Isolation and Characterization of Low-Temperature and High-Salinity Amylase from Halomonas sp. KS41843
Abstract
1. Introduction
2. Materials and Methods
2.1. Amylase-Producing Bacteria Screening and Identification
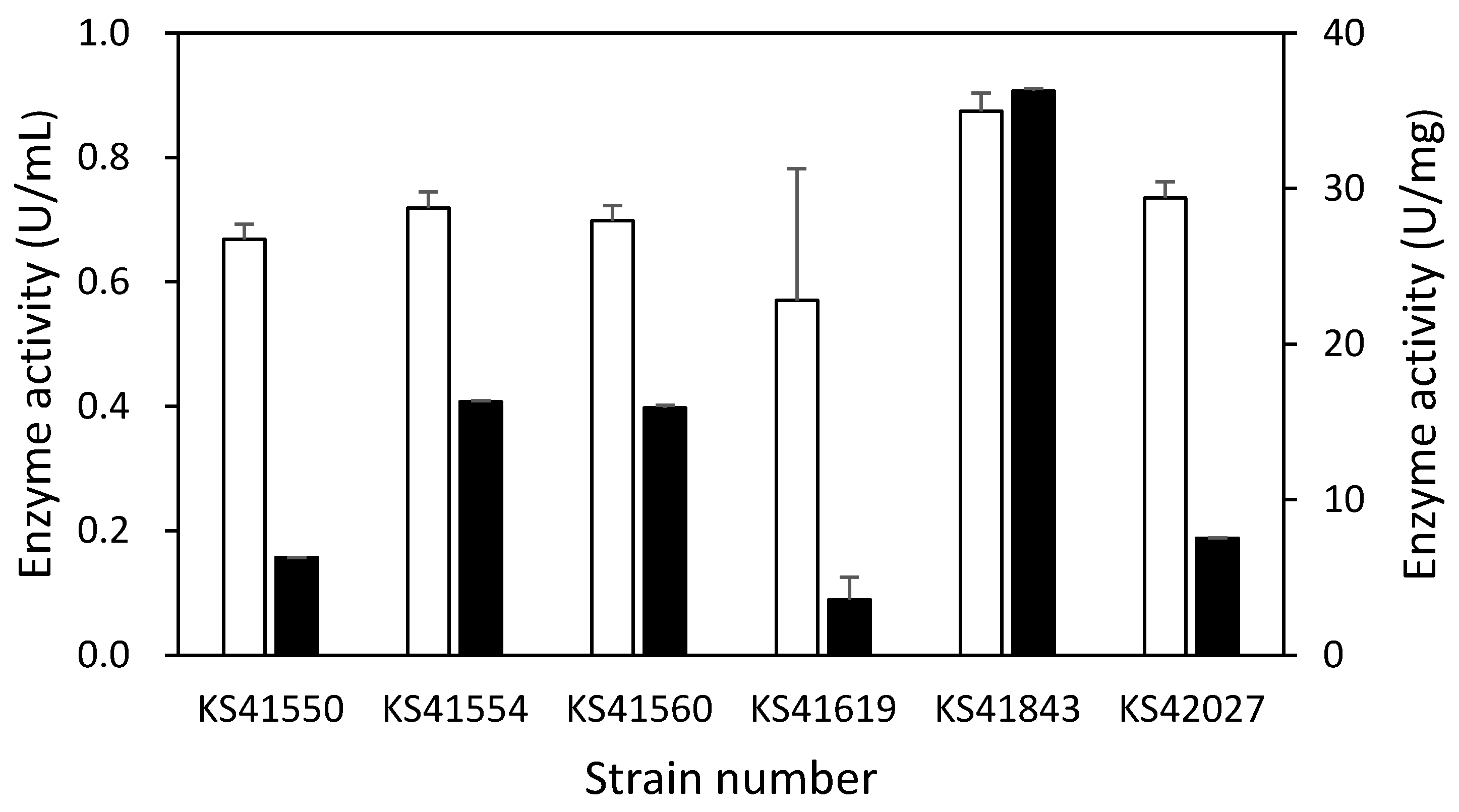
2.2. Amylase Production and Activity Quantification of Six Strains
2.3. Liquid Culture of Halomonas sp. KS41843
2.4. Purification of Amylase
2.5. Characterization of Purified Amylase Enzyme
2.5.1. Effect of Temperature
2.5.2. Effect of pH
2.5.3. Effect of Metal Ions
2.5.4. Effect of NaCl
3. Results
3.1. Identification and Selection of Bacterial Strains Capable of Enzyme Production
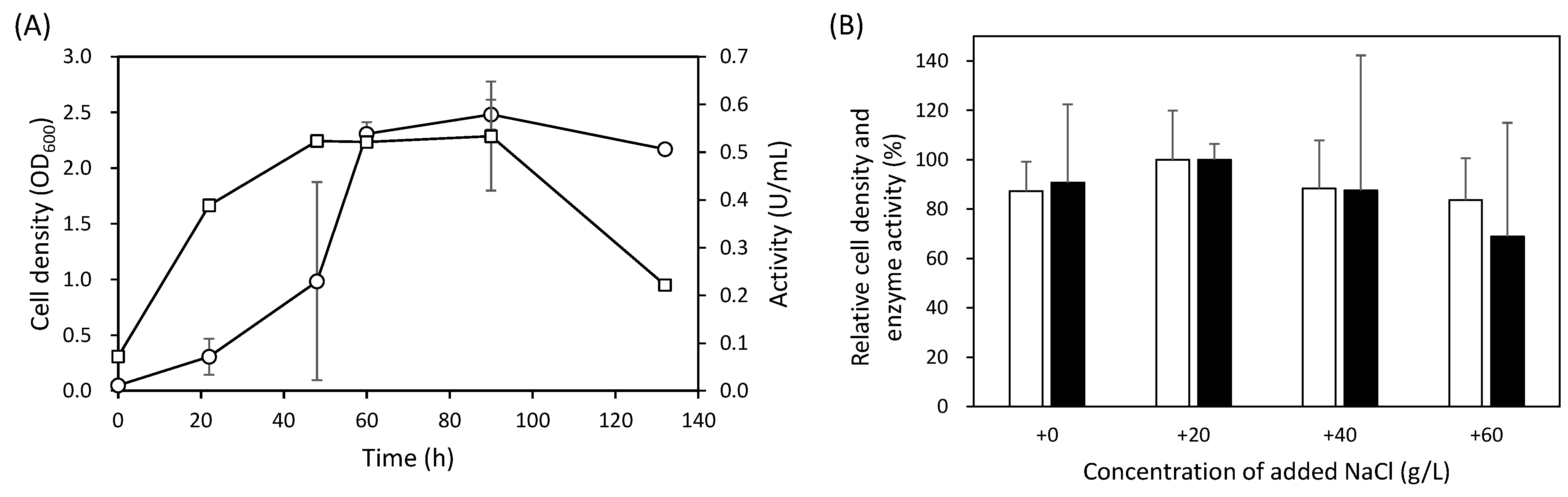
3.2. Effect of Salt Concentration on Cell Growth and Amylase Production by KS41843
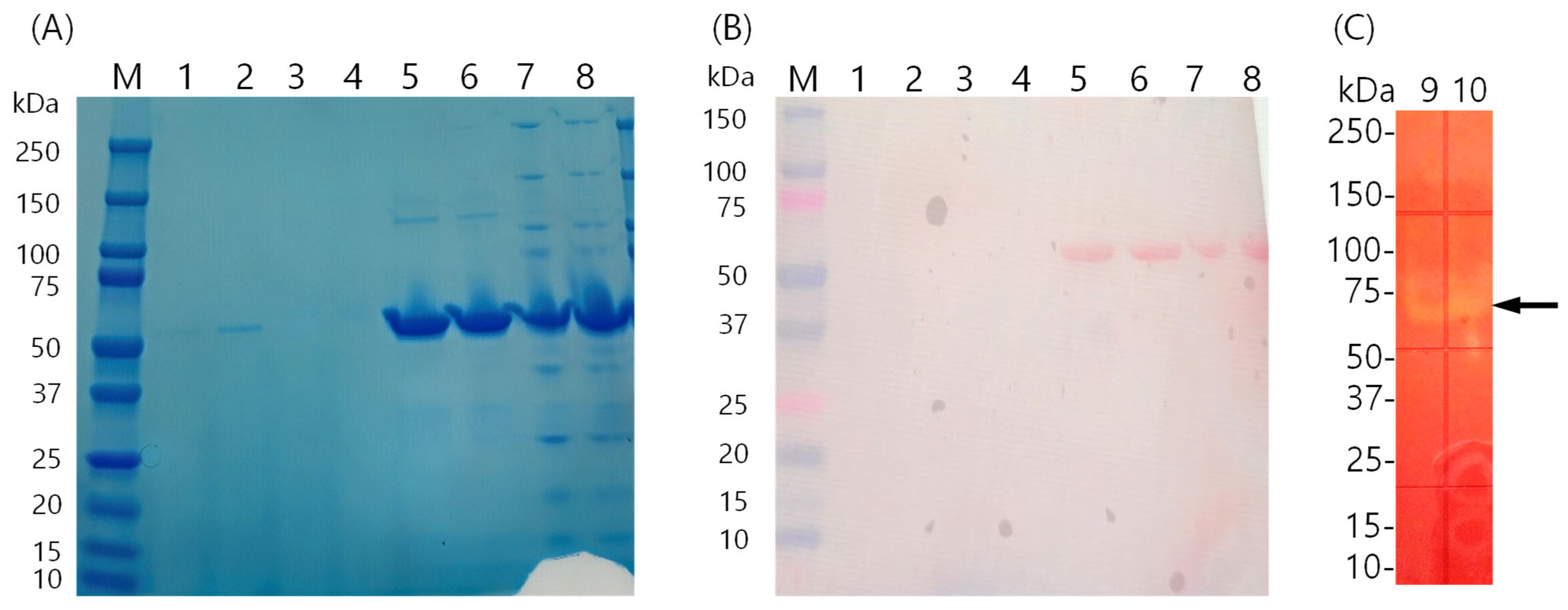
3.3. Production and Purification of Amylase KS41843
3.4. Effect of Temperature and pH on Amylase Activity and Stability
3.5. Effect of Metal Ions on Amylase Activity
3.6. Salt Tolerance of Amylase
4. Discussion
| Enzyme | Microbial Source | Optimum Temperature (°C) | Optimum pH | Specific Activity (U/mg) | Molecular Weight (kDa) | Cations Activators | Reference |
|---|---|---|---|---|---|---|---|
| KS41843 | Halomonas sp. KS41843 | 30 | 5.0 | 110.32 | 60 | Co2+, Mn2+, Mb2+, Fe2+, Na+ | This study |
| AmyD-1 | Bacillus sp. dsh19-1 | 20 | 6.0 | 16.4 | 50.1 | Na+, Ca2+ | [26] |
| KS7913 | Alteromonas sp. KS7913 | 25 | 7.0 | 200.34 | 70 | Mn2+, Ba2+, Ca2+ | [27] |
| α-Amylase | Pseudoalteromonas sp. 2-3 | 20 | 8.0 | 51.7 | 68.8 | Ca2+ | [40] |
| AmyZ | Zunongwangia profunda | 35 | 7.0 | 284.4 | 66 | Sr2+, Fe3+, Mg2+, Ba2+, NH4+, K+ | [41] |
| Ef-Amy I | Eisenia fedida | 40 | 5.5 | 174 | 57 | - | [43] |
| Ef-Amy II | Eisenia fedida | 35 | 5.0 | 65 | 57 | Ca2+ | [43] |
| amylase | Nocardiopsis sp. 7326 | 35 | 8 | 548 | 55 | Ca2+, Mn2+, Mg2+, Cu2+, Co2+ | [44] |
| ParAmy | Pseudoalteromonas arctica GS230 | 30 | 7.5 | 25.5 | 55 | Mn2+, K+, Na+ | [45] |
| GA2 | Microbacterium foliorum | 20 | 9 | - | - | Mg2+ | [47] |
| GA6 | Bacillus cereus | 20 | 10 | - | - | Ca2+ | [47] |
