Microbial Propionic Acid Production
Abstract
:1. Introduction
2. Overview of Developments in the Fermentation Process
3. Biological Propionic Acid Biosynthesis
3.1. Fermentation Routes for Propionate Production
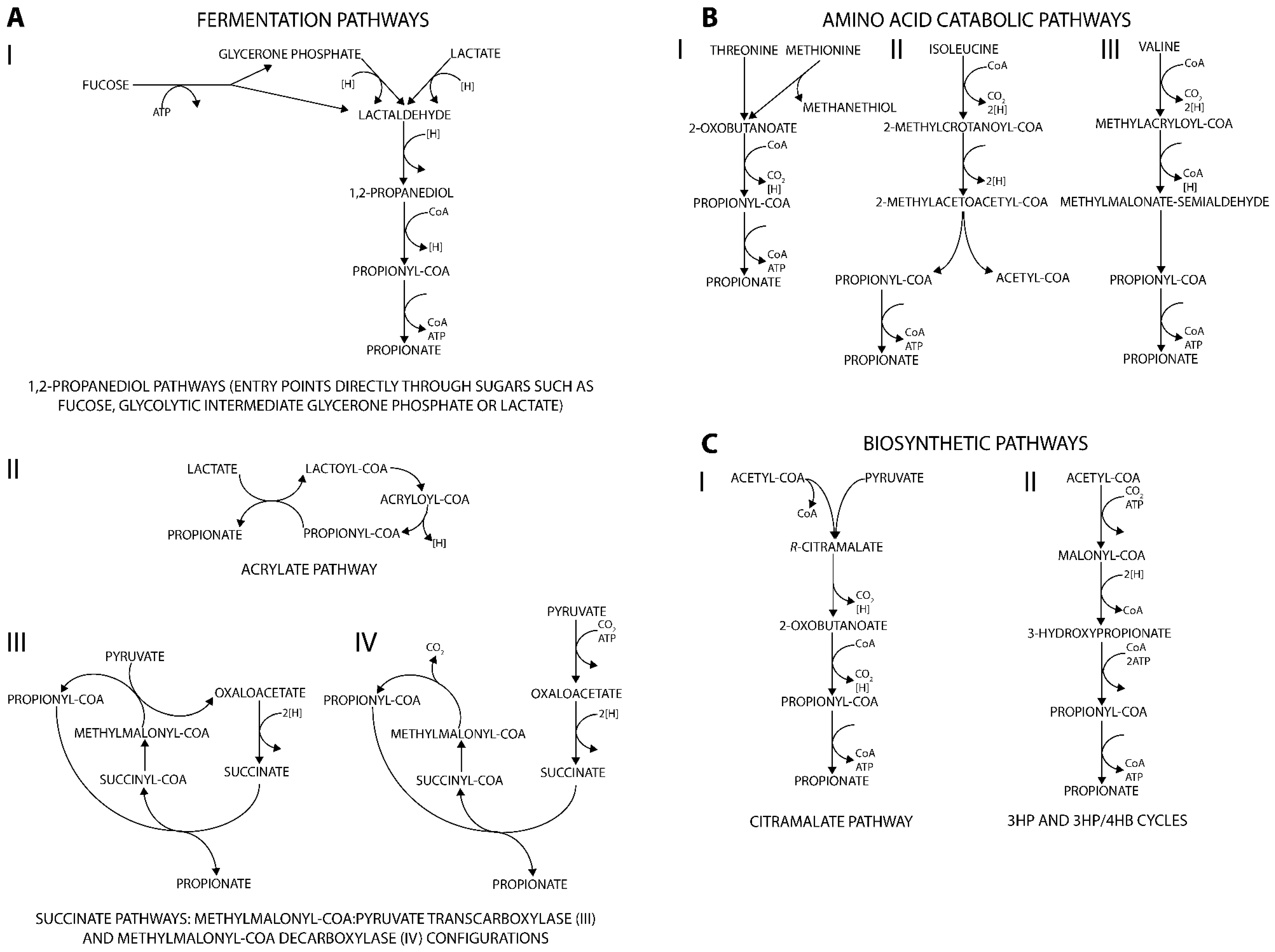
3.1.1. 1,2-propanediol Associated Pathways
3.1.2. Acrylate Pathway
3.1.3. Succinate Pathway
3.2. Degradation of Amino Acids to Produce Propionate
3.3. Biosynthetic Routes via Propionyl-CoA
3.3.1. Citramalate Pathway
3.3.2. 3HP/4HB Cycles
4. Genetic Engineering to Overcome the Current Challenges for Propionate Production
4.1. Empirical Strain Design
4.2. Rational Strain Engineering
4.3. Gene Knockouts
4.4. Gene Overexpression
4.5. Propionic Acid Biosynthesis by Non-Native Producers
5. Concluding Remarks and Future Directions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zidwick, M.J.; Chen, J.S.; Rogers, P. Organic acid and solvent production: Propionic and butyric acids and ethanol. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2013; pp. 135–167. [Google Scholar]
- Wemmenhove, E.; van Valenberg, H.J.; Zwietering, M.H.; van Hooijdonk, T.C.; Wells-Bennik, M.H. Minimal inhibitory concentrations of undissociated lactic, acetic, citric and propionic acid for Listeria monocytogenes under conditions relevant to cheese. Food Microbiol. 2016, 58, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Chávez, C.R.; Edwards, S.; Moure-Eraso, R.; Geiser, K. Sustainability of bio-based plastics: General comparative analysis and recommendations for improvement. J. Clean Prod. 2012, 23, 47–56. [Google Scholar] [CrossRef]
- Hebert, R.F.; Hebert Sam-E, L. Stable Indole-3-Propionate Salts of S-adenosyl-L-Methionine. U.S. Patent 9534010, 3 January 2017. Available online: https://www.google.com/patents/US9534010 (accessed on 25 February 2017).
