A Novel Mycoprotein Candidate: Neurospora intermedia FF171 from Pu-Erh Tea with Genomics-Based Safety Profiling
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Isolation and Purification of Neurospora
2.3. Identification of Neurospora intermedia
2.3.1. Identification by Internal Transcribed Spacer (ITS)
2.3.2. Identification by QMA, TMI, TML, and DMG Sequences
2.4. Screening of Neurospora intermedia Strains with High Protein Yield and Analysis of Protein Yield Under Different Fermentation Conditions
2.4.1. Screening High-Protein-Yield Strains
2.4.2. Genetic Stability Experiment
2.4.3. Fermentation with Different Carbon and Nitrogen Sources
2.5. Mycotoxin Content Determination
2.6. Whole-Genome Sequencing and Annotation
2.7. Safety Assessment of N. intermedia FF171 Based on Bioinformatics Analysis
2.7.1. Overview of Bioinformatics Analysis
2.7.2. Antibiotic Resistance Assessment
2.7.3. Toxigenicity Analysis
2.7.4. Allergenicity Analysis
- Preliminary screening using the AlgPred 2.0 hybrid model: The Algpred 2.0 hybrid model (Random Forest + BLAST + MERCI) was used for preliminary screening of allergenic proteins in N. intermedia FF171. AlgPred 2.0 is a web server designed to predict whether a protein is an allergen. The AlgPred 2.0 dataset contains a total of 10,075 allergens, 10,075 non-allergens, and 10,451 IgE epitopes [54]. The Algpred 2.0 hybrid model refers to an integrated approach that combines and performs weighted calculation of multiple methods (a Random Forest (RF) model based on machine learning (ML) techniques, BLAST similarity search, and MERCI motif analysis) to generate a hybrid score, aiming to improve the classification accuracy of allergenic proteins. In this study, the threshold for the hybrid score was set at 0.3 to identify allergenic proteins.
- Refinement against manually curated non-allergenic proteins: To refine the allergenic protein prediction results, the allergenic proteins predicted by AlgPred 2.0 were subjected to sequence homology searches against manually annotated non-allergenic proteins from the UniProtKB/Swiss-Prot database. A comparison between predicted allergenic proteins and non-allergenic proteins from the UniProt/Swiss-Prot database (downloaded on 19 February 2025) revealed significant sequence homology for 1000 entries through BLASTP. All subsequent BLASTP analyses in this section employed the same default parameters.