| wtAmy175 | Pseudoalteromonas sp. M175 | 30 | 7.5 | 289.79 | 61 | - | [48] |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fang, W.; Xue, S.; Deng, P.; Zhang, X.; Wang, X.; Xiao, Y.; Fang, Z. AmyZ1: A novel alpha-amylase from marine bacterium Pontibacillus sp. ZY with high activity toward raw starches. Biotechnol. Biofuels 2019, 12, 95. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Chu, X.; Lv, J.; Li, Q.; Tian, J.; Wu, N. Improving the thermostability of acidic pullulanase from Bacillus naganoensis by rational design. PLoS ONE 2016, 11, e0165006. [Google Scholar] [CrossRef]
- Thakur, M.; Sharma, N.; Rai, A.K.; Singh, S.P. A novel cold-active type I pullulanase from a hot-spring metagenome for effective debranching and production of resistant starch. Bioresour. Technol. 2021, 320, 124288. [Google Scholar] [CrossRef]
- Zhang, S.Y.; Guo, Z.W.; Wu, X.L.; Ou, X.Y.; Zong, M.H.; Lou, W.Y. Recombinant expression and characterization of a novel cold-adapted type I pullulanase for efficient amylopectin hydrolysis. J. Biotechnol. 2020, 313, 39–47. [Google Scholar] [CrossRef]
- Mahmoudnia, F. Comparison of the synthesis of the alpha-amylase enzyme by the native strain Bacillus licheniformis in immobilized and immersed cells. Iran. J. Microbiol. 2024, 16, 827–834. [Google Scholar] [CrossRef]
- Siddiqui, K.S.; Cavicchioli, R. Cold-adapted enzymes. Ann. Rev. Biochem. 2006, 75, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Javed, A.; Qazi, J.I. Psychrophilic microbial enzymes implications in coming biotechnological processes. Am. Sci. Res. J. Eng. Technol. Sci. 2016, 23, 103–120. [Google Scholar]
- Gocheva, Y.G.; Krumova, E.T.; Slokoska, L.S.; Miteva, J.G.; Vassilev, S.V.; Angelova, M.B. Cell response of Antarctic and temperate strains of Penicillium spp. to different growth temperature. Mycol. Res. 2006, 110 Pt 11, 1347–1354. [Google Scholar] [CrossRef]
- Goncalves, V.N.; Vaz, A.B.; Rosa, C.A.; Rosa, L.H. Diversity and distribution of fungal communities in lakes of Antarctica. FEMS Microbiol. Ecol. 2012, 82, 459–471. [Google Scholar] [CrossRef]
- de Souza, L.M.D.; Ogaki, M.B.; Teixeira, E.A.A.; de Menezes, G.C.A.; Convey, P.; Rosa, C.A.; Rosa, L.H. Communities of culturable freshwater fungi present in Antarctic lakes and detection of their low-temperature-active enzymes. Braz. J. Microbiol. 2023, 54, 1923–1933. [Google Scholar] [CrossRef] [PubMed]