- Market Research Store. Propionic Acid Market for Animal Feed & Grain Preservatives, Calcium & Sodium Propionates, Cellulose Acetate Propionate and Other Applications: Global Industry Perspective, Comprehensive Analysis, Size, Share, Growth, Segment, Trends and Forecast, 2014–2020. Available online: http://www.marketresearchstore.com/report/propionic-acid-market-for-animal-feed-grain-z39993 (accessed on 25 February 2017).
- Stowers, C.C.; Cox, B.M.; Rodriguez, B.A. Development of an industrializable fermentation process for propionic acid production. J. Ind. Microbiol. Biotechnol. 2014, 41, 837–852. [Google Scholar] [CrossRef] [PubMed]
- Fitz, A. über spaltpilzgärungen, IV Bericht der Deutsch. Chem. Ges. 1878, 11, 1890. [Google Scholar] [CrossRef]
- Swick, R.W.; Wood, H.G. The role of transcarboxylation in propionic acid fermentation. Proc. Natl. Acad. Sci. USA 1960, 46, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, B.A.; Stowers, C.C.; Pham, V.; Cox, B.M. The production of propionic acid, propanol and propylene via sugar fermentation: An industrial perspective on the progress, technical challenges and future outlook. Green Chem. 2014, 16, 1066–1076. [Google Scholar] [CrossRef]
- Luna-Flores, C.H.; Cox, B.M.; Stowers, C.C.; Nielsen, L.K.; Marcellin, E. Isolated Propionibacterium strain used for producing propionic acid comprises a modified gene, relative to strain Propionibacterium acidipropionici selected from the group consisting of the ABC polar amino acid transporter gene. Patent WO2017055932-A2; Filed on September 30, 2015 (US234900P), Available online: http://apps.webofknowledge.com.ezproxy.library.uq.edu.au/full_record.do?product=DIIDW&search_mode=GeneralSearch&qid=1&SID=Q2PwF9TQkv7q9hDEN4a&page=1&doc=1&colname=DIIDW (accessed on 10 May 2017).
- Liu, L.; Zhu, Y.; Li, J.; Wang, M.; Lee, P.; Du, G.; Chen, J. Microbial production of propionic acid from propionibacteria: Current state, challenges and perspectives. Crit. Rev. Biotechnol. 2012, 32, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.T.; Zhu, H.; Li, Y.; Hong, G. Continuous propionate production from whey permeate using a novel fibrous bed bioreactor. Biotechnol. Bioeng. 1994, 43, 1124–1130. [Google Scholar] [CrossRef] [PubMed]
- Barbirato, F.; Chedaille, D.; Bories, A. Propionic acid fermentation from glycerol: Comparison with conventional substrates. Appl. Microbiol. Biotechnol. 1997, 47, 441–446. [Google Scholar] [CrossRef]
- Coral, J.; Karp, S.G.; de Souza Vandenberghe, L.P.; Parada, J.L.; Pandey, A.; Soccol, C.R. Batch fermentation model of propionic acid production by Propionibacterium acidipropionici in different carbon sources. Appl. Biochem. Biotechnol. 2008, 151, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Jin, Y.; Yang, S.T. High cell density propionic acid fermentation with an acid tolerant strain of Propionibacterium acidipropionici. Biotechnol. Bioeng. 2015, 112, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Eş, I.; Khaneghah, A.M.; Hashemi, S.M.; Koubaa, M. Current advances in biological production of propionic acid. Biotechnol. Lett. 2017. [Google Scholar] [CrossRef] [PubMed]
- Falentin, H.; Deutsch, S.M.; Jan, G.; Loux, V.; Thierry, A.; Parayre, S.; Maillard, M.B.; Dherbécourt, J.; Cousin, F.J.; Jardin, J.; et al. The complete genome of Propionibacterium freudenreichii CIRM-BIA1 T, a hardy Actinobacterium with food and probiotic applications. PLoS ONE 2010, 235, e11748. [Google Scholar]
- Parizzi, L.P.; Grassi, M.C.; Llerena, L.A.; Carazzolle, M.F.; Queiroz, V.L.; Lunardi, I.; Zeidler, A.F.; Teixeira, P.J.; Mieczkowski, P.; Rincones, J.; et al. The genome sequence of Propionibacterium acidipropionici provides insights into its biotechnological and industrial potential. BMC Genom. 2012, 13, 562. [Google Scholar] [CrossRef] [PubMed]
- Tufvesson, P.; Ekman, A.; Sardari, R.R.; Engdahl, K.; Tufvesson, L. Economic and environmental assessment of propionic acid production by fermentation using different renewable raw materials. Bioresour. Technol. 2013, 149, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Sun, J.; Zhang, A.; Yang, S.T. Propionic acid fermentation. In Bioprocessing Technologies in Biorefinery for Sustainable Production of Fuels, Chemicals, and Polymers; Wiley & Sons: New York, 2013; pp. 331–350. [Google Scholar]
