Molecular and Phenotypic Characterization of Metschnikowia pulcherrima Strains from Douro Wine Region
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeasts Strains and Maintenance Conditions
2.2. Molecular Characterization
DNA Extraction and Amplification
2.3. Phenotypic Characterization
2.3.1. Stress Resistance Assays
2.3.2. Evaluation of Enzymatic Activities
2.4. Single and Mixed-Culture Fermentations
2.4.1. Grape-Juice Preparation
2.4.2. Inocula Preparation and Fermentation Conditions
2.4.3. Determination of Yeast Growth and Fermentation Parameters
2.5. Analytical Determinations
2.6. Statistical Analysis
3. Results and Discussion
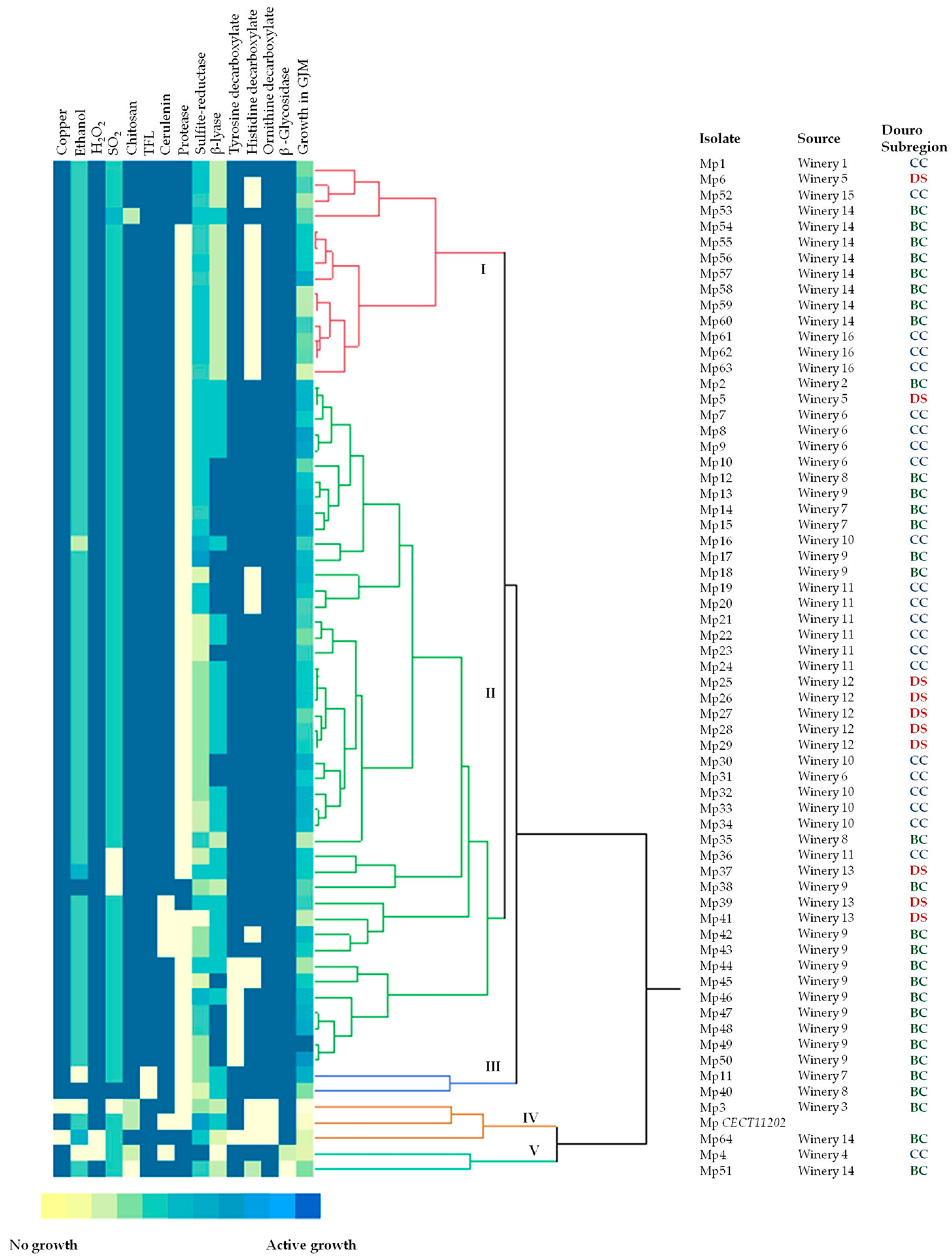
3.1. Molecular Characterization
3.2. Phenotypic Characterization
3.3. S. cerevisiae and M. pulcherrima Single and Mixed Fermentations
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Heard, G.M.; Fleet, G.H. Growth of natural yeast flora during the fermentation of inoculated wines. Appl. Environ. Microbiol. 1985, 50, 727–728. [Google Scholar] [PubMed]
- Henick-Kling, T.; Edinger, W.; Daniel, P.; Monk, P. Selective effects of sulfur dioxide and yeast starter culture addition on indigenous yeast populations and sensory characteristics of wine. J. Appl. Microbiol. 1998, 84, 865–876. [Google Scholar] [CrossRef]
- Jolly, N.P.; Augustyn, O.P.R.; Pretorius, I.S. The effect of non-Saccharomyces yeasts on fermentation and wine quality. S. Afr. J. Enol. Vitic. 2003, 24, 55–62. [Google Scholar] [CrossRef]
- Comitini, F.; Gobbi, M.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 2011, 28, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Caruso, M.; Fiore, C.; Contursi, M.; Salzano, G.; Paparella, A.; Romano, P. Formation of biogenic amines as criteria for the selection of wine yeasts. World J. Microbiol. Biotechnol. 2002, 18, 159–163. [Google Scholar] [CrossRef]
- Pretorius, I.S. Tailoring wine yeast for the new millennium: Novel approaches to the ancient art of winemaking. Yeast 2000, 16, 675–729. [Google Scholar] [CrossRef]
- Romano, P.; Fiore, C.; Paraggio, M.; Caruso, M.; Capece, A. Function of yeast species and strains in wine flavour. Int. J. Food Microbiol. 2003, 86, 169–180. [Google Scholar] [CrossRef]
- Jolly, N.; Augustyn, O.P.H.; Pretorius, I.S. The use of Candida pulcherrima in combination with Saccharomyces cerevisiae for the production of chenin blanc wine. S. Afr. J. Enol. Vitic. 2003, 24, 63–69. [Google Scholar] [CrossRef]
- Varela, C.; Siebert, T.; Cozzolino, D.; Rose, L.; McLean, H.; Henschke, P.A. Discovering a chemical basis for differentiating wines made by fermentation with ‘wild’ indigenous and inoculated yeasts: Role of yeast volatile compounds. Aust. J. Grape Wine Res. 2009, 15, 238–248. [Google Scholar] [CrossRef]
- Mateo, J.J.; Di Stefano, R. Description of the β-glucosidase activity of wine yeasts. Food Microbiol. 1997, 14, 583–591. [Google Scholar] [CrossRef]
- Esteve-Zarzoso, B.; Manzanares, P.; Ramón, D.; Querol, A. The role of non-Saccharomyces yeasts in industrial winemaking. Int. Microbiol. 1998, 1, 143–148. [Google Scholar] [PubMed]