- Cavicchioli, R.; Charlton, T.; Ertan, H.; Mohd Omar, S.; Siddiqui, K.S.; Williams, T.J. Biotechnological uses of enzymes from psychrophiles. Microb. Biotechnol. 2011, 4, 449–460. [Google Scholar] [CrossRef]
- Zucconi, L.; Canini, F.; Temporiti, M.E.; Tosi, S. Extracellular enzymes and bioactive compounds from Antarctic terrestrial fungi for bioprospecting. Int. J. Environ. Res. Public Health 2020, 17, 6459. [Google Scholar] [CrossRef]
- Margesin, R.; Feller, G. Biotechnological applications of psychrophiles. Environ. Technol. 2010, 31, 835–844. [Google Scholar] [CrossRef]
- Ramasamy, K.P.; Mahawar, L.; Rajasabapathy, R.; Rajeshwari, K.; Miceli, C.; Pucciarelli, S. Comprehensive insights on environmental adaptation strategies in Antarctic bacteria and biotechnological applications of cold adapted molecules. Front. Microbiol. 2023, 14, 1197797. [Google Scholar] [CrossRef]
- Yavari-Bafghi, M.; Amoozegar, M.A. Pharmaceutical applications of halophilic enzymes. Heliyon 2025, 11, e42754. [Google Scholar] [CrossRef]
- Wang, G.; Luo, M.; Lin, J.; Lin, Y.; Yan, R.; Streit, W.R.; Ye, X. A new extremely halophilic, calcium-independent and surfactant-resistant alpha-amylase from Alkalibacterium sp. SL3. J. Microbiol. Biotechnol. 2019, 29, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Bandal, J.N.; Tile, V.A.; Sayyed, R.Z.; Jadhav, H.P.; Azelee, N.I.W.; Danish, S.; Datta, R. Statistical based bioprocess design for improved production of amylase from halophilic Bacillus sp. H7 isolated from marine water. Molecules 2021, 26, 2833. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Khare, S.K. Purification and characterization of maltooligosaccharide-forming alpha-amylase from moderately halophilic Marinobacter sp. EMB8. Bioresour. Technol. 2012, 116, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Khare, S.K. Chloride activated halophilic alpha-amylase from Marinobacter sp. EMB8: Production optimization and nanoimmobilization for efficient starch hydrolysis. Enzym. Res. 2015, 2015, 859485. [Google Scholar] [CrossRef]
- Kumar, S.; Karan, R.; Kapoor, S.; Singh, S.P.; Khare, S.K. Screening and isolation of halophilic bacteria producing industrially important enzymes. Braz. J. Microbiol. 2012, 43, 1595–1603. [Google Scholar] [CrossRef]