- Scott, K.P.; Martin, J.C.; Campbell, G.; Mayer, C.D.; Flint, H.J. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium “Roseburia. inulinivorans”. J. Bacteriol. 2006, 188, 4340–4349. [Google Scholar] [CrossRef] [PubMed]
- Zhuge, X.; Liu, L.; Shin, H.D.; Chen, R.R.; Li, J.; Du, G.; Chen, J. Development of a Propionibacterium-Escherichia. coli shuttle vector for metabolic engineering of Propionibacterium jensenii, an efficient producer of propionic acid. Appl. Environ. Microbiol. 2013, 79, 4595–4602. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.-H.; Chen, F.; Xu, H.; Wu, B.; Yao, J.; Ying, H.-J.; Ouyang, P.-K. Propionic acid fermentation by Propionibacterium freudenreichii CCTCC M207015 in a multi-point fibrous-bed bioreactor. Bioprocess. Biosyst. Eng. 2010, 33, 1077–1085. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, Y.-G.; Zhang, R.-B.; Zhang, F.; Zhu, J. Glycerol/glucose co-fermentation: One more proficient process to produce propionic acid by Propionibacterium acidipropionici. Curr. Microbial. 2011, 62, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Goswami, V.; Srivastava, A. Propionic acid production in an in situ cell retention bioreactor. Appl. Micriobiol. Biotechnol. 2001, 56, 676–680. [Google Scholar] [CrossRef]
- Gu, Z.; Rickert, D.A.; Glatz, B.A.; Glatz, C.E. Feasibility of propionic acid production by extractive fermentation. Le Lait 1999, 79, 137–148. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, J.; Tan, M.; Liu, L.; Jiang, L.; Sun, J.; Lee, P.; Du, G.; Chen, J. Optimization and scale-up of propionic acid production by propionic acid-tolerant Propionibacterium acidipropionici with glycerol as the carbon source. Bioresour. Technol. 2010, 101, 8902–8906. [Google Scholar] [CrossRef] [PubMed]
- Dishisha, T.; Ståhl, Å.; Lundmark, S.; Hatti-Kaul, R. An economical biorefinery process for propionic acid production from glycerol and potato juice using high cell density fermentation. Bioresour. Technol. 2013, 135, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ma, C.; Gao, C.; Xu, P. Efficient utilization of hemicellulose hydrolysate for propionic acid production using Propionibacterium acidipropionici. Bioresour. Technol. 2012, 114, 711–714. [Google Scholar] [CrossRef] [PubMed]
- Dishisha, T.; Alvarez, M.T.; Hatti-Kaul, R. Batch- and continuous propionic acid production from glycerol using free and immobilized cells of Propionibacterium acidipropionici. Bioresour. Technol. 2012, 118, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Wei, P.; Cai, J.; Zhu, X.; Wang, Z.; Huang, L.; Xu, Z. Improving the productivity of propionic acid with FBB-immobilized cells of an adapted acid-tolerant Propionibacterium acidipropionici. Bioresour. Technol. 2012, 112, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, S.-T. Propionic acid production in glycerol/glucose co-fermentation by Propionibacterium freudenreichii subsp shermanii. Bioresour. Technol. 2013, 137, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Chen, F.; Xu, H.; Wu, B.; Li, H.; Li, S.; Ouyang, P. Green and economical production of propionic acid by Propionibacterium freudenreichii CCTCC M207015 in plant fibrous-bed bioreactor. Bioresour. Technol. 2011, 102, 6141–6146. [Google Scholar] [CrossRef] [PubMed]
- Navone, L.; McCubbin, T.; Gonzalez-Garcia, R.A.; Nielsen, L.K.; Marcellin, E. Genome-scale model guided design of Propionibacterium for enhanced propionic acid production. Microb. Cell Fact. 2017. Manuscript submitted to Metabolic Engineering. [Google Scholar]
- Ibarra, R.U.; Edwards, J.S.; Palsson, B.O. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 2002, 420, 186–189. [Google Scholar] [CrossRef] [PubMed]
- Staib, L.; Fuchs, T.M. Regulation of fucose and 1, 2-propanediol utilization by Salmonella enterica serovar Typhimurium. Front. Microbial. 2006. [Google Scholar] [CrossRef] [PubMed]