- Comparison with allergens in the AllergenOnline database: The remaining allergenic proteins were compared with allergens in the AllergenOnline database. Following the guidelines issued by the Food and Agriculture Organization/World Health Organization (FAO/WHO) [55], Codex Alimentarius Commission [56,57], and EFSA [48,58], sequences showing >35% homology were identified as allergenic proteins using an 80-amino acid sliding window. Given that many conserved proteins easily reach this threshold, matches with an E-value < 1 × 10−7 and full-length sequence homology > 50% were further filtered, as these criteria are considered more predictive [59].
- Homology analysis with proteins from GRAS strains: The putative allergenic proteins obtained in the previous step were subjected to BLASTP analysis (default search parameters) against the Swiss-Prot annotated proteins derived from GRAS strains. The criteria for significant sequence homology were based on the functional thresholds established by Pearson (>40% identity, E-value < 0.001, and bit score > 50) [46]. Based on the significant homologous sequences previously identified between FF171 and GRAS strains, potential allergenic proteins were further screened to identify those unique to FF171. N. intermedia FF171 proteins exhibiting significant homology to GRAS strain proteomes were considered functionally similar and were not classified as allergenic proteins of FF171.
- Homology investigation with proteins from common foods and humans: The predicted allergenic proteins were further investigated for sequence homology with proteins from widely consumed food sources and humans (Homo sapiens) [60] by conducting BLASTP searches within the NCBI non-redundant protein sequence database (default search parameters). The widely consumed foods selected in this study included chicken (Gallus gallus), beef (Bos taurus), pork (Sus scrofa), rice (Oryza sativa), wheat (Triticum aestivum), soybean (Glycine max), corn (Zea mays), pepper (Capsicum annuum), and sweet orange (Citrus sinensis). Each allergenic protein in the AllergenOnline database sharing at least 50% homology with N. intermedia FF171 proteins was subjected to sequence homology searches against human proteins. The comparative analyses applied the standard thresholds for functional sequence homology, which were based on the parameters reported by Pearson [46].