- Swiegers, J.H.; Capone, D.L.; Pardon, K.H.; Elsey, G.M.; Sefton, M.A.; Francis, I.L.; Pretorius, I.S. Engineering volatile thiol release in Saccharomyces cerevisiae for improved wine aroma. Yeast 2007, 24, 561–574. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.; Maicas, S. Application of non-Saccharomyces yeasts to wine-making process. Fermentation 2016, 2, 14. [Google Scholar] [CrossRef]
- Capece, A.; Romaniello, R.; Siesto, G.; Pietrafesa, R.; Massari, C.; Poeta, C.; Romano, P. Selection of indigenous Saccharomyces cerevisiae strains for nero d’avola wine and evaluation of selected starter implantation in pilot fermentation. Int. J. Food Microbiol. 2010, 144, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Tristezza, M.; Fantastico, L.; Vetrano, C.; Bleve, G.; Corallo, D.; Grieco, F.; Mita, G.; Grieco, F. Molecular and technological characterization of Saccharomyces cerevisiae strains isolated from natural fermentation of susumaniello grape must in apulia, southern italy. Int. J. Microbiol. 2014, 2014, 897428. [Google Scholar] [CrossRef] [PubMed]
- Mestre Furlani, M.V.; Maturano, Y.P.; Combina, M.; Mercado, L.A.; Toro, M.E.; Vazquez, F. Selection of non-Saccharomyces yeasts to be used in grape musts with high alcoholic potential: A strategy to obtain wines with reduced ethanol content. FEMS Yeast Res. 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Ciani, M.; Maccarelli, F. Oenological properties of non-Saccharomyces yeasts associated with wine-making. World J. Microbiol. Biotechnol. 1997, 14, 199–203. [Google Scholar] [CrossRef]
- Fleet, G.H. Wine yeasts for the future. FEMS Yeast Res. 2008, 8, 979–995. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Lepe, J.A.; Morata, A. New trends in yeast selection for winemaking. Trends Food Sci. Technol. 2012, 23, 39–50. [Google Scholar]
- Ciani, M.; Beco, L.; Comitini, F. Fermentation behaviour and metabolic interactions of multistarter wine yeast fermentations. Int. J. Food Microbiol. 2006, 108, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Rantsiou, K.; Dolci, P.; Giacosa, S.; Torchio, F.; Tofalo, R.; Torriani, S.; Suzzi, G.; Rolle, L.; Cocolin, L. Candida zemplinina can reduce acetic acid produced by Saccharomyces cerevisiae in sweet wine fermentations. Appl. Environ. Microbiol. 2012, 78, 1987–1994. [Google Scholar] [CrossRef] [PubMed]
- Lage, P.; Barbosa, C.; Mateus, B.; Vasconcelos, I.; Mendes-Faia, A.; Mendes-Ferreira, A. H. guilliermondii impacts growth kinetics and metabolic activity of S. cerevisiae: The role of initial nitrogen concentration. Int. J. Food Microbiol. 2014, 172, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Rossouw, D.; Bauer, F.F. Exploring the phenotypic space of non-Saccharomyces wine yeast biodiversity. Food Microbiol. 2016, 55, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Jolly, N.P.; Varela, C.; Pretorius, I.S. Not your ordinary yeast: Non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 2014, 14, 215–237. [Google Scholar] [CrossRef] [PubMed]
- Ciani, M.; Comitini, F. Non-Saccharomyces wine yeasts have a promising role in biotechnological approaches to winemaking. Ann. Microbiol. 2011, 61, 25–32. [Google Scholar] [CrossRef]