- Saxena, R.K.; Anand, P.; Saran, S.; Isar, J.; Agarwal, L. Microbial production and applications of 1, 2-propanediol. Indian J. Microbiol. 2010, 50, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Cameron, D.C.; Cooney, C.L. A Novel Fermentation: The Production of R (–)–1, 2–Propanediol and Acetol by Clostridium thermosaccharolyticum. Nat. Biotechnol. 1986, 4, 651–654. [Google Scholar] [CrossRef]
- Zhu, L.; Guan, X.; Xie, N.; Wang, L.; Yu, B.; Ma, Y. Fermentative production of enantiomerically pure S-1, 2-propanediol from glucose by engineered E. coli strain. Appl. Microbiol. Biotechnol. 2016, 100, 1241–1251. [Google Scholar] [CrossRef] [PubMed]
- Elferink, S.J.O.; Krooneman, J.; Gottschal, J.C.; Spoelstra, S.F.; Faber, F.; Driehuis, F. Anaerobic conversion of lactic acid to acetic acid and 1, 2-propanediol by Lactobacillus buchneri. Appl. Environ. Microbiol. 2001, 67, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.; Guo, J. Stereospecific microbial conversion of lactic acid into 1, 2-propanediol. ACS Synth. Biol. 2014, 4, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Altaras, N.E.; Cameron, D.C. Metabolic engineering of a 1, 2-propanediol pathway in Escherichia coli. Appl. Environ. Microbiol. 1999, 65, 1180–1185. [Google Scholar] [PubMed]
- Hino, T.; Shimada, K.; Maruyama, T. Substrate preference in a strain of Megasphaera. elsdenii, a ruminal bacterium, and its implications in propionate production and growth competition. Appl. Environ. Microbiol. 1994, 60, 1827–1831. [Google Scholar] [PubMed]
- Ladd, J.N.; Walker, D.J. The fermentation of lactate and acrylate by the rumen micro-organism LC. Biochem. J. 1959, 71, 364. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, V.; Vaidyanathan, H.; Djurdjevic, I.; Jayamani, E.; Ramachandran, K.B.; Buckel, W.; Jayaraman, G.; Ramalingam, S. Engineering Escherichia coli with acrylate pathway genes for propionic acid synthesis and its impact on mixed-acid fermentation. Appl. Environ. Microbiol. 2013, 97, 1191–1200. [Google Scholar] [CrossRef] [PubMed]
- Hilpert, W.; Dimroth, P. On the mechanism of sodium ion translocation by methylmalonyl-CoA decarboxylase from Veillonella. alcalescens. Eur. J. Biochem. 1991, 195, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Dimroth, P.; Jockel, P.; Schmid, M. Coupling mechanism of the oxaloacetate decarboxylase Na+ pump. BBA-Bioenergetics 2001, 1505, 1–14. [Google Scholar] [CrossRef]
- Di Berardino, M.; Dimroth, P. Aspartate 203 of the oxaloacetate decarboxylase β-subunit catalyses both the chemical and vectorial reaction of the Na+ pump. EMBO J. 1996, 15, 1842. [Google Scholar] [PubMed]
- Shen, C.R.; Liao, J.C. Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via the keto-acid pathways. Metab. Eng. 2008, 10, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Park, J.H.; Kim, T.Y.; Lee, S.Y. Metabolic engineering of Escherichia coli for the production of 1-propanol. Metab. Eng. 2012, 14, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Pfeifer, B.A. Metabolic and pathway engineering to influence native and altered erythromycin production through E. coli. Metab. Eng. 2013, 19, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Tseng, H.C.; Harwell, C.L.; Martin, C.H.; Prather, K.L. Biosynthesis of chiral 3-hydroxyvalerate from single propionate-unrelated carbon sources in metabolically engineered E. coli. Microb. Cell Fact. 2010, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.R.; Liao, J.C. Synergy as design principle for metabolic engineering of 1-propanol production in Escherichia coli. Metab. Eng. 2013, 17, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Hou, J.; Zhang, F.; Ai, G.; Li, M.; Cai, S.; Liu, H.; Wang, L.; Wang, Z.; Zhang, S.; et al. Multiple propionyl coenzyme A-supplying pathways for production of the bioplastic poly (3-hydroxybutyrate-co-3-hydroxyvalerate) in Haloferax. mediterranei. Appl. Environ. Microbiol. 2013, 79, 2922–2931. [Google Scholar] [CrossRef] [PubMed]
- Berg, I.A.; Kockelkorn, D.; Buckel, W.; Fuchs, G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 2007, 318, 1782–1786. [Google Scholar] [CrossRef] [PubMed]