- Dalmaso, G.Z.; Ferreira, D.; Vermelho, A.B. Marine extremophiles: A source of hydrolases for biotechnological applications. Mar. Drugs 2015, 13, 1925–1965. [Google Scholar] [CrossRef] [PubMed]
- Oliva, G.; Di Stasio, L.; Vigliotta, G.; Guarino, F.; Cicatelli, A.; Castiglione, S. Exploring the potential of four novel halotolerant bacterial strains as Plant-Growth-Promoting Rhizobacteria (PGPR) under saline conditions. Appl. Sci. 2023, 13, 4320. [Google Scholar] [CrossRef]
- Dutta, B.; Bandopadhyay, R. Biotechnological potentials of halophilic microorganisms and their impact on mankind. Beni Suef Univ. J. Basic Appl. Sci. 2022, 11, 75. [Google Scholar] [CrossRef]
- Ben Hmad, I.; Gargouri, A. Halophilic filamentous fungi and their enzymes: Potential biotechnological applications. J. Biotechnol. 2024, 381, 11–18. [Google Scholar] [CrossRef]
- Ahmad, A.; Rahamtullah; Mishra, R. Structural and functional adaptation in extremophilic microbial alpha-amylases. Biophys. Rev. 2022, 14, 499–515. [Google Scholar] [CrossRef]
- Dou, S.; Chi, N.; Zhou, X.; Zhang, Q.; Pang, F.; Xiu, Z. Molecular cloning, expression, and biochemical characterization of a novel cold-active alpha-amylase from Bacillus sp. dsh19-1. Extremophiles 2018, 22, 739–749. [Google Scholar] [CrossRef]
- Kim, M.J.; Kim, J.A.; Park, H.J.; Yim, J.H.; Kim, I.C.; Han, S.J. Novel cold-active amylase-producing bacterium from Chukchi Sea and its enzyme properties. Pol. Polar Res. 2024, 45, 215–230. [Google Scholar] [CrossRef]
- Sarnthima, R.; Khammaung, S.; Sa-Ard, P. Culture broth of Ganoderma lucidum exhibited antioxidant, antibacterial and alpha-amylase inhibitory activities. J. Food Sci. Technol. 2017, 54, 3724–3730. [Google Scholar] [CrossRef] [PubMed]
- Jalil, V.; Khan, M.; Haider, S.Z.; Shamim, S. Investigation of the Antibacterial, Anti-Biofilm, and Antioxidative Effect of Piper betle Leaf Extract against Bacillus gaemokensis MW067143 Isolated from Dental Caries, an In Vitro-In Silico Approach. Microorganisms 2022, 10, 2485. [Google Scholar] [CrossRef] [PubMed]
