [121] |
| Azelaic acid | 2.71 mg/L | Leuconostoc citreum L123 | Supernatant | Antifungal | Organic extraction | [122] |
| Bacteriocins | 0.72 g/L | Staphylococcus gallinarum and S. epidermidis | Supernatant | Antimicrobial | Salting-out, solvent extraction, ultrafiltration, adsorption–desorption, ion exchange, and size exclusion chromatography | [123,124] |
| Caproic acid | 102 mg/L | Lactobacillus sanfrancisco CB1 | Supernatant | Antimicrobial, Flavor, and fuel precursor | Organic extraction | [125] |
| Conjugated linoleic acid | 40 g/L | Bifidobacterium spp., Propionibacterium freudenreichii, Lactobacillus plantarum AKU 1009a | Intracellular or cell-associated | Reduces carcinogenesis, atherosclerosis, and body fat | Intracellular (or associated with cells) and extracellular; urea treatment after organic extraction | [116,117] |
| Cyclic dipeptides | - | Lactobacillus spp., Leuconostoc spp., Weissella spp., and Lactococcus lactis | Supernatant | Antiviral, antifungal | Selective precipitation (ethanol, trichloroacetic acid, or ammonium sulfate); ultra and nano-filtration; chromatographic methods. | [126,127,128] |
| Diacetyl and acetoin | DC 3.5 mg/L AMC 2.6 g/L | Leuconostoc sp., Streptococcus diacerylactis | Supernatant | Flavor and fragrance | Distillation at 86–87 °C and reactive distillation | [129,130] |
| Exopoly-saccharides | 5.12 g/L | Lactobacillus acidophilus | Supernatant | Antioxidant, antibacterial, antiulcer, antitumor, immunostimulatory | Ethanol precipitation | [131] |
| Lipoteichoic acid | - | Staphylococcus aureus Lactobacillus rhamnosus GG | Cell-associated | Immunomodulator | Organic extraction, hydrophobic interaction, chromatography | [110,111] |
| Mevalonic acid Mevalonolactone | - | Lactobacillus plantarum VTT E-78076 | Supernatant | Antifungal | Ultra and nano-filtration | [132] |
| Phenyl lactic and p-hydroxyphenyl acetic acid | - | Lactobacillus plantarum strain 21B | Supernatant | Antifungal | Ultra and nano-filtration | [133] |
| Reuterin (3-hydroxypropionaldehyde) | 8 mg/L | Lactobacillus reuteri | Supernatant | Antimicrobial | Alcoholic extraction, organic extraction, size filtration, and ion exchange; distillation | [133,134] |
| Sweeteners (mannitol, tagatose, sorbitol, trehalose) | - | Supernatant | Food industry | Ultra and nano-filtration, chromatographic methods | [135] | |
| Vitamins (B-group) | - | Supernatant | Food supplement | Ultra and nano-filtration, chromatographic methods | [136,137] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mora-Villalobos, J.A.; Montero-Zamora, J.; Barboza, N.; Rojas-Garbanzo, C.; Usaga, J.; Redondo-Solano, M.; Schroedter, L.; Olszewska-Widdrat, A.; López-Gómez, J.P. Multi-Product Lactic Acid Bacteria Fermentations: A Review. Fermentation 2020, 6, 23. https://doi.org/10.3390/fermentation6010023
Mora-Villalobos JA, Montero-Zamora J, Barboza N, Rojas-Garbanzo C, Usaga J, Redondo-Solano M, Schroedter L, Olszewska-Widdrat A, López-Gómez JP. Multi-Product Lactic Acid Bacteria Fermentations: A Review. Fermentation. 2020; 6(1):23. https://doi.org/10.3390/fermentation6010023
Chicago/Turabian StyleMora-Villalobos, José Aníbal, Jéssica Montero-Zamora, Natalia Barboza, Carolina Rojas-Garbanzo, Jessie Usaga, Mauricio Redondo-Solano, Linda Schroedter, Agata Olszewska-Widdrat, and José Pablo López-Gómez. 2020. "Multi-Product Lactic Acid Bacteria Fermentations: A Review" Fermentation 6, no. 1: 23. https://doi.org/10.3390/fermentation6010023
APA StyleMora-Villalobos, J. A., Montero-Zamora, J., Barboza, N., Rojas-Garbanzo, C., Usaga, J., Redondo-Solano, M., Schroedter, L., Olszewska-Widdrat, A., & López-Gómez, J. P. (2020). Multi-Product Lactic Acid Bacteria Fermentations: A Review. Fermentation, 6(1), 23. https://doi.org/10.3390/fermentation6010023