- Mattozzi, M.D.; Ziesack, M.; Voges, M.J.; Silver, P.A.; Way, J.C. Expression of the sub-pathways of the Chloroflexus. aurantiacus 3-hydroxypropionate carbon fixation bicycle in E. coli: Toward horizontal transfer of autotrophic growth. Metab. Eng. 2013, 16, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.H.; Barry, K.; Chertkov, O.; Dalin, E.; Han, C.S.; Hauser, L.J.; Honchak, B.M.; Karbach, L.E.; Land, M.L.; Lapidus, A.; et al. Complete genome sequence of the filamentous anoxygenic phototrophic bacterium Chloroflexus. aurantiacus. BMC Genom. 2011, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- McCubbin, T.; Palfreyman, R.W.; Stowers, C.; Nielsen, L.K.; Marcellin, E. A pan-genome guided Propionibacterium genome scale metabolic network reconstruction. Microbiome 2017. Submitted. [Google Scholar]
- Jore, J.P.M.; Van Luijk, N.; Luiten, R.G.M.; Van der Werf, M.J.; Pouwels, P.H. Efficient Transformation System for Propionibacterium freudenreichii Based on a Novel Vector. Appl. Environ. Microbiol. 2001, 67, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Kiatpapan, P.; Hashimoto, Y.; Nakamura, H.; Piao, Y.Z.; Ono, H.; Yamashita, M.; Murooka, Y. Characterization of pRGO1, a plasmid from Propionibacterium acidipropionici, and its use for development of a host-vector system in propionibacteria. Appl. Environ. Microbiol. 2000, 66, 4688–4695. [Google Scholar] [CrossRef] [PubMed]
- Kiatpapan, P.; Murooka, Y. Genetic manipulation system in propionibacteria. J. Biosci. Bioeng. 2002, 93, 1–8. [Google Scholar] [CrossRef]
- O’Connell Motherway, M.; O’Driscoll, J.; Fitzgerald, G.F.; Van Sinderen, D. Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium. breve UCC2003. Microb. Biotechnol. 2009, 2, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Wang, W.; Deng, A.; Sun, Z.; Zhang, Y.; Liang, Y.; Che, Y.; Wen, T. A mimicking-of-DNA-methylation-patterns pipeline for overcoming the restriction barrier of bacteria. PLoS Genet. 2012, 8, e1002987. [Google Scholar] [CrossRef] [PubMed]
- Suwannakham, S.; Huang, Y.; Yang, S.T. Construction and characterization of ack knock-out mutants of Propionibacterium acidipropionici for enhanced propionic acid fermentation. Biotechnol. Bioeng. 2006, 94, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Guan, N.; Zhu, G.; Li, J.; Shin, H.D.; Du, G.; Chen, J. Pathway engineering of Propionibacterium jensenii for improved production of propionic acid. Sci. Rep. 2016, 6, 19963. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Cui, H.; Zhu, L.; Hu, Y.; Xu, X.; Li, S.; Huang, H. Enhanced propionic acid production from whey lactose with immobilized Propionibacterium acidipropionici and the role of trehalose synthesis in acid tolerance. Green Chem. 2015, 17, 250–259. [Google Scholar] [CrossRef]
- Guan, N.; Li, J.; Shin, H.D.; Du, G.; Chen, J.; Liu, L. Metabolic engineering of acid resistance elements to improve acid resistance and propionic acid production of Propionibacterium jensenii. Biotechnol. Bioeng. 2015, 113, 1294–1304. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ammar, E.M.; Zhang, A.; Wang, L.; Lin, M.; Yang, S.-T. Engineering Propionibacterium freudenreichii subsp. shermanii for enhanced propionic acid fermentation: Effects of overexpressing propionyl-CoA: Succinate CoA transferase. Metab. Eng. 2015, 27, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Lin, M.; Wang, L.; Ammar, E.M.; Yang, S.-T. Metabolic engineering of Propionibacterium freudenreichii subsp. shermanii for enhanced propionic acid fermentation: Effects of overexpressing three biotin-dependent carboxylases. Process. Biochem. 2015, 50, 194–204. [Google Scholar] [CrossRef]
- Ammar, E.M.; Jin, Y.; Wang, Z.; Yang, S.T. Metabolic engineering of Propionibacterium freudenreichii: Effect of expressing phosphoenolpyruvate carboxylase on propionic acid production. Appl. Microbiol. Biotechnol. 2014, 98, 7761–7772. [Google Scholar] [CrossRef] [PubMed]
- Guan, N.; Liu, L.; Zhuge, X.; Xu, Q.; Li, J.; Du, C.; Chen, J. Genome-shuffling improves acid tolerance of Propionibacterium acidipropionici. In Advances in Chemistry Research; Taylor, J.C., Ed.; Nova: New York, NY, USA, 2012; Volume 15, pp. 143–152. [Google Scholar]