- Zottich, U.; Da Cunha, M.; Carvalho, A.O.; Dias, G.B.; Silva, N.C.; Santos, I.S.; do Nacimento, V.V.; Miguel, E.C.; Machado, O.L.; Gomes, V.M. Purification, biochemical characterization and antifungal activity of a new lipid transfer protein (LTP) from Coffea canephora seeds with alpha-amylase inhibitor properties. Biochim. Biophys. Acta 2011, 1810, 375–383. [Google Scholar] [CrossRef]
- Bhanja, T.; Rout, S.; Banerjee, R.; Bhattacharyya, B.C. Comparative profiles of alpha-amylase production in conventional tray reactor and GROWTEK bioreactor. Bioprocess Biosyst. Eng. 2007, 30, 369–376. [Google Scholar] [CrossRef]
- Xian, L.; Wang, F.; Luo, X.; Feng, Y.L.; Feng, J.X. Purification and characterization of a highly efficient calcium-independent alpha-amylase from Talaromyces pinophilus 1-95. PLoS ONE 2015, 10, e0121531. [Google Scholar] [CrossRef]
- Zeb, A.; Khan, Y.; He, H.; Zhang, D.; Shen, S. Molecular identification of Halomonas AZ07 and its multifunctional enzymatic activities to degrade Pyropia yezoensis under high-temperature condition. Mol. Biol. Rep. 2024, 51, 816. [Google Scholar] [CrossRef] [PubMed]
- Carlson, R.P.; Oshota, O.; Shipman, M.; Caserta, J.A.; Hu, P.; Saunders, C.W.; Xu, J.; Jay, Z.J.; Reeder, N.; Richards, A.; et al. Integrated molecular, physiological and in silico characterization of two Halomonas isolates from industrial brine. Extremophiles 2016, 20, 261–274. [Google Scholar] [CrossRef]
- Abosamaha, A.; Williamson, M.P.; Gilmour, D.J. Utilization and accumulation of compatible solutes in Halomonas pacifica: A species of moderately halophilic bacteria isolated from a saline lake in South Libya. Access Microbiol. 2022, 4, 000359. [Google Scholar] [CrossRef]
- Zhang, T.; Cui, T.; Cao, Y.; Li, Y.; Li, F.; Zhu, D.; Xing, J. Whole genome sequencing of the halophilic Halomonas qaidamensis XH36, a novel species strain with high ectoine production. Antonie Leeuwenhoek 2022, 115, 545–559. [Google Scholar] [CrossRef]
- Vreeland, R.H.; Litchfield, C.D.; Martin, E.L.; Elliot, E. Halomonas elongata, a new genus and species of extremely salt-tolerant bacteria. Int. J. Syst. Bacteriol. 1980, 30, 485–495. [Google Scholar] [CrossRef]