- Sadoudi, M.; Tourdot-Maréchal, R.; Rousseaux, S.; Steyer, D.; Gallardo-Chacón, J.-J.; Ballester, J.; Vichi, S.; Guérin-Schneider, R.; Caixach, J.; Alexandre, H. Yeast–yeast interactions revealed by aromatic profile analysis of Sauvignon blanc wine fermented by single or co-culture of non-Saccharomyces and Saccharomyces yeasts. Food Microbiol. 2012, 32, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Quiros, M.; Rojas, V.; Gonzalez, R.; Morales, P. Selection of non-Saccharomyces yeast strains for reducing alcohol levels in wine by sugar respiration. Int. J. Food Microbiol. 2014, 181, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Contreras, A.; Hidalgo, C.; Henschke, P.A.; Chambers, P.J.; Curtin, C.; Varela, C. Evaluation of non-Saccharomyces yeasts for the reduction of alcohol content in wine. Appl. Environ. Microbiol. 2014, 80, 1670–1678. [Google Scholar] [CrossRef] [PubMed]
- Morales, P.; Rojas, V.; Quiros, M.; Gonzalez, R. The impact of oxygen on the final alcohol content of wine fermented by a mixed starter culture. Appl. Microbiol. Biotechnol. 2015, 99, 3993–4003. [Google Scholar] [CrossRef] [PubMed]
- Ciani, M.; Morales, P.; Comitini, F.; Tronchoni, J.; Canonico, L.; Curiel, J.A.; Oro, L.; Rodrigues, A.J.; Gonzalez, R. Non-conventional yeast species for lowering ethanol content of wines. Front. Microbiol. 2016, 7, 642. [Google Scholar] [CrossRef] [PubMed]
- Alonso-del-Real, J.; Lairón-Peris, M.; Barrio, E.; Querol, A. Effect of temperature on the prevalence of Saccharomyces non cerevisiae species against a S. cerevisiae wine strain in wine fermentation: Competition, physiological fitness, and influence in final wine composition. Front. Microbiol. 2017, 8, 150. [Google Scholar] [CrossRef] [PubMed]
- Escott, C.; Del Fresno, J.M.; Loira, I.; Morata, A.; Tesfaye, W.; González, M.C.; Suárez-Lepe, J.A. Formation of polymeric pigments in red wines through sequential fermentation of flavanol-enriched musts with non-Saccharomyces yeasts. Food Chem. 2018, 239, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Loira, I.; Vejarano, R.; Bañuelos, M.A.; Morata, A.; Tesfaye, W.; Uthurry, C.; Villa, A.; Cintora, I.; Suárez-Lepe, J.A. Influence of sequential fermentation with Torulaspora delbrueckii and Saccharomyces cerevisiae on wine quality. LWT-Food Sci. Technol. 2014, 59, 915–922. [Google Scholar] [CrossRef]
- Morata, A.; Benito, S.; Loira, I.; Palomero, F.; González, M.C.; Suárez-Lepe, J.A. Formation of pyranoanthocyanins by Schizosaccharomyces pombe during the fermentation of red must. Int. J. Food Microbiol. 2012, 159, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Petruzzi, L.; Capozzi, V.; Berbegal, C.; Corbo, M.R.; Bevilacqua, A.; Spano, G.; Sinigaglia, M. Microbial resources and enological significance: Opportunities and benefits. Front. Microbiol. 2017, 8, 995. [Google Scholar] [CrossRef] [PubMed]
- Belda, I.; Navascues, E.; Marquina, D.; Santos, A.; Calderon, F.; Benito, S. Dynamic analysis of physiological properties of Torulaspora delbrueckii in wine fermentations and its incidence on wine quality. Appl. Microbiol. Biotechnol. 2015, 99, 1911–1922. [Google Scholar] [CrossRef] [PubMed]