- Guan, N.; Liu, L.; Shin, H.-D.; Chen, R.R.; Zhang, J.; Li, J.; Du, G.; Shi, Z.; Chen, J. Systems-level understanding of how Propionibacterium acidipropionici respond to propionic acid stress at the microenvironment levels: Mechanism and application. J. Biotechnol. 2013, 167, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Suwannakham, S.; Huang, Y.; Yang, S.T. Enhanced propionic acid fermentation by Propionibacterium acidipropionici mutant obtained by adaptation in a fibrous-bed bioreactor. Biotechnol. Bioeng. 2005, 91, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Yang, S.T. Propionic acid production from glycerol by metabolically engineered Propionibacterium acidipropionici. Process. Biochem. 2009, 44, 1346–1351. [Google Scholar] [CrossRef]
- Luna-Flores, C.H.; Palfreyman, R.W.; Krömer, J.O.; Nielsen, L.K.; Marcellin, E. Improved production of propionic acid using genome shuffling. Biotechnol. J. 2016, 12, 1600120. [Google Scholar] [CrossRef] [PubMed]
- Luna-Flores, C.H.; Stowers, C.C.; Cox, B.; Nielsen, L.K.; Marcellin, E. Scalable and economical process for propionic acid biosynthesis. Biotechnol. Biofuels 2017. Submitted. [Google Scholar]
- Biot-Pelletier, D.; Martin, V.J. Evolutionary engineering by genome shuffling. Appl. Microbiol. Biotechnol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, J.-Z.; Huang, J.-S.; Mao, Z.W. Genome shuffling of Propionibacterium shermanii for improving vitamin B12 production and comparative proteome analysis. J. Biotechnol. 2010, 148, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhuge, X.; Shin, H.D.; Chen, R.R.; Li, J.; Du, G.; Chen, J. Improved production of propionic acid via combinational overexpression of glycerol dehydrogenase and malate dehydrogenase from Klebsiella. pneumoniae in Propionibacterium jensenii. Appl. Environ. Microbiol. 2015, 81, 2256–2264. [Google Scholar] [CrossRef] [PubMed]
- Stowers, C.; Dow AgroSciences LLC. Personal Comunication, Indianapolis, IN, USA, 2017.
- Haller, T.; Buckel, T.; Rétey, J.; Gerlt, J.A. Discovering new enzymes and metabolic pathways: Conversion of succinate to propionate by Escherichia coli. Biochemistry 2000, 39, 4622–4629. [Google Scholar] [CrossRef] [PubMed]
- Dellomonaco, C.; Rivera, C.; Campbell, P.; Gonzalez, R. Engineered respiro-fermentative metabolism for the production of biofuels and biochemicals from fatty acid-rich feedstocks. Appl. Environ. Micriobiol. 2010, 76, 5067–5078. [Google Scholar] [CrossRef] [PubMed]
- Srirangan, K.; Akawi, L.; Liu, X.; Westbrook, A.; Blondeel, E.J.; Aucoin, M.G.; Moo-Young, M.; Chou, C.P. Manipulating the sleeping beauty mutase operon for the production of 1-propanol in engineered Escherichia coli. Biotechnol. Biofuels 2013. [Google Scholar] [CrossRef] [PubMed]
- Akawi, L.; Srirangan, K.; Liu, X.; Moo-Young, M.; Chou, C.P. Engineering Escherichia coli for high-level production of propionate. J. Ind. Microbiol. Biotechnol. 2015, 42, 1057–1072. [Google Scholar] [CrossRef] [PubMed]
| Year | Event |
|---|---|
| 1854 | Adolph Strecker observed the formation of propionic acid from sugar in a mixture of calcium carbonate-sugar [1]. |
| 1861–1879 | Pasteur showed that fermentation occurs due to the activity of microbes. |
| 1878 | Fist work on propionic acid production by Propionibacteria. Albert Fitz predicted that 3 moles of lactic acid would lead to the production of 2 moles of propionic acid, 1 mole acetic acid, 1 mole CO2 and 1 mole H2O [7]. |
| 1906 | 11 species of propionibacteria were identified as propionic acid producers during cheese making [1]. |
| 1928 | First mention of glycerol as carbon source for propionic acid production [1]. |
| 1937 | First complete study on propionibacteria metabolism during propionic acid fermentation by Wood [1]. |
| 1949 | A complete review of the factors affecting propionic acid fermentation was published [1]. |
| 1920–1953 | 17 patents for propionic acid production by different Propionibacterium strains were approved [1]. |
| 1961 | Immobilized cells are first used to reach higher production yields [1]. |