- Rathour, R.; Gupta, J.; Tyagi, B.; Thakur, I.S. Production and characterization of psychrophilic α-amylase from a psychrophilic bacterium, Shewanella sp. ISTPL2. Amylase 2020, 4, 1–10. [Google Scholar] [CrossRef]
- Rathod, B.G.; Pandala, S.; Poosarla, V.G. A novel halo-acid-alkali-tolerant and surfactant stable amylase secreted from halophile Bacillus siamensis F2 and Its application in waste valorization by bioethanol production and food industry. Appl. Biochem. Biotechnol. 2023, 195, 4775–4795. [Google Scholar] [CrossRef]
- Sanchez, A.C.; Ravanal, M.C.; Andrews, B.A.; Asenjo, J.A. Heterologous expression and biochemical characterization of a novel cold-active alpha-amylase from the Antarctic bacteria Pseudoalteromonas sp. 2-3. Protein Expr. Purif. 2019, 155, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Huang, Z.; Liu, Z. A novel cold-active and salt-tolerant alpha-amylase from marine bacterium Zunongwangia profunda: Molecular cloning, heterologous expression and biochemical characterization. Extremophiles 2014, 18, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Akbulut, K.; Taranaci, S.; Ozkok, S.; Varan, N.E.; Yildirim, D.; Binay, B. Heterologous expression of calcium-independent mesophilic alpha-amylase from Priestia megaterium: Immobilization on genipin-modified multi-walled carbon nanotubes and silica supports to enhance thermostability and catalytic activity. Bioorg. Chem. 2025, 155, 108151. [Google Scholar] [CrossRef]
- Tsukamoto, K.; Ariki, S.; Nakazawa, M.; Sakamoto, T.; Ueda, M. Novel cold-adapted raw-starch digesting alpha-amylases from Eisenia fetida: Gene cloning, expression, and characterization. Biotechnol. Rep. 2021, 31, e00662. [Google Scholar] [CrossRef]
- Zhang, J.W.; Zeng, R.Y. Purification and characterization of a cold-adapted alpha-amylase produced by Nocardiopsis sp. 7326 isolated from Prydz Bay, Antarctic. Mar. Biotechnol. 2008, 10, 75–82. [Google Scholar] [CrossRef]
- Lu, M.; Wang, S.; Fang, Y.; Li, H.; Liu, S.; Liu, H. Cloning, expression, purification, and characterization of cold-adapted alpha-amylase from Pseudoalteromonas arctica GS230. Protein J. 2010, 29, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Ottoni, J.R.; Rodrigues e Silva, T.; Maia de Oliveira, V.; Zambrano Passarini, M.R. Characterization of amylase produced by cold-adapted bacteria from Antarctic samples. Biocatal. Agric. Biotechnol. 2020, 23, 101452. [Google Scholar] [CrossRef]