2.8. Statistical Analysis
3. Results and Discussion
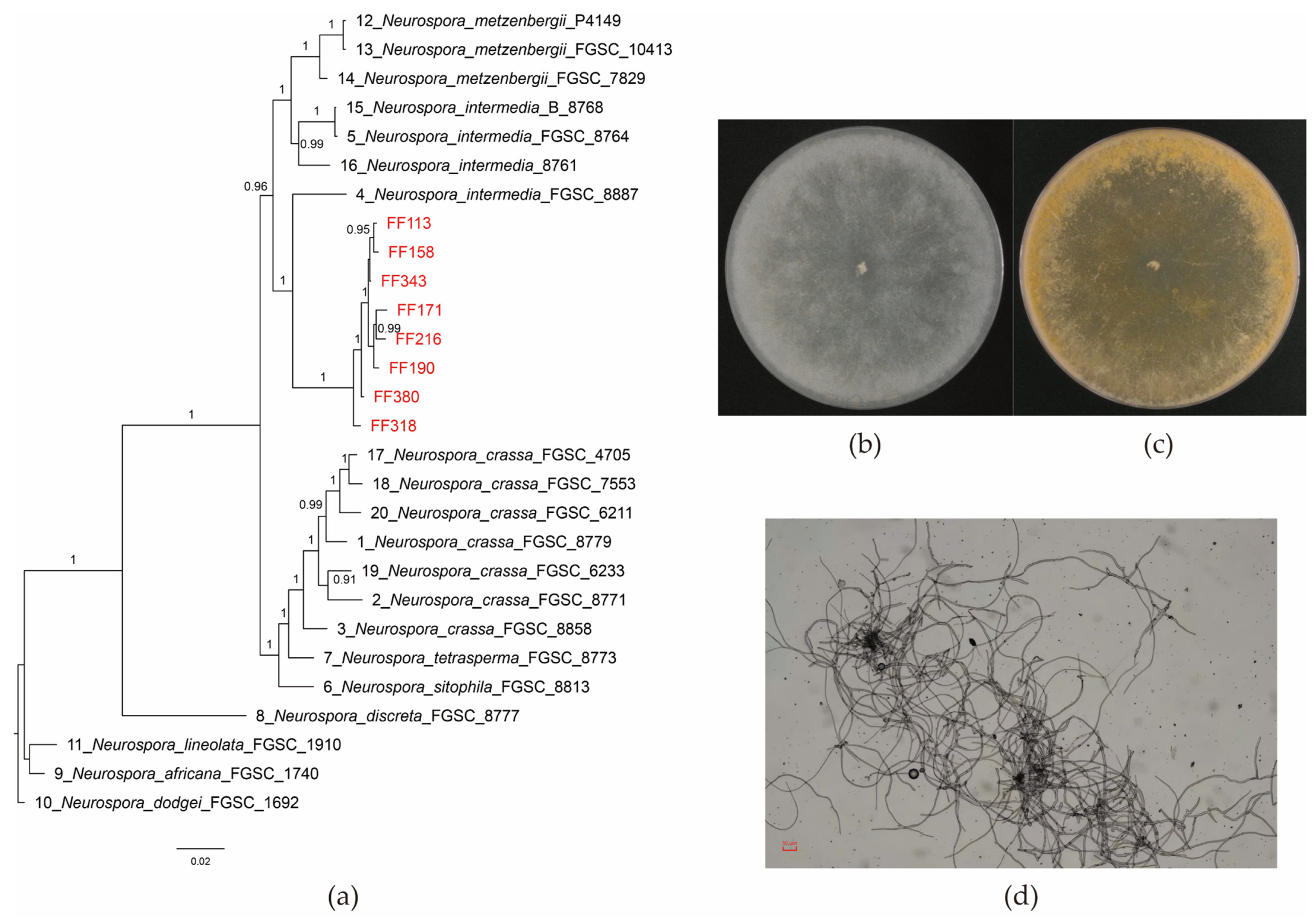
3.1. Isolation, Purification, and Identification of Neurospora intermedia
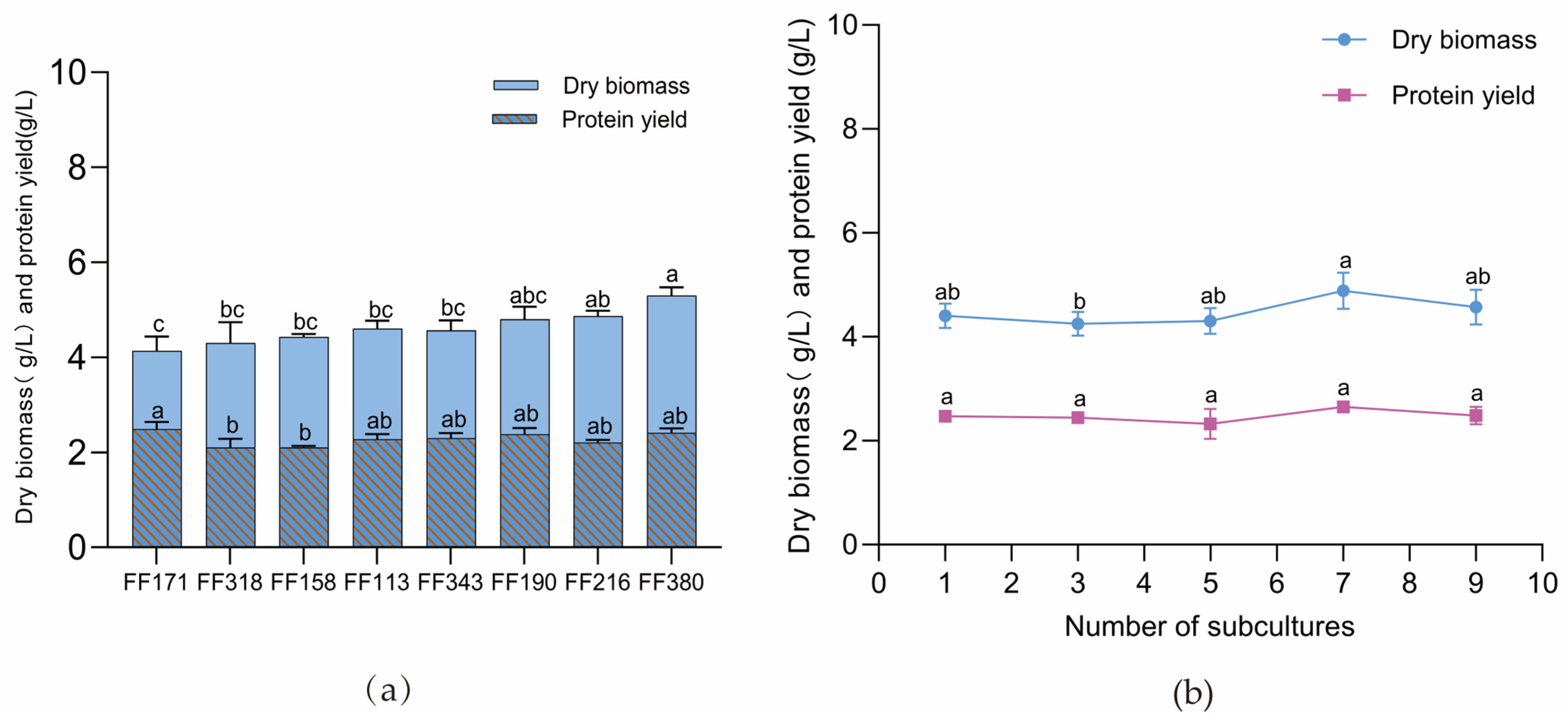
3.2. Protein Production Capacity and Genetic Stability of N. intermedia FF171
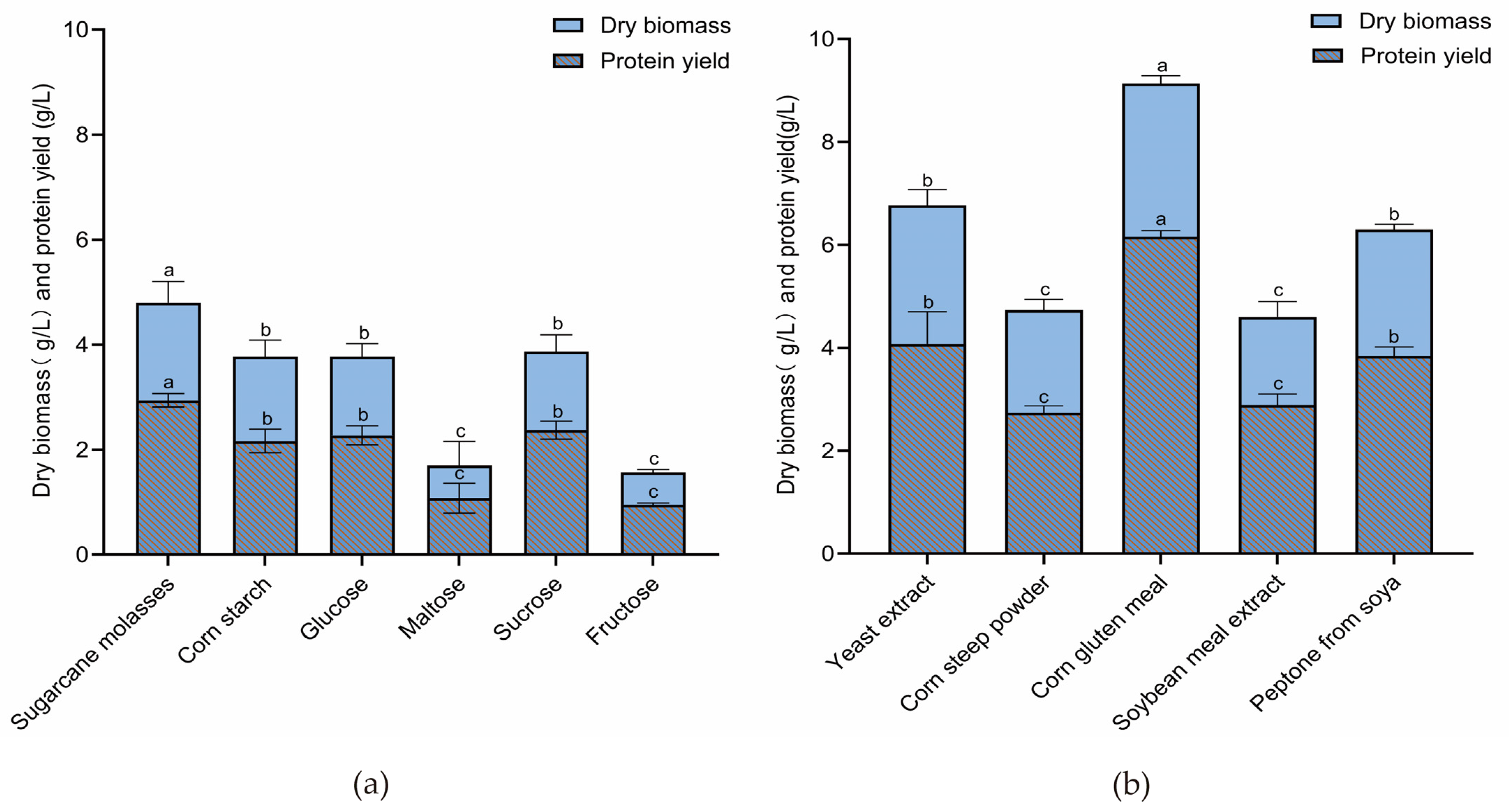
3.3. Effects of Carbon and Nitrogen Sources on Protein Yield of N. intermedia FF171
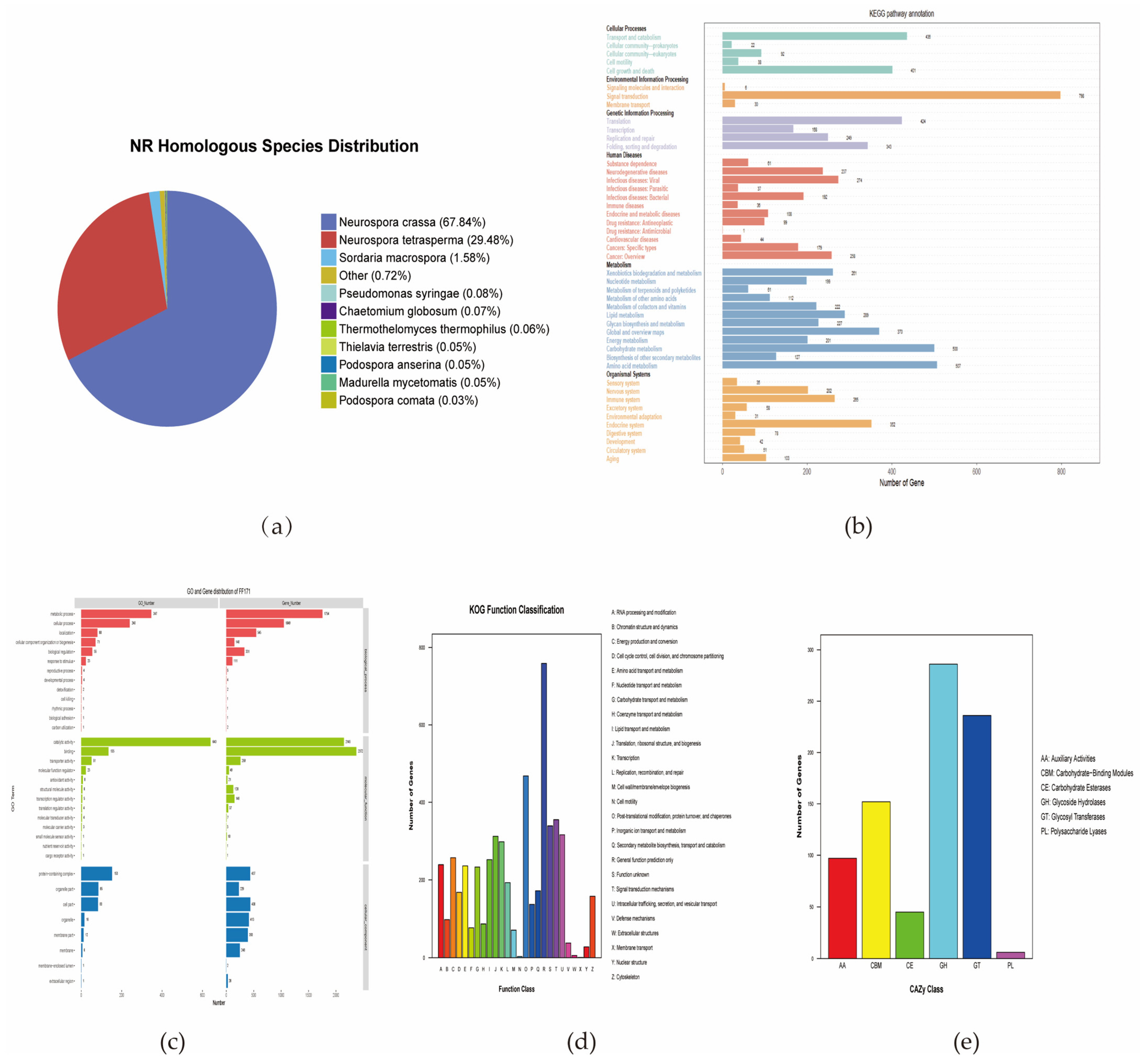
3.4. Genome Sequencing, Annotation, and Metabolic Pathway Analysis of N. intermedia FF171
3.5. Bioinformatics-Based Safety Analysis of the N. intermedia FF171
3.5.1. Antibiotic Resistance
3.5.2. Toxigenicity and Pathogenicity
3.5.3. Assessment of Allergenicity
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Furlan, O.; de Oliveira, N.S.; de Paula, R.C.; Rosa, R.T.; Michelotto, P.V.; Weber, S.H.; Bianchini, L.F.; Rosa, E.A.R. Pilot scale production of high-content mycoprotein using Rhizopus microsporus var. oligosporus by submerged fermentation and agro-industrial by-products. Bioresour. Technol. 2024, 413, 131515. [Google Scholar] [CrossRef]
- Hayes, M. Challenges Regarding Protein Provision for the Growing Global Population: Improving the Environmental Impact of Traditional Protein Supply Chains and Maximising use of Coproducts and Alternative, New Resources. Glob. Chall. 2023, 7, 2300059. [Google Scholar] [CrossRef]
- Lumsden, C.L.; Jägermeyr, J.; Ziska, L.; Fanzo, J. Critical overview of the implications of a global protein transition in the face of climate change: Key unknowns and research imperatives. One Earth 2024, 7, 1187–1201. [Google Scholar] [CrossRef]
- Aimutis, W.R.; Shirwaiker, R. A perspective on the environmental impact of plant-based protein concentrates and isolates. Proc. Natl. Acad. Sci. USA 2024, 121, e2319003121. [Google Scholar] [CrossRef]
- Lima, M.; Costa, R.; Rodrigues, I.; Lameiras, J.; Botelho, G. A Narrative Review of Alternative Protein Sources: Highlights on Meat, Fish, Egg and Dairy Analogues. Foods 2022, 11, 2053. [Google Scholar] [CrossRef]
- Qian, F.; Riddle, M.C.; Wylie-Rosett, J.; Hu, F.B. Red and Processed Meats and Health Risks: How Strong Is the Evidence? Diabetes Care 2020, 43, 265–271. [Google Scholar] [CrossRef]