| 1962 | The Wayman process was developed. It consisted of a continuous system with immobilised cells of P. acidipropionici [1]. |
| 1960–2010 | Selection of overproducer strains and new production strategies. |
| 2011–2013 | Complete genome of P. shermanii [17] and P. acidipropionici [18] were sequenced and published. |
| 2013–2014 | Techno economic studies suggest the fermentation of sugar to propionate can be profitable if productivity reaches 1–2 g/L/h, yield reaches 0.6 g/g and final titre reaches ≈100 g/L [9,19]. |
| Strain | Fermentation Approach | Substrate (s) | Titre (g/L) | PA Yield (g/g) | Productivity (g/L/h) | References |
|---|---|---|---|---|---|---|
| P. acidipropionici | Batch | Glucose/Glycerol | 22 | 0.57 | 0.152 | [24] |
| Fed-batch | Glucose/Glycerol | 30 | 0.54 | 0.152 | [24] | |
| Sequential batch | Glucose | 35 | 0.62 | 1.28 | [15] | |
| Fed-batch | Glucose | 56 | 0.43 | 2.23 | [15] | |
| Continuous | Lactose | 19 | 0.4 | 0.9 | [25] | |
| Fed-batch | Glucose | 71 | - | - | [26] | |
| Fed-batch | Glycerol | 48 | 0.59 | 0.2 | [27] | |
| Batch | Glucose | 45 | 0.45 | 2 | [6] | |
| Batch | Corn mash | 24 | 0.6 | 0.5 | [9] | |
| Sequential batch (with cell recycle) | Glycerol | 27 | 0.78 | 0.22 | [28] | |
| Fed-batch | Xylose | 53 | 0.35 | 0.23 | [29] | |
| Fed-batch | Corncob molasses | 72 | - | 0.28 | [29] | |
| PEI-Poraver bioreactor (Continuous) | Glycerol | 14 | 0.86 | 1.4 | [30] | |
| Fibrous-bed bioreactor (Fed-batch) | Glucose | 51 | 0.43 | 0.71 | [31] | |
| Fibrous-bed bioreactor (Fed-batch) | Sugarcane bagasse hydrolysate | 59 | 0.37 | 0.38 | [31] | |
| P. shermanii | Fibrous-bed bioreactor (Repeated-batch) | Glucose/Glycerol | 75 | 0.57 | 0.25 | [32] |
| P. freudenreichii | Multi-point fibrous-bed bioreactor (Fed-batch) | Glucose | 67 | 0.43 | 0.14 | [23] |
| Plant fibrous-bed bioreactor (Fed-batch) | Hydrolysed cane molasses | 92 | 0.46 | 0.36 | [33] | |
| Plant fibrous-bed bioreactor (Fed-batch) | Hydrolysate of cane molasses & waste Propionibacterium cells | 80 | 0.4 | 0.26 | [33] |
| Microorganism | Substrates | Products | Pathway |
|---|---|---|---|
| Propionibacteria acidipropionici P. freudenreichii 1 P. shermanii 2 | Glucose, sucrose, lactate, glycerol | Propionate, acetate, succinate, CO2 | Wood-Werkman cycle (Figure 1(AIII)) |
| Clostridia propionicum | Glycerol, lactate, alanine, serine, threonine | Propionate, succinate, formate, acetate, n-propanol | Acrylate pathway (Figure 1(AII)) |
| Bacteroides fragilis B. ruminicola | Glucose | Acetate, lactate propionate, succinate, formate, CO2 | Succinate pathway (Figure 1(AIV)) |
| Veillonella parvula V. alcalescens | Lactate, succinate | Propionate, acetate, CO2, H2 | Succinate pathway (Figure 1(AIV)) |
| Propionigenum modestum | Succinate | Propionate, CO2 | Succinate pathway (Figure 1(AIV)) |
| Selenomonas ruminantium S. sputigena | Lactate Glucose | Propionate, lactate, acetate, CO2 | Succinate pathway (Figure 1(AIV)) |
| Megasphaera elsdenii | Lactate | Acetate, propionate, butyrate | Acrylate pathway (Figure 1(AII)) |
| Salmonella typhimurium | Deoxy sugars, glucose, 1,2-propanediol | 1,2-propanediol, propanol, propionate, acetate, formate, lactate, CO2 | 1,2-propanediol pathway (Figure 1(AI)) |
| PDO pathway | Maximum Yields (mol/mol Glc) | Expected Yields (mol/mol Glc) | ||
|---|---|---|---|---|
| PA | ATP | PA | ATP | |
| Deoxy sugar 1 | 1 | 2.5 | 1 | 3 2 |
| DHAP | 1.71 | 0 | 0 | 3 |
| Lactate | 1.71 | 3.43 | 1.33 | 4 |
| Engineered lactate pathway | 1.71 | 0 3 | 1 | 3 |
| All | 1.71 | 3.43 | 0 | 3 |
| Products | Pathway Yields (mol/mol Glc) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Catabolic Pathways | Biosynthetic Pathways | Fermentation Routes | Overall 1 | |||||||
| Val/Iso | Thr | Met | Citramalate | 3HP/4HB | Propanediol | Acrylate | Na+ Pumping | Wood-Werkman | ||
| ATP | 2.29 | 0 | 1 | 2.4 | 0 | 3.43 | 1.71 | 2.57 | 3.43 | 3.43 |
| Propionate | 0.29 | 1.33 | 1 | 0.4 | 1.33 | 1.71 | 1.71 | 1.71 | 1.71 | 1.71 |
| Acetate | 0 | 0.67 | 1 | 0 | 0.67 | 0 | 0 | 0 | 0 | 0 |