- Kuddus, M.; Roohi; Saima; Ahmad, I.Z. Cold-active extracellular α-amylase production from novel bacteria Microbacterium foliorum GA2 and Bacillus cereus GA6 isolated from Gangotri glacier, Western Himalaya. J. Gen. Eng. Biotechnol. 2012, 10, 151–159. [Google Scholar] [CrossRef]
- Wang, X.; Kan, G.; Shi, C.; Xie, Q.; Ju, Y.; Wang, R.; Qiao, Y.; Ren, X. Purification and characterization of a novel wild-type alpha-amylase from Antarctic sea ice bacterium Pseudoalteromonas sp. M175. Protein Expr. Purif. 2019, 164, 105444. [Google Scholar] [CrossRef]
- Yang, C.; Wang, Z.; Li, Y.; Niu, Y.; Du, M.; He, X.; Ma, C.; Tang, H.; Xu, P. Metabolic versatility of halotolerant and alkaliphilic strains of Halomonas isolated from alkaline black liquor. Bioresour. Technol. 2010, 101, 6778–6784. [Google Scholar] [CrossRef]
- Mukhtar, S.; Mehnaz, S.; Mirza, M.S.; Malik, K.A. Isolation and characterization of bacteria associated with the rhizosphere of halophytes (Salsola stocksii and Atriplex amnicola) for production of hydrolytic enzymes. Braz. J. Microbiol. 2019, 50, 85–97. [Google Scholar] [CrossRef]
- Murakami, S.; Nishimoto, H.; Toyama, Y.; Shimamoto, E.; Takenaka, S.; Kaulpiboon, J.; Prousoontorn, M.; Limpaseni, T.; Pongsawasdi, P.; Aoki, K. Purification and characterization of two alkaline, thermotolerant alpha-amylases from Bacillus halodurans 38C-2-1 and expression of the cloned gene in Escherichia coli. Biosci. Biotechnol. Biochem. 2007, 71, 2393–2401. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Ahmed, S.; Fang, Y. Cloning, expression and characterization of a novel alpha-amylase from Salinispora arenicola CNP193. Protein J. 2019, 38, 716–722. [Google Scholar] [CrossRef]
- Jaiswal, N.; Jaiswal, P. Thermostable α-amylases and laccases: Paving the way for sustainable industrial applications. Processes 2024, 12, 1341. [Google Scholar] [CrossRef]
- Gomez-Villegas, P.; Vigara, J.; Romero, L.; Gotor, C.; Raposo, S.; Goncalves, B.; Leon, R. Biochemical characterization of the amylase activity from the new haloarchaeal strain Haloarcula sp. HS isolated in the Odiel Marshlands. Biology 2021, 10, 337. [Google Scholar] [CrossRef] [PubMed]
