Enhancing Nutritional Value and Sensory Quality of Spirulina (Arthrospira platensis) Through Preharvest Co-Cultivation with Yeast Saccharomyces cerevisiae
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganism Strains and Pre-Culture Conditions
2.2. Co-Culture Experiments
2.3. Profiling of Yeast Extracellular Organic Acids
2.4. Color Evaluation and Pigment Analysis
2.5. Sensory and Instrumental Volatile Flavor Analysis
2.6. Genome Sequencing and Transcriptomic Analysis
2.7. qRT-PCR Analysis
2.8. Statistical Analysis
3. Results and Discussion
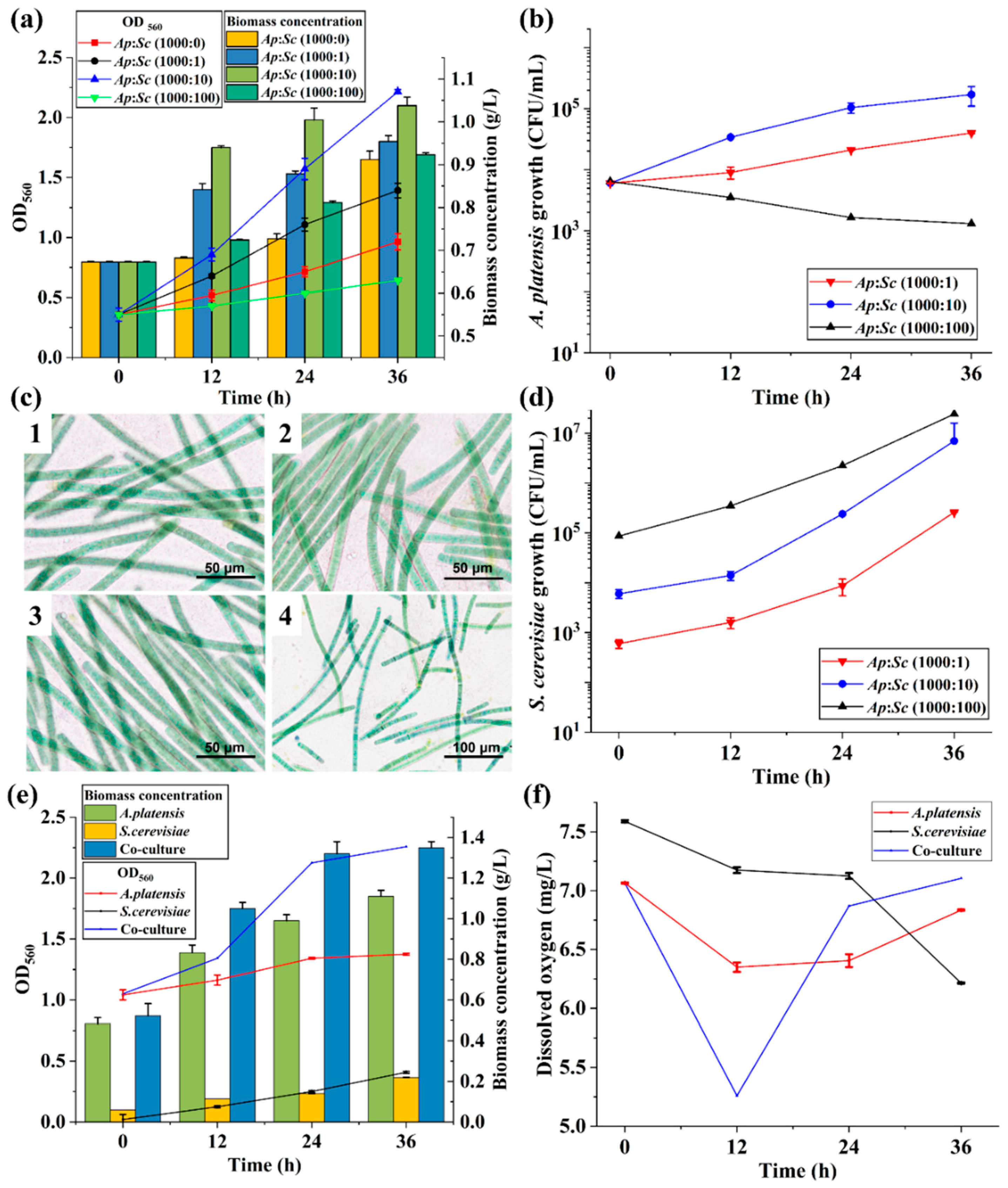
3.1. Establishment of the Microalgae–Yeast Co-Culture System
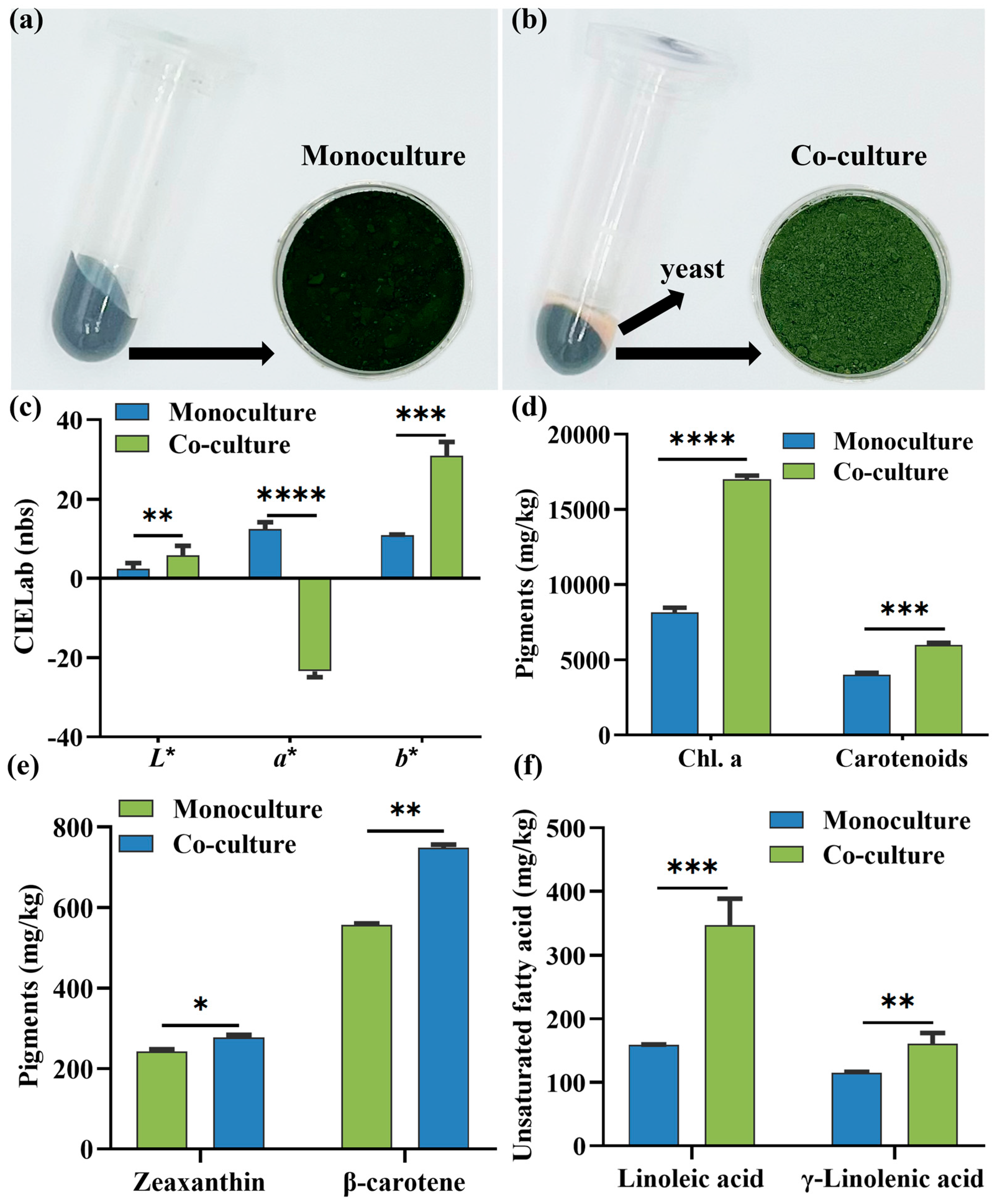
3.2. Color and Compositional Variations in Co-Cultured Microalgae Biomass
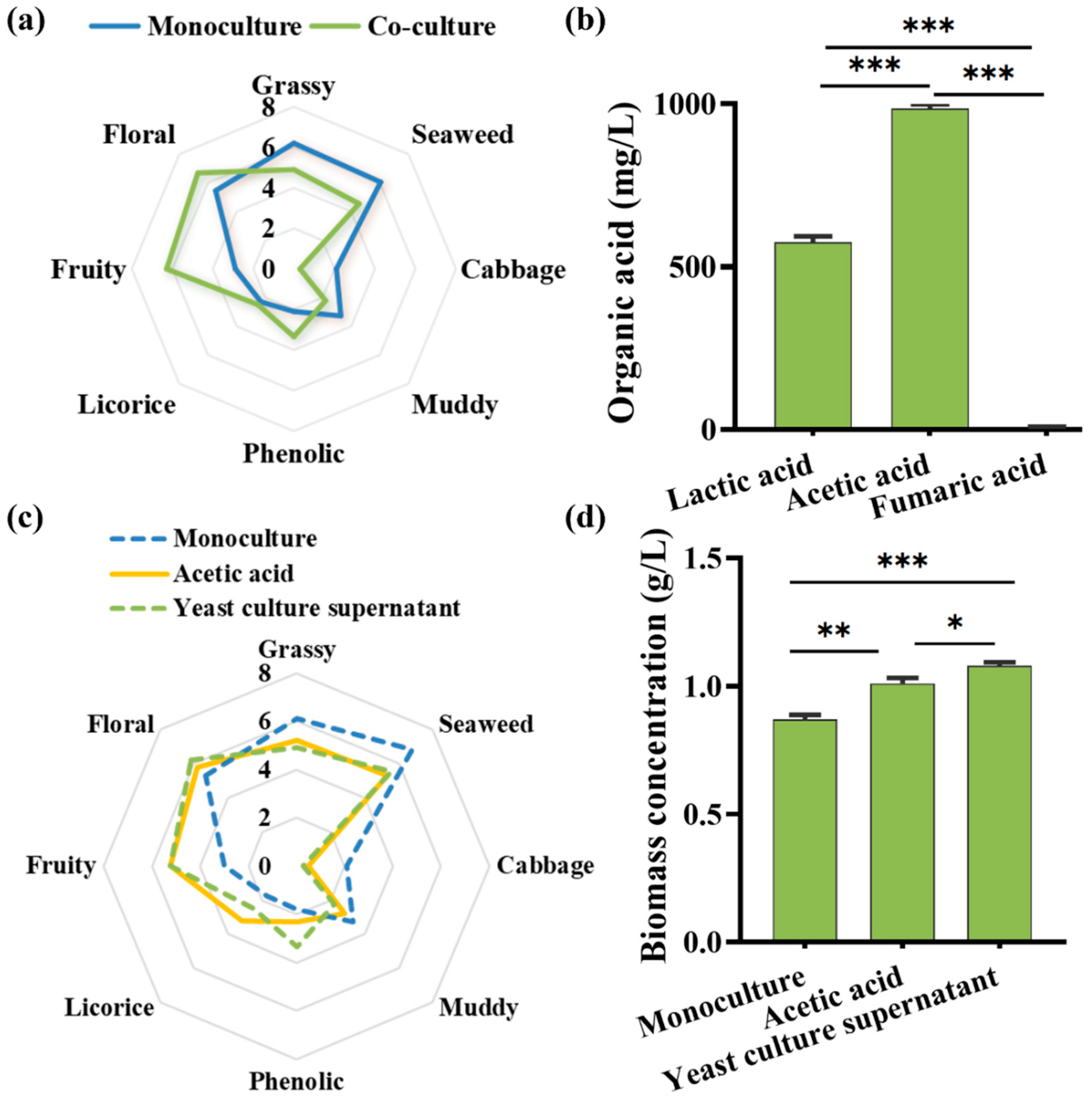
3.3. Volatile Flavor Profile of Co-Cultured Microalgae Biomass
3.4. Acetic Acid as a Central Effector and Mediator in the Co-Culture System