- Capozzi, V.; Garofalo, C.; Chiriatti, M.A.; Grieco, F.; Spano, G. Microbial terroir and food innovation: The case of yeast biodiversity in wine. Microbiol. Res. 2015, 181, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, C.; Russo, P.; Beneduce, L.; Massa, S.; Spano, G.; Capozzi, V. Non-Saccharomyces biodiversity in wine and the ‘microbial terroir’: A survey on nero di troia wine from the Apulian region, Italy. Ann. Microbiol. 2016, 66, 143–150. [Google Scholar] [CrossRef]
- Tristezza, M.; Tufariello, M.; Capozzi, V.; Spano, G.; Mita, G.; Grieco, F. The oenological potential of Hanseniaspora uvarum in simultaneous and sequential co-fermentation with Saccharomyces cerevisiae for industrial wine production. Front. Microbiol. 2016, 7, 670. [Google Scholar] [CrossRef] [PubMed]
- Looke, M.; Kristjuhan, K.; Kristjuhan, A. Extraction of genomic DNA from yeasts for pcr-based applications. BioTechniques 2011, 50, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Huey, B.; Hall, J. Hypervariable DNA fingerprinting in Escherichia coli: Minisatellite probe from bacteriophage M13. J. Bacteriol. 1989, 171, 2528–2532. [Google Scholar] [CrossRef] [PubMed]
- Lieckfeldt, E.; Meyer, W.; Borner, T. Rapid identification and differentiation of yeasts by DNA and pcr fingerprinting. J. Basic Microbiol. 1993, 33, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, V.A.; Vicente, M.A.; Fietto, L.G.; de Miranda Castro, I.; Coutrim, M.X.; Schüller, D.; Alves, H.; Casal, M.; de Oliveira Santos, J.; Araújo, L.D.; et al. Biochemical and molecular characterization of Saccharomyces cerevisiae strains obtained from sugar-cane juice fermentations and their impact in cachaça production. Appl. Environ. Microbiol. 2008, 74, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Franco-Duarte, R.; Umek, L.; Zupan, B.; Schuller, D. Computational approaches for the genetic and phenotypic characterization of a Saccharomyces cerevisiae wine yeast collection. Yeast 2009, 26, 675–692. [Google Scholar] [CrossRef] [PubMed]
- Jiranek, V.; Langridge, P.; Henschke, P.A. Regulation of hydrogen sulfide liberation in wine-producing Saccharomyces cerevisiae strains by assimilable nitrogen. Appl. Environ. Microbiol. 1995, 61, 461–467. [Google Scholar] [PubMed]
- Mendes-Ferreira, A.; Mendes-Faia, A.; Leao, C. Survey of hydrogen sulphide production by wine yeasts. J. Food Prot. 2002, 65, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, C.; Lage, P.; Vilela, A.; Mendes-Faia, A.; Mendes-Ferreira, A. Phenotypic and metabolic traits of commercial Saccharomyces cerevisiae yeasts. AMB Express 2014, 4, 39. [Google Scholar] [CrossRef] [PubMed]
- Belda, I.; Ruiz, J.; Alastruey Izquierdo, A.; Navascues, E.; Marquina, D.; Santos, A. Unraveling the enzymatic basis of wine “flavorome”: A phylo-functional study of wine related yeast species. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- González, J.; Gallardo, C.; Pombar, A.; Rego, P.; A Rodríguez, L. Determination of enzymatic activities in ecotypic Saccharomyces and non-Saccharomyces yeast. Electron. J. Environ. Agric. Food Chem. 2004, 3, 743–750. [Google Scholar]