- Gamarra-Castillo, O.; Echeverry-Montaña, N.; Marbello-Santrich, A.; Hernández-Carrión, M.; Restrepo, S. Meat Substitute Development from Fungal Protein (Aspergillus oryzae). Foods 2022, 11, 2940. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, L.; Panaite, S.-A.; Bertazzo, A.; Visioli, F. Animal- and Plant-Based Protein Sources: A Scoping Review of Human Health Outcomes and Environmental Impact. Nutrients 2022, 14, 5115. [Google Scholar] [CrossRef] [PubMed]
- Hadi, J.; Brightwell, G. Safety of Alternative Proteins: Technological, Environmental and Regulatory Aspects of Cultured Meat, Plant-Based Meat, Insect Protein and Single-Cell Protein. Foods 2021, 10, 1226. [Google Scholar] [CrossRef]
- Lee, D.J.; Na Kang, A.; Lee, J.; Kwak, M.-J.; Mun, D.; Lee, D.; Oh, S.; Kim, Y. Molecular characterization of Fusarium venenatum-based microbial protein in animal models of obesity using multi-omics analysis. Commun. Biol. 2024, 7, 133. [Google Scholar] [CrossRef]
- Bartholomai, B.M.; Ruwe, K.M.; Thurston, J.; Jha, P.; Scaife, K.; Simon, R.; Abdelmoteleb, M.; Goodman, R.E.; Farhi, M. Safety evaluation of Neurospora crassa mycoprotein for use as a novel meat alternative and enhancer. Food Chem. Toxicol. 2022, 168, 113342. [Google Scholar] [CrossRef]
- Lee, D.; Pan, J.H.; Kim, D.; Heo, W.; Shin, E.C.; Kim, Y.J.; Shim, Y.Y.; Reaney, M.J.T.; Ko, S.; Hong, S.; et al. Mycoproteins and their health-promoting properties: Fusarium species and beyond. Compr. Rev. Food Sci. Food Saf. 2024, 23, e13365. [Google Scholar] [CrossRef]
- Molfetta, M.; Morais, E.G.; Barreira, L.; Bruno, G.L.; Porcelli, F.; Dugat-Bony, E.; Bonnarme, P.; Minervini, F. Protein Sources Alternative to Meat: State of the Art and Involvement of Fermentation. Foods 2022, 11, 2065. [Google Scholar] [CrossRef]
- Li, K.; Qiao, K.; Xiong, J.; Guo, H.; Zhang, Y. Nutritional Values and Bio-Functional Properties of Fungal Proteins: Applications in Foods as a Sustainable Source. Foods 2023, 12, 4388. [Google Scholar] [CrossRef]
- U.S. FDA. Agency Response Letter GRAS Notice No. GRN 91. Marlow Foods Ltd., North Yorkshire, UK. Mycoprotein. Available online: https://hfpappexternal.fda.gov/scripts/fdcc/index.cfm?set=GRASNotices&id=91&sort=GRN_No&order=DESC&startrow=1&type=basic&search=GRN%2091 (accessed on 10 January 2025).
- U.S. FDA. Agency Response Letter GRAS Notice No. GRN 904. Natures Fynd, Pershing Rd, Chicago. Fungal Protein from Fusarium sp. Mycelia. Available online: https://www.fda.gov/media/142277/download (accessed on 10 January 2025).