| Ethanol | 1.43 | 0 | 0 | 1.2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Formate | 1.43 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| CO2 | 0.86 | 0.67 | 0 | 0.4 | 0.67 | 0.86 | 0.86 | 0.86 | 0.86 | 0.86 |
| Products | Pathway Yields (mol/mol Glc) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Catabolic Pathways | Biosynthetic Pathways | Fermentation Routes | Overall 1 | |||||||
| Val/Iso | Thr | Met | Citramalate | 3HP/4HB | Propanediol | Acrylate | Na+ Pumping | Wood-Werkman | ||
| ATP | 3 | 3 | 3 | 3 | 3 | 4 | 3 | 3.25 2 | 4 | 4 |
| Propionate | 0 | 0 | 0 | 0 | 0 | 1.33 | 1 | 1 2 | 1.33 | 1.33 |
| Acetate | 1 | 1 | 1 | 1 | 1 | 0.67 | 1 | 1 2 | 0.67 | 0.67 |
| Ethanol | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Formate | 2 | 2 | 2 | 2 | 2 | 0 | 1 | 1 2 | 0 | 0 |
| CO2 | 0 | 0 | 0 | 0 | 0 | 0 | 0.67 | 0 2 | 0 | 0.67 |
| Amino Acid | Valine | Isoleucine | Threonine | Methionine | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Substrate | Pyr | ATP | NADH | NADPH | Pyr | ATP | NADH | NADPH | Pyr | ATP | NADH | NADPH | Pyr | ATP | NADH | NADPH |
| Degradation | 0 | 1 | 5 | 0 | 0 | 2 | 3 | 1 | 0 | 1 | 0 | 1(0) | 0 | 1 | 0 | 1(0) |
| Biosynthesis | −2 | 0 | −1 | −1 | −3 | 0 | 1 | −2(−1) | −1 | −3 | −1 | −3 | −2 | −2 | −1 | −2(−1) |
| Combined | −2 | 1 | 4 | −1 | −2 | 1 | 4 | −1(0) | −1 | −2 | −1 | −1(−2) | −2 | −1 | −1 | −1 |
| Amino Acid Pathway | Maximum Yields (mol/mol Glc) | Expected Yields 1 (mol/mol Glc) | ||
|---|---|---|---|---|
| PA | ATP | PA | ATP | |
| Valine/Isoleucine | 0.29 | 2.29 | 0 | 3 |
| Threonine | 1.33 | 0 | 0 | 3 |
| Methionine | 1 2 | 1 2 | 0 | 3 |
| All | 1.45 | 0 | 0 | 3 |
| Aim | Strategy | Strain | Results | Reference |
|---|---|---|---|---|
| Decrease by-products | Genome editing | P. acidipropionici ACK-Tet strain | Acetate production reduced ~14%. ~13% improvement of propionate production. | [64] |
| Genome editing, overexpression | P. jensenii poxB or ldh knock-out and ppc overexpression | Maximum 30% improvement in titre and 24% improvement productivity | [65] | |
| Improve acid tolerance | Overexpression | P. acidipropionici otsA overexpression strain | Propionic acid yield 11% higher. | [66] |
| Overexpression | P. jensenii strains overexpressing gadB, arcA, arc, gdh or ybaS | Up to a 1.5-fold increase in yield and 5.4-fold increase in titre, in shake flasks | [67] | |
| Increase of metabolic flux towards propionate production | Overexpression | P. shermanii CoAT overexpression strain | Increase yield and productivity, maximum 10% and 46%, respectively. | [68] |
| Overexpression of heterologous enzymes from P. acidipropionici | P. shermanii overexpressing mmc, pyc or mmd | Strongest phenotype observed with mmc overexpressing strain with 14% increase in yield from glucose and 17% increase in productivity from glucose/glycerol co-fermentation. Performed in serum bottles. | [69] | |
| Overexpression of heterologous enzymes from E. coli | P. shermanii overexpressing ppc strain | Improved productivity on glycerol only, no improvement in yield. | [70] | |
| Overexpression of heterologous enzymes from E. coli and Klebsiella pneumoniae | P. jensenii co-expression of gdh and mdh | Increase in propionate synthesis, but slow growth of the mutant strain. | [71] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez-Garcia, R.A.; McCubbin, T.; Navone, L.; Stowers, C.; Nielsen, L.K.; Marcellin, E. Microbial Propionic Acid Production. Fermentation 2017, 3, 21. https://doi.org/10.3390/fermentation3020021
Gonzalez-Garcia RA, McCubbin T, Navone L, Stowers C, Nielsen LK, Marcellin E. Microbial Propionic Acid Production. Fermentation. 2017; 3(2):21. https://doi.org/10.3390/fermentation3020021
Chicago/Turabian StyleGonzalez-Garcia, R. Axayacatl, Tim McCubbin, Laura Navone, Chris Stowers, Lars K. Nielsen, and Esteban Marcellin. 2017. "Microbial Propionic Acid Production" Fermentation 3, no. 2: 21. https://doi.org/10.3390/fermentation3020021
APA StyleGonzalez-Garcia, R. A., McCubbin, T., Navone, L., Stowers, C., Nielsen, L. K., & Marcellin, E. (2017). Microbial Propionic Acid Production. Fermentation, 3(2), 21. https://doi.org/10.3390/fermentation3020021