- Arévalo Villena, M.; Úbeda Iranzo, J.F.; Cordero Otero, R.R.; Briones Pérez, A.I. Optimization of a rapid method for studying the cellular location of β-glucosidase activity in wine yeasts. J. Appl. Microbiol. 2005, 99, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Granchi, L.; Romano, P.; Mangani, S.; Guerrini, S.; Vincenzini, M. Production of biogenic amines by wine microorganisms. Bull. l’OIV Off. Int. Vigne Vin 2005, 78, 595–609. [Google Scholar]
- Mendes-Ferreira, A.; Barbosa, C.; Falco, V.; Leao, C.; Mendes-Faia, A. The production of hydrogen sulphide and other aroma compounds by wine strains of Saccharomyces cerevisiae in synthetic media with different nitrogen concentrations. J. Ind. Microbiol. Biotechnol. 2009, 36, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Aerny, J. Composés azotés des moûts et des vins. Rev. Suisse Vitic. Arboric. Hortic. 1996, 28, 161–165. [Google Scholar]
- Ugliano, M.; Henschke, P.A. Yeasts and wine flavour. In Wine Chemistry and Biochemistry; Moreno-Arribas, M.V., Polo, M.C., Eds.; Springer: New York, NY, USA, 2009; pp. 313–392. [Google Scholar]
- Libkind, D.; Ruffini, A.; van Broock, M.; Alves, L.; Sampaio, J.P. Biogeography, host specificity, and molecular phylogeny of the basidiomycetous yeast phaffia rhodozyma and its sexual form, xanthophyllomyces dendrorhous. Appl. Environ. Microbiol. 2007, 73, 1120–1125. [Google Scholar] [CrossRef] [PubMed]
- Dunn, B.; Richter, C.; Kvitek, D.J.; Pugh, T.; Sherlock, G. Analysis of the Saccharomyces cerevisiae pan-genome reveals a pool of copy number variants distributed in diverse yeast strains from differing industrial environments. Genome Res. 2012, 22, 908–924. [Google Scholar] [CrossRef] [PubMed]
- Aponte, M.; Blaiotta, G. Potential role of yeast strains isolated from grapes in the production of taurasi docg. Front. Microbiol. 2016, 7, 809. [Google Scholar] [CrossRef] [PubMed]
- Berbegal, C.; Spano, G.; Tristezza, M.; Grieco, F.; Capozzi, V. Microbial resources and innovation in the wine production sector. S. Afr. J. Enol. Vitic. 2017, 38, 156–166. [Google Scholar] [CrossRef]
- Bagder Elmaci, S.; Gulgor, G.; Tokatli, M.; Erten, H.; Isci, A.; Ozcelik, F. Effectiveness of chitosan against wine-related microorganisms. Antonie Van Leeuwenhoek 2015, 107, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Cavazza, A.; Guzzon, R.; Malacarne, M.; Larcher, R. The influence of the copper content in grape must on alcoholic fermentation kinetics and wine quality. A survey on the performance of 50 commercial active dry yeasts. Vitis 2013, 52, 149–155. [Google Scholar]
- Gava, A.; Ficagna, E.; Rossato, S.B.; Cisilotto, B.; Miotto, S.P.S.; Gabbi, H.T. Change in kinetic parameters of commercial yeast in the presence of copper fungicides. BIO Web Conf. 2016, 7, 02029. [Google Scholar] [CrossRef]
- Fay, J.C.; McCullough, H.L.; Sniegowski, P.D.; Eisen, M.B. Population genetic variation in gene expression is associated with phenotypic variation in Saccharomyces cerevisiae. Genome Biol. 2004, 5, R26. [Google Scholar] [CrossRef] [PubMed]