- U.S. FDA. Agency Response Letter GRAS Notice No. GRN 1117. The Better Meat Co, Promenade Street, West Sacramento. Mycelial Biomass from Neurospora crassa. Available online: https://www.fda.gov/media/175166/download (accessed on 10 January 2025).
- Haas, D.; Pfeifer, B.; Reiterich, C.; Partenheimer, R.; Reck, B.; Buzina, W. Identification and quantification of fungi and mycotoxins from Pu-erh tea. Int. J. Food Microbiol. 2013, 166, 316–322. [Google Scholar] [CrossRef]
- Tian, J.; Zhu, Z.; Wu, B.; Wang, L.; Liu, X. Bacterial and fungal communities in Pu’er tea samples of different ages. J. Food Sci. 2013, 78, M1249–M1256. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Skaar, I.; Sulyok, M.; Liu, X.; Rao, M.; Taylor, J.W. The Microbiome and Metabolites in Fermented Pu-erh Tea as Revealed by High-Throughput Sequencing and Quantitative Multiplex Metabolite Analysis. PLoS ONE 2016, 11, e0157847. [Google Scholar] [CrossRef] [PubMed]
- Perkins, D.D.; Davis, R.H. Evidence for safety of Neurospora species for academic and commercial uses. Appl. Environ. Microbiol. 2000, 66, 5107–5109. [Google Scholar] [CrossRef] [PubMed]
- Shahryari, Z.; Fazaelipoor, M.H.; Ghasemi, Y.; Lennartsson, P.R.; Taherzadeh, M.J. Amylase and Xylanase from Edible Fungus Neurospora intermedia: Production and Characterization. Molecules 2019, 24, 721. [Google Scholar] [CrossRef]
- Lozy, F.; Meetro, J.; Simon, R.; Calabrese, P.; Whiteley, J.M. Genotoxicity, acute, and subchronic toxicity evaluation of dried Neurospora crassa protein-rich biomass. Toxicol. Res. 2022, 11, 1003–1017. [Google Scholar] [CrossRef]
- Gmoser, R.; Ferreira, J.A.; Taherzadeh, M.J.; Lennartsson, P.R. Post-treatment of Fungal Biomass to Enhance Pigment Production. Appl. Biochem. Biotechnol. 2019, 189, 160–174. [Google Scholar] [CrossRef]
- Nair, R.B.; Taherzadeh, M.J. Valorization of sugar-to-ethanol process waste vinasse: A novel biorefinery approach using edible ascomycetes filamentous fungi. Bioresour. Technol. 2016, 221, 469–476. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, L.; Zhang, H.; Li, H.; Yang, Y.; Yu, J.; Xiang, H. First Report of Root Rot of Tobacco Caused by Fusarium concentricum in China. Plant Disease 2024, 109, 719. [Google Scholar] [CrossRef]
- Dettman, J.R.; Jacobson, D.J.; Taylor, J.W. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 2003, 57, 2703–2720. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- AQSIQ. Determination of Aflatoxins, Ochratoxin, Fumonisin B1, Deoxynivalenol, T-2 and HT-2 Toxins in Peanut, Grain and Their Products for Export. Available online: https://www.nhc.gov.cn/sps/c100088/201704/e8b91e9057a84df7a30875a69bf7de12/files/1732844732475_44027.zip (accessed on 10 January 2025).
- Zerbino, D.R.; McEwen, G.K.; Margulies, E.H.; Birney, E. Pebble and Rock Band: Heuristic Resolution of Repeats and Scaffolding in the Velvet Short-Read de Novo Assembler. PLoS ONE 2009, 4, e8407. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef]
- Hunt, M.; Newbold, C.; Berriman, M.; Otto, T.D. A comprehensive evaluation of assembly scaffolding tools. Genome Biol. 2014, 15, R42. [Google Scholar] [CrossRef]
- Boetzer, M.; Pirovano, W. Toward almost closed genomes with GapFiller. Genome Biol. 2012, 13, R56. [Google Scholar] [CrossRef]
- Harris, M.A.; Clark, J.; Ireland, A.; Lomax, J.; Ashburner, M.; Foulger, R.; Eilbeck, K.; Lewis, S.; Marshall, B.; Mungall, C.; et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- U.S. FDA. Agency Response Letter GRAS Notice No. GRN 945. 3F BIO Limited, Glasgow, UK. Fungal Protein from Fusarium sp. Mycelia. Available online: https://www.fda.gov/media/145554/download (accessed on 10 January 2025).
- U.S. FDA. Agency Response Letter GRAS Notice No. GRN 807. BLIS Technologies Ltd., South Dunedin, New Zealand, Streptococcus salivarius M18. Available online: https://www.fda.gov/media/133874/download (accessed on 10 January 2025).
- U.S. FDA. Agency Response Letter GRAS Notice No. GRN 725. Ganeden Biotech, Inc., Inactivated Bacillus coagulans GBI-30, 6086, Mayfield Heights (OH). Available online: https://www.fda.gov/media/111306/download (accessed on 10 January 2025).
- U.S. FDA. Agency Response Letter GRAS Notice No. GRN 660. Ganeden Biotech, Inc., Bacillus coagulans GBI-30, 6086 Spores, Mayfield (OH). Available online: https://www.fda.gov/media/100025/download (accessed on 10 January 2025).
- U.S. FDA. Agency Response Letter GRAS Notice No. GRN 281. Fonterra Co operative Group, Auckland, New Zealand. Lactobacillus rhamnosus Strain HN001 Produced in a Milk-Based Medium. Available online: https://hfpappexternal.fda.gov/scripts/fdcc/index.cfm?set=GRASNotices&id=281 (accessed on 10 January 2025).
- U.S. FDA. Agency Response Letter GRAS Notice No. GRN 531. Biosearch Life, S.A, Lactobacillus fermentum CECT5716, Granada, Spain. Available online: https://hfpappexternal.fda.gov/scripts/fdcc/index.cfm?set=GRASNotices&id=531 (accessed on 10 January 2025).
- U.S. FDA. Agency Response Letter GRAS Notice No. GRN 758. Lallemand Health Solutions, Lactobacillus helveticus R0052, Bifidobacterium longum subsp. Infantis R0033, and Bifidobacterium bifidum R0071, Mirabel (QC). Available online: https://www.fda.gov/media/122558/download (accessed on 10 January 2025).