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Singh, S.K.; Shukla, L.; Yadav, N.; Singh, P.K.; Singh, S.M.; Yadav, M.K.; Kaushalendra; Kumar, A. Spirulina: From ancient food to innovative super nutrition of the future and its market scenario as a source of nutraceutical. In Cyanobacterial Biotechnology in the 21st Century; Springer Nature: Singapore, 2023; pp. 51–61. [Google Scholar]
- Subashchandrabose, S.R.; Ramakrishnan, B.; Megharaj, M.; Venkateswarlu, K.; Naidu, R. Consortia of cyanobacteria/microalgae and bacteria: Biotechnological potential. Biotechnol. Adv. 2011, 29, 896–907. [Google Scholar] [CrossRef] [PubMed]
- Padmaperuma, G.; Kapoore, R.V.; Gilmour, D.J.; Vaidyanathan, S. Microbial consortia: A critical look at microalgae co-cultures for enhanced biomanufacturing. Crit. Rev. Biotechnol. 2018, 38, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Pandey, J.P.; Pathak, N.; Tiwari, A.S. Standardization of pH and light intensity for the biomass production of Spirulina platensis. J. Algal Biomass Util. 2010, 1, 93–102. [Google Scholar]
- Naidoo, R.K.; Simpson, Z.F.; Oosthuizen, J.R.; Bauer, F.F. Nutrient exchange of carbon and nitrogen promotes the formation of stable mutualisms between Chlorella sorokiniana and Saccharomyces cerevisiae: Under engineered synthetic growth conditions. Front. Microbiol. 2019, 10, 00609. [Google Scholar] [CrossRef]
- Shu, C.H.; Tsai, C.C.; Chen, K.Y.; Liao, W.H.; Huang, H.C. Enhancing high quality oil accumulation and carbon dioxide fixation by a mixed culture of Chlorella sp. and Saccharomyces cerevisiae. J. Taiwan Inst. Chem. Eng. 2013, 44, 936–942. [Google Scholar] [CrossRef]
- Naseema Rasheed, R.; Pourbakhtiar, A.; Mehdizadeh Allaf, M.; Baharlooeian, M.; Rafiei, N.; Alishah Aratboni, H.; Morones-Ramírez, J.R.; Winck, F.V. Microalgal co-cultivation-recent methods, trends in omic-studies, applications, and future challenges. Front. Bioeng. Biotechnol. 2