- Warringer, J.; Zörgö, E.; Cubillos, F.A.; Zia, A.; Gjuvsland, A.; Simpson, J.T.; Forsmark, A.; Durbin, R.; Omholt, S.W.; Louis, E.J.; et al. Trait variation in yeast is defined by population history. PLoS Genet. 2011, 7, e1002111. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Bakalinsky, A.T. Ssu1 mediates sulphite efflux in Saccharomyces cerevisiae. Yeast 2000, 16, 881–888. [Google Scholar] [CrossRef]
- Pérez-Ortín, J.E.; Querol, A.; Puig, S.; Barrio, E. Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res. 2002, 12, 1533–1539. [Google Scholar] [CrossRef] [PubMed]
- Mendes-Ferreira, A.; Barbosa, C.; Jimenez-Marti, E.; Del Olmo, M.L.; Mendes-Faia, A. The wine yeast strain-dependent expression of genes implicated in sulfide production in response to nitrogen availability. J. Microbiol. Biotechnol. 2010, 20, 1314–1321. [Google Scholar] [CrossRef] [PubMed]
- Landolfo, S.; Politi, H.; Angelozzi, D.; Mannazzu, I. Ros accumulation and oxidative damage to cell structures in Saccharomyces cerevisiae wine strains during fermentation of high-sugar-containing medium. Biochim. Biophys. Acta 2008, 1780, 892–898. [Google Scholar] [CrossRef] [PubMed]
- Bisson, L.F.; Karpel, J.E.; Ramakrishnan, V.; Joseph, L. Functional genomics of wine yeast Saccharomyces cerevisiae. Adv. Food Nutr. Res. 2007, 53, 65–121. [Google Scholar] [PubMed]
- Ashida, S.; Ichikawa, E.; Suginami, K.; Imayasu, S. Isolation and application of mutants producing sufficient isoamyl acetate, a sake flavor component. Agric. Biol. Chem. 1987, 51, 2061–2065. [Google Scholar]
- Ichikawa, E.; Hosokawa, N.; Hata, Y.; Abe, Y.; Suginami, K.; Imayasu, S. Breeding of a sake yeast with improved ethyl caproate productivity. Agric. Biol. Chem. 1991, 55, 2153–2154. [Google Scholar]
- Mendes Ferreira, A.; Clímaco, M.C.; Mendes Faia, A. The role of non-Saccharomyces species in releasing glycosidic bound fraction of grape aroma components—A preliminary study. J. Appl. Microbiol. 2001, 91, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Zott, K.; Thibon, C.; Bely, M.; Lonvaud-Funel, A.; Dubourdieu, D.; Masneuf-Pomarede, I. The grape must non-Saccharomyces microbial community: Impact on volatile thiol release. Int. J. Food Microbiol. 2011, 151, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Jiranek, V.; Langridge, P.; Henschke, P.A. Validation of bismuth-containing indicator media for predicting H2S-producing potential of Saccharomyces cerevisiae wine yeasts under enological conditions. Am. J. Enol. Vitic. 1995, 46, 269–273. [Google Scholar]
- Van Rensburg, P.; Pretorius, I.S. Enzymes in winemaking: Harnessing natural catalysts for efficient biotransformations-A review. S. Afr. J. Enol. Vitic. 2000, 21, 52–73. [Google Scholar]
- Charoenchai, C.; Fleet, G.H.; Henschke, P.A.; Todd, B.E.N.T. Screening of non-Saccharomyces wine yeasts for the presence of extracellular hydrolytic enzymes. Aust. J. Grape Wine Res. 1997, 3, 2–8. [Google Scholar] [CrossRef]
- Theron, L.W.; Divol, B. Microbial aspartic proteases: Current and potential applications in industry. Appl. Microbiol. Biotechnol. 2014, 98, 8853–8868. [Google Scholar] [CrossRef] [PubMed]
- Bisson, L.F. Stuck and sluggish fermentations. Am. J. Enol. Vitic. 1999, 50, 107–119. [Google Scholar]
- Salmon, J.M. Effect of sugar transport inactivation in Saccharomyces cerevisiae on sluggish and stuck enological fermentations. Appl. Environ. Microbiol. 1989, 55, 953–958. [Google Scholar] [PubMed]