- U.S. FDA. Agency Response Letter GRAS Notice No. GRN 865. Dupont Nutrition and Biosciences (Danisco USA, Inc), Madison (WI). Lactobacillus acidophilus NCFM. Available online: https://www.fda.gov/media/134208/download (accessed on 10 January 2025).
- U.S. FDA. Agency Response Letter GRAS Notice No. GRN 814. BIFIDO Co., Ltd, Gangwon Province, Republic of Korea. Bifidobacterium bifidum BGN4. Available online: https://www.fda.gov/media/133875/download (accessed on 10 January 2025).
- Pearson, W.R. An introduction to sequence similarity (“homology”) searching. Curr. Protoc. Bioinform. 2013, 42, 3.1.1–3.1.8. [Google Scholar] [CrossRef]
- Scaife, K.; Vo, T.D.; Dommels, Y.; Leune, E.; Albermann, K.; Pařenicová, L. In silico and in vitro safety assessment of a fungal biomass from Rhizomucor pusillus for use as a novel food ingredient. Food Chem. Toxicol. 2023, 179, 113972. [Google Scholar] [CrossRef]
- EFSA. Scientific Guidance for the Submission of Dossiers on Food Enzymes. Available online: https://doi.org/10.2903/j.efsa.2021.6851 (accessed on 25 January 2025). [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- EFSA. EFSA Statement on the Requirements for Whole Genome Sequence Analysis of Microorganisms Intentionally Used in the Food Chain. Efsa J. Available online: https://doi.org/10.2903/j.efsa.2024.8912 (accessed on 25 January 2025). [CrossRef]
- Pariza, M.W.; Gillies, K.O.; Kraak-Ripple, S.F.; Leyer, G.; Smith, A.B. Determining the safety of microbial cultures for consumption by humans and animals. Regul. Toxicol. Pharmacol. 2015, 73, 164–171. [Google Scholar] [CrossRef]
- van der Vlugt, C.J. Overview of sixteen scientific opinions on genetically modified plants obtained by new genomic techniques. EFSA Support. Publ. 2021, 18, 1973E. [Google Scholar] [CrossRef]
- Cosentino, S.; Voldby Larsen, M.; Møller Aarestrup, F.; Lund, O. PathogenFinder—Distinguishing Friend from Foe Using Bacterial Whole Genome Sequence Data. PLoS ONE 2013, 8, e77302. [Google Scholar] [CrossRef]
- Sharma, N.; Patiyal, S.; Dhall, A.; Pande, A.; Arora, C.; Raghava, G.P.S. AlgPred 2.0: An improved method for predicting allergenic proteins and mapping of IgE epitopes. Brief. Bioinform. 2021, 22, bbaa294. [Google Scholar] [CrossRef] [PubMed]
- FAO/WHO. Evaluation of Allergenicity of Genetically Modified Foods: Report of a Joint FAO/WHO Expert Consultation on Allergenicity of Foods Derived from Biotechnology, 22–25 January 2001. Available online: https://iris.who.int/handle/10665/340572 (accessed on 23 January 2025).
- Commission, C.A. Guideline for the Conduct of Food Safety Assessment of Foods Derived from Recombinant-DNA Plants. Available online: https://www.fao.org/4/y5819e/y5819e03.htm (accessed on 23 January 2025).
- Commission, C.A. Foods Derived from Modern Biotechnology. Available online: https://openknowledge.fao.org/bitstreams/3229fca1-ff5a-4561-8e89-cd4a48a6438a/download (accessed on 23 January 2025).
- EFSA Panel on Genetically Modified Organisms (GMO); Naegeli, H.; Birch, A.N.; Casacuberta, J.; De Schrijver, A.; Gralak, M.A.; Guerche, P.; Jones, H.; Manachini, B.; Messéan, A.; et al. Guidance on allergenicity assessment of genetically modified plants. EFSA J. 2017, 15, e04862. [Google Scholar] [CrossRef] [PubMed]
- Abdelmoteleb, M.; Zhang, C.; Furey, B.; Kozubal, M.; Griffiths, H.; Champeaud, M.; Goodman, R.E. Evaluating potential risks of food allergy of novel food sources based on comparison of proteins predicted from genomes and compared to www.AllergenOnline.org. Food Chem. Toxicol. 2021, 147, 111888. [Google Scholar] [CrossRef]
- Scaife, K.; Taylor, S.L.; Pařenicová, L.; Goodman, R.E.; Vo, T.D.; Leune, E.; Abdelmoteleb, M.; Dommels, Y. In silico evaluation of the potential allergenicity of a fungal biomass from Rhizomucor pusillus for use as a novel food ingredient. Regul. Toxicol. Pharmacol. 2024, 150, 105629. [Google Scholar] [CrossRef]
- Coelho, M.O.C.; Monteyne, A.J.; Dunlop, M.V.; Harris, H.C.; Morrison, D.J.; Stephens, F.B.; Wall, B.T. Mycoprotein as a possible alternative source of dietary protein to support muscle and metabolic health. Nutr. Rev. 2020, 78, 486–497. [Google Scholar] [CrossRef] [PubMed]
- Furey, B.; Slingerland, K.; Bauter, M.R.; Dunn, C.; Goodman, R.E.; Koo, S. Safety evaluation of Fy Protein™ (Nutritional Fungi Protein), a macroingredient for human consumption. Food Chem. Toxicol. 2022, 166, 113005. [Google Scholar] [CrossRef]
- Hashemi, S.S.; Karimi, K.; Taherzadeh, M.J. Integrated process for protein, pigments, and biogas production from baker’s yeast wastewater using filamentous fungi. Bioresour. Technol. 2021, 337, 125356. [Google Scholar] [CrossRef]
- He, H.; Cao, Z.; Wang, T.; Tang, C.; Li, Y.; Li, X. Metabolomics Combined with Physiology and Transcriptomics Reveal the Response of Samsoniella hepiali to Key Metabolic Pathways and Its Degradation Mechanism during Subculture. Antioxidants 2024, 13, 780. [Google Scholar] [CrossRef]