- Masasa, M.; Kushmaro, A.; Chernova, H.; Shashar, N.; Guttman, L. Carbohydrate-active enzymes of a novel halotolerant Alkalihalobacillus species for hydrolysis of starch and other algal polysaccharides. Microbiol. Spectr. 2022, 10, e0107822. [Google Scholar] [CrossRef] [PubMed]



| Sample No. | Closest Match | Identification |
|---|---|---|
| KS41550 | Alteromonas addita | Alteromonas KS41550 |
| KS41554 | Alteromonas naphthalenivorans | Alteromonas KS41554 |
| KS41560 | Halomonas titanicae | Halomonas KS41560 |
| KS41619 | Pseudoalteromonas agrivorans | Pseudoalteromonas KS41619 |
| KS41843 | Halomonas titanicae | Halomonas sp. KS41843 |
| KS42027 | Subtercola vilae | Subtercola KS42027 |
| Stage | Total Activity | Total Protein Content (mg) | Specific Activity (U/mg) | Fold | Yield (%) |
|---|---|---|---|---|---|
| Cell culture broth | 162.89 | 13.80 | 11.80 | 1.00 | 100.00 |
| 10 kDa cut-off | 38.89 | 4.21 | 9.23 | 0.78 | 23.87 |
| DEAE column | 22.50 | 0.93 | 24.19 | 2.05 | 13.81 |
| Superdex 75 | 9.82 | 0.09 | 110.32 | 9.35 | 6.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.A.; Kim, M.J.; Yim, J.H.; Kim, I.-C.; Rhee, J.-S.; Han, S.J. Isolation and Characterization of Low-Temperature and High-Salinity Amylase from Halomonas sp. KS41843. Fermentation 2025, 11, 465. https://doi.org/10.3390/fermentation11080465
Kim JA, Kim MJ, Yim JH, Kim I-C, Rhee J-S, Han SJ. Isolation and Characterization of Low-Temperature and High-Salinity Amylase from Halomonas sp. KS41843. Fermentation. 2025; 11(8):465. https://doi.org/10.3390/fermentation11080465
Chicago/Turabian StyleKim, Jin A, Min Ju Kim, Joung Han Yim, Il-Chan Kim, Jae-Sung Rhee, and Se Jong Han. 2025. "Isolation and Characterization of Low-Temperature and High-Salinity Amylase from Halomonas sp. KS41843" Fermentation 11, no. 8: 465. https://doi.org/10.3390/fermentation11080465
APA StyleKim, J. A., Kim, M. J., Yim, J. H., Kim, I.-C., Rhee, J.-S., & Han, S. J. (2025). Isolation and Characterization of Low-Temperature and High-Salinity Amylase from Halomonas sp. KS41843. Fermentation, 11(8), 465. https://doi.org/10.3390/fermentation11080465