- Barbosa, C.; Mendes-Faia, A.; Lage, P.; Mira, N.P.; Mendes-Ferreira, A. Genomic expression program of Saccharomyces cerevisiae along a mixed-culture wine fermentation with Hanseniaspora guilliermondii. Microb. Cell Fact. 2015, 14, 124. [Google Scholar] [CrossRef] [PubMed]
- Mendes-Ferreira, A.; Mendes-Faia, A.; Leão, C. Growth and fermentation patterns of Saccharomyces cerevisiae under different ammonium concentrations and its implications in winemaking industry. J. Appl. Microbiol. 2004, 97, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Oro, L.; Ciani, M.; Comitini, F. Antimicrobial activity of Metschnikowia pulcherrima on wine yeasts. J. Appl. Microbiol. 2014, 116, 1209–1217. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Mas, A.; Esteve-Zarzoso, B. The interaction between Saccharomyces cerevisiae and non-Saccharomyces yeast during alcoholic fermentation is species and strain specific. Front. Microbiol. 2016, 7, 502. [Google Scholar] [CrossRef] [PubMed]
- Holm Hansen, E.; Nissen, P.; Sommer, P.; Nielsen, J.C.; Arneborg, N. The effect of oxygen on the survival of non-Saccharomyces yeasts during mixed culture fermentations of grape juice with Saccharomyces cerevisiae. J. Appl. Microbiol. 2001, 91, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Sipiczki, M. Metschnikowia strains isolated from botrytized grapes antagonize fungal and bacterial growth by iron depletion. Appl. Environ. Microbiol. 2006, 72, 6716–6724. [Google Scholar] [CrossRef] [PubMed]
- Canonico, L.; Agarbati, A.; Comitini, F.; Ciani, M. Torulaspora delbrueckii in the brewing process: A new approach to enhance bioflavour and to reduce ethanol content. Food Microbiol. 2016, 56, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Benito, S.; Hofmann, T.; Laier, M.; Lochbühler, B.; Schüttler, A.; Ebert, K.; Fritsch, S.; Röcker, J.; Rauhut, D. Effect on quality and composition of Riesling wines fermented by sequential inoculation with non-Saccharomyces and Saccharomyces cerevisiae. Eur. Food Res. Technol. 2015, 241, 707–717. [Google Scholar] [CrossRef]
- Spiropoulos, A.; Tanaka, J.; Flerianos, I.; Bisson, L.F. Characterization of hydrogen sulfide formation in commercial and natural wine isolates of Saccharomyces. Am. J. Enol. Vitic. 2000, 51, 233–248. [Google Scholar]
- Huang, C.; Roncoroni, M.; Gardner, R.C. MET2 affects production of hydrogen sulfide during wine fermentation. Appl. Microbiol. Biotechnol. 2014, 98, 7125–7135. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.; Surdin-Kerjan, Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1997, 61, 503–532. [Google Scholar] [PubMed]


| Douro Subregion | Source | Strain Code |
|---|---|---|
| BC | Winery 14 | Mp51, Mp53, Mp54, Mp55, Mp56, Mp57, Mp58, Mp59, Mp60, Mp64 |
| Winery 2 | Mp2 | |
| Winery 3 | Mp3 | |
| Winery 7 | Mp11, Mp14, Mp15 | |
| Winery 8 | Mp12, Mp35, Mp40 | |
| Winery 9 | Mp13, Mp17, Mp18, Mp38, Mp42, Mp43, Mp44, Mp45, Mp46, Mp47, Mp48, Mp49, Mp50 | |
| CC | Winery 1 | Mp1 |
| Winery 4 | Mp4 | |
| Winery 6 | Mp7, Mp8, Mp9, Mp10, Mp31 | |
| Winery 10 | Mp16, Mp30, Mp32, Mp33, Mp34 | |