- Renno, D.V.; Saunders, G.; Bull, A.T.; Holt, G. The genetic stability of Penicillium chrysogenum transformants in a fermentor. Appl. Microbiol. Biotechnol. 1990, 34, 364–367. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.W.; Branco, S.; Gao, C.; Hann-Soden, C.; Montoya, L.; Sylvain, I.; Gladieux, P. Sources of Fungal Genetic Variation and Associating It with Phenotypic Diversity. Microbiol. Spectr. 2017, 5, 635–655. [Google Scholar] [CrossRef]
- Kim, G.Y.; Yang, J.; Han, Y.H.; Seo, S.W. Synthetic redesign of Escherichia coli W for faster metabolism of sugarcane molasses. Microb. Cell Factories 2024, 23, 242. [Google Scholar] [CrossRef] [PubMed]
- González-Torres, M.; Hernández-Rosas, F.; Pacheco, N.; Salinas-Ruiz, J.; Herrera-Corredor, J.A.; Hernández-Martínez, R. Levan Production by Suhomyces kilbournensis Using Sugarcane Molasses as a Carbon Source in Submerged Fermentation. Molecules 2024, 29, 1105. [Google Scholar] [CrossRef]
- Ortiz, M.E.; Fornaguera, M.J.; Raya, R.R.; Mozzi, F. Lactobacillus reuteri CRL 1101 highly produces mannitol from sugarcane molasses as carbon source. Appl. Microbiol. Biotechnol. 2012, 95, 991–999. [Google Scholar] [CrossRef]
- Naseem, A.; Rasul, I.; Raza, Z.A.; Muneer, F.; Rehman, A.U.; Nadeem, H. Bacterial production of polyhydroxyalkanoates (PHAs) using various waste carbon sources. PeerJ 2024, 12, e17936. [Google Scholar] [CrossRef]
- Ye, Q.; Li, X.; Yan, M.; Cao, H.; Xu, L.; Zhang, Y.; Chen, Y.; Xiong, J.; Ouyang, P.; Ying, H. High-level production of heterologous proteins using untreated cane molasses and corn steep liquor in Escherichia coli medium. Appl. Microbiol. Biotechnol. 2010, 87, 517–525. [Google Scholar] [CrossRef]
- Tateno, T.; Fukuda, H.; Kondo, A. Direct production of L-lysine from raw corn starch by Corynebacterium glutamicum secreting Streptococcus bovis alpha-amylase using cspB promoter and signal sequence. Appl. Microbiol. Biotechnol. 2007, 77, 533–541. [Google Scholar] [CrossRef]
- Finnigan, T.; Theobald, H.; Bajka, B. Mycoprotein: A Healthy and Sustainable Source of Alternative Protein-Based Foods. Annu. Rev. Food Sci. Technol. 2025, 16, 105–125. [Google Scholar] [CrossRef] [PubMed]
- Wiebe, M. Myco-protein from Fusarium venenatum: A well-established product for human consumption. Appl. Microbiol. Biotechnol. 2002, 58, 421–427. [Google Scholar] [CrossRef]
- Ge, H.; Zhang, Z.; Xiao, J.; Tan, H.; Huang, G. Release of Leu-Pro-Pro from corn gluten meal by fermentation with a Lactobacillus helveticus strain. J. Sci. Food Agric. 2022, 102, 1095–1104. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, S.S.; Karimi, K.; Taherzadeh, M.J. Valorization of vinasse and whey to protein and biogas through an environmental fungi-based biorefinery. J. Environ. Manag. 2022, 303, 114138. [Google Scholar] [CrossRef] [PubMed]
- Svedberg, J.; Hosseini, S.; Chen, J.; Vogan, A.A.; Mozgova, I.; Hennig, L.; Manitchotpisit, P.; Abusharekh, A.; Hammond, T.M.; Lascoux, M.; et al. Convergent evolution of complex genomic rearrangements in two fungal meiotic drive elements. Nat. Commun. 2018, 9, 4242. [Google Scholar] [CrossRef]
- Dubey, M.K.; Aamir, M.; Kaushik, M.S.; Khare, S.; Meena, M.; Singh, S.; Upadhyay, R.S. PR Toxin—Biosynthesis, Genetic Regulation, Toxicological Potential, Prevention and Control Measures: Overview and Challenges. Front. Pharmacol. 2018, 9, 288. [Google Scholar] [CrossRef]
- Rekdal, V.M.; Villalobos-Escobedo, J.M.; Rodriguez-Valeron, N.; Garcia, M.O.; Vásquez, D.P.; Rosales, A.; Sörensen, P.M.; Baidoo, E.E.K.; de Carvalho, A.C.; Riley, R.; et al. Neurospora intermedia from a traditional fermented food enables waste-to-food conversion. Nat. Microbiol. 2024, 9, 2666–2683. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Chen, Y.; Chou, H.; Lee, S.; Tai, H.; Shen, H.D. cDNA cloning and immunologic characterization of a novel EF-1beta allergen from Penicillium citrinum. Allergy 2005, 60, 366–371. [Google Scholar] [CrossRef] [PubMed]
| Anno-Database | Annotated Number | Percentage (%) |
|---|---|---|
| NR | 8806 | 98.81 |
| KEGG | 4899 | 54.97 |
| GO | 4582 | 51.41 |
| KOG | 4717 | 52.93 |
| CAZy | 822 | 9.22 |
| Pfam | 6185 | 69.40 |
| Swiss-Prot | 5792 | 64.99 |
| Total | 8912 | 100% |
| N. intermedia FF171 Protein from Genome | Allergens Matched in Allergen Online | Description |
|---|---|---|
| Overall Conclusion of Risks for Cross-Reactivity for N. intermedia FF171 Protein | ||
| g3197.t1_1 Elongation factor 1-beta | gi|38326693|gid|246|elongation factor 1 beta-like | It was further evaluated. |
| g2102.t1_1 Superoxide dismutase [Mn], mitochondrial | gi|83305645|gid|330|Superoxide dismutase [Mn], mitochondrial | It shares significant sequence homology (54.10% identity) with the superoxide dismutase [Mn](mitochondrial) from a GRAS strain (GRN No. 1117). It is highly unlikely to pose a risk of food allergy. |
| gi|1648970|gid|330|manganese superoxide dismutase | ||
| gi|529279957|gid|1885|manganese superoxide dismutase | ||
| gi|348137|gid|590|superoxide dismutase (manganese) | ||
| gi|149786150|gid|1092|manganese superoxide dismutase-like protein | ||
| gi|10862818|gid|590|IgE-binding protein MnSOD | ||
| gi|5777414|gid|590|MnSOD | ||
| g7030.t1_1 Heat shock 70 kDa protein | gi|14423733|gid|250|Heat shock 70 kDa protein | It shares significant sequence homology (99.59% identity) with the heat shock 70 kDa protein from a GRAS strain (GRN No. 1117). It is highly unlikely to pose a risk of food allergy. |
| gi|729764|gid|519|Heat shock 70 kDa protein | ||
| gi|442565876|gid|2076|heat shock protein 70 | ||
| gi|1055365842|gid|2591|heat shock-like protein | ||
| gi|1561006361|gid|3077|allergen Der p 28 | ||
| gi|685432788|gid|2076|Der f 28 allergen | ||
| gi|94468818|gid|2708|heat shock cognate 70 | ||
| g2853.t1_1 Translationally controlled tumor protein homolog | gi|112824341|gid|1337|TCTP | It shares significant sequence homology (99.41% identity) with the translationally controlled tumor protein homolog from a GRAS strain (GRN No. 1117). It is highly unlikely to pose a risk of food allergy. |
| gi|1679357707|gid|2864|RecName: Full = Translationally-cont | ||
| g513.t1_1 Alcohol dehydrogenase 1 | gi|86278351|gid|2582|alcohol dehydrogenase | It shares significant sequence homology (65.04% identity) with the alcohol dehydrogenase 1 from a GRAS strain (GRN No. 1117). It is highly unlikely to pose a risk of food allergy. |
| gi|608690|gid|395|alcohol dehydrogenase | ||
| g4129.t1_1 Protein argonaute-2 | gi|291197394|gid|1338|ragweed homologue of Art v 1 precursor | It shares significant sequence homology (59.40% identity) with the ATP-dependent RNA helicase ded1 from a GRAS strain (GRN No. 1117). It is highly unlikely to pose a risk of food allergy. |
| gi|285005079|gid|1338|ragweed homologue of Art v 1 precur | ||
| gi|291482310|gid|1338|ragweed homologue of Art v 1 precur | ||
| gi|291482308|gid|1338|ragweed homologue of Art v 1 precur | ||
| g816.t1_1 60S acidic ribosomal protein P1 | gi|371537645|gid|1983|60S acidic ribosomal phosphoprotein P1 | It shares significant sequence homology (50.00% identity) with the large ribosomal subunit protein uL10 from a GRAS strain (GRN No. 1117). It is highly unlikely to pose a risk of food allergy. |
| gi|1350779|gid|73|60S acidic ribosomal protein P1 | ||
| gi|59894749|gid|848|60S acidic ribosomal P1 phosphoprotein Pen b 26 | ||
| gi|83305635|gid|331|60S acidic ribosomal protein P2 | ||
| gi|6686524|gid|331|rAsp f 8 | ||
| gi|5777795|gid|520|minor allergen, ribosomal protein P2 | ||
| g1058.t1_1 Nuclear transport factor 2 | gi|21748151|gid|489|putative nuclear transport factor 2 | It shares significant sequence homology (100.00% identity) with the nuclear transport factor 2 from a GRAS strain (GRN No. 1117). It is highly unlikely to pose a risk of food allergy. |
| gi|21748153|gid|72|putative nuclear transport factor 2 | ||
| g5256.t1_1 Peptidyl-prolyl cis-trans isomerase H | gi|4138173|gid|651|allergen | It shares significant sequence homology (98.90% identity) with the peptidyl-prolyl cis-trans isomerase H from a GRAS strain (GRN No. 1117). It is highly unlikely to pose a risk of food allergy. |
| gi|37958141|gid|951|Der f Mal f 6 allergen | ||
| gi|5019414|gid|325|PPIase | ||
| gi|1220142|gid|1926|cyclophilin | ||
| gi|91680605|gid|863|cyclophilin | ||
| gi|373939374|gid|1941|cyclophilin | ||
| gi|2325204258|gid|3374|Per a 18 allergen | ||
| gi|1432030624|gid|3310|peptidyl-prolyl cis-trans isomerase | ||
| gi|1373739558|gid|2869|cyclophilin 0101 | ||
| gi|946715057|gid|2304|cyclophilin A | ||
| gi|21886603|gid|344|peptidylprolyl isomerase | ||
| g5315.t1_1 Cuticle-degrading protease | gi|23894240|gid|313|tri m 2 allergen | It shares significant sequence homology (99.24% identity) with the cuticle-degrading protease from a GRAS strain (GRN No. 1117). It is highly unlikely to pose a risk of food allergy. |
| gi|2612389725|gid|2227|Chain C, Alkaline protease 1 | ||
| gi|2612389721|gid|2227|Chain D, Alkaline protease 1 | ||
| gi|2612389719|gid|2227|Chain C, Alkaline protease 1 | ||
| gi|2612389715|gid|2227|Chain C, Alkaline protease 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Hong, C.; Kang, D.; Zhou, F.; Dong, L.; Yang, G.; Li, M.; Wang, L.; Zhao, H.; Zhang, W.; Cui, Y.; et al. A Novel Mycoprotein Candidate: Neurospora intermedia FF171 from Pu-Erh Tea with Genomics-Based Safety Profiling. Fermentation 2026, 12, 27. https://doi.org/10.3390/fermentation12010027
Hong C, Kang D, Zhou F, Dong L, Yang G, Li M, Wang L, Zhao H, Zhang W, Cui Y, et al. A Novel Mycoprotein Candidate: Neurospora intermedia FF171 from Pu-Erh Tea with Genomics-Based Safety Profiling. Fermentation. 2026; 12(1):27. https://doi.org/10.3390/fermentation12010027
Chicago/Turabian StyleHong, Chengzhen, Dingrong Kang, Furong Zhou, Lichao Dong, Guofei Yang, Mingxia Li, Li Wang, Haifeng Zhao, Wei Zhang, Yinshan Cui, and et al. 2026. "A Novel Mycoprotein Candidate: Neurospora intermedia FF171 from Pu-Erh Tea with Genomics-Based Safety Profiling" Fermentation 12, no. 1: 27. https://doi.org/10.3390/fermentation12010027
APA StyleHong, C., Kang, D., Zhou, F., Dong, L., Yang, G., Li, M., Wang, L., Zhao, H., Zhang, W., Cui, Y., Cao, J., & Zhao, W. (2026). A Novel Mycoprotein Candidate: Neurospora intermedia FF171 from Pu-Erh Tea with Genomics-Based Safety Profiling. Fermentation, 12(1), 27. https://doi.org/10.3390/fermentation12010027