| Winery 11 | Mp19, Mp20, Mp21, Mp22, Mp23, Mp24, Mp36 | |
| Winery 15 | Mp52 | |
| Winery 16 | Mp61, Mp62, Mp63 | |
| DS | Winery 5 | Mp5, Mp6 |
| Winery 12 | Mp25, Mp26, Mp27, Mp28, Mp29 | |
| Winery 13 | Mp37, Mp39, Mp41 | |
| CECT | Mp CECT11202 |
| Grape Juice | Starter Culture | Maximum Fermentation Rate (g h−1) | Specific Growth Rate (h−1) | DW (g L−1) | |
|---|---|---|---|---|---|
| Sc UCD522 | Mp39 | ||||
| GJ | Sc UCD522 | 0.206 ± 0.009 c | 0.094 ± 0.021 a | 2.64 ± 0.03 b | |
| Sc UCD522 + Mp39 | 0.167 ± 0.006 d | 0.092 ± 0.008 a,b | 0.056 ± 0.004 c,d | 2.18 ± 0.76 b,c | |
| Mp39 | 0.050 ± 0.006 f | 0.067 ± 0.014 b,c,d | 1.32 ± 0.00 c | ||
| GJ + DAP | Sc UCD522 | 0.494 ± 0.009 a | 0.075 ± 0.009 a,b,c | 5.07 ± 0.12 a | |
| Sc UCD522 + Mp39 | 0.467 ± 0.017 b | 0.060 ± 0.002 c,d | 0.044 ± 0.009 d | 2.90 ± 0.26 b | |
| Mp39 | 0.083 ± 0.001 e | 0.080 ± 0.003 a,b,c | 1.53 ± 0.29 c | ||
| Grape Juice | Starter Culture | Fermentation Length (h) | pH | Volatile Acidity (g L−1) * | Total SO2 (mg L−1) | Titratable Acidity (g L−1) ** | Ethanol (% v/v) | Total H2S (µg L−1) | Final YAN (mg L−1) |
|---|---|---|---|---|---|---|---|---|---|
| GJ | Sc UCD522 | 312 | 3.11 ± 0.01 a | 0.29 ± 0.02 a | 8.19 ± 0.72 c | 7.63 ± 0.13 a | 11.40 ± 0.00 a,b | 301.86 ± 21.18 b | 8.75 ± 2.47 b |
| Sc UCD522 + Mp39 | 456 | 2.98 ± 0.18 a,b | 0.18 ± 0.04 b | 10.50 ± 1.08 b,c | 7.37 ± 0.40 a | 10.60 ± 0.42 b | 278.75 ± 82.02 b | 1.75 ± 2.47 b | |
| Mp39 | - | 2.92 ± 0.01 a,b | 0.18 ± 0.00 b | 10.24 ± 0.00 b,c | 7.76 ± 0.05 a | 3.75 ± 0.64 c | 40.35 ± 4.38 c | 10.50 ± 0.00 b | |
| GJ + DAP | Sc UCD522 | 144 | 2.91 ± 0.02 a,b | 0.09 ± 0.00 c | 9.98 ± 1.81 b,c | 8.55 ± 0.42 a | 12.10 ± 0.28 a | 397.11 ± 13.17 a | 8.75 ± 2.47 b |
| Sc UCD522 + Mp39 | 192 | 2.90 ± 0.06 b | 0.17 ± 0.02 b | 11.78 ± 0.73 a,b | 7.80 ± 0.21 a | 10.75 ± 0.07 b | 46.51 ± 9.19 c | 10.50 ± 4.95 b | |
| Mp39 | - | 2.94 ± 0.07 a,b | 0.11 ± 0.02 c | 13.32 ± 1.45 a | 9.10 ± 1.83 a | 3.50 ± 0.01 c | 25.03 ± 8.92 c | 126.00 ± 29.69 a |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbosa, C.; Lage, P.; Esteves, M.; Chambel, L.; Mendes-Faia, A.; Mendes-Ferreira, A. Molecular and Phenotypic Characterization of Metschnikowia pulcherrima Strains from Douro Wine Region. Fermentation 2018, 4, 8. https://doi.org/10.3390/fermentation4010008
Barbosa C, Lage P, Esteves M, Chambel L, Mendes-Faia A, Mendes-Ferreira A. Molecular and Phenotypic Characterization of Metschnikowia pulcherrima Strains from Douro Wine Region. Fermentation. 2018; 4(1):8. https://doi.org/10.3390/fermentation4010008
Chicago/Turabian StyleBarbosa, Catarina, Patrícia Lage, Marcos Esteves, Lélia Chambel, Arlete Mendes-Faia, and Ana Mendes-Ferreira. 2018. "Molecular and Phenotypic Characterization of Metschnikowia pulcherrima Strains from Douro Wine Region" Fermentation 4, no. 1: 8. https://doi.org/10.3390/fermentation4010008
APA StyleBarbosa, C., Lage, P., Esteves, M., Chambel, L., Mendes-Faia, A., & Mendes-Ferreira, A. (2018). Molecular and Phenotypic Characterization of Metschnikowia pulcherrima Strains from Douro Wine Region. Fermentation, 4(1), 8. https://doi.org/10.3390/fermentation4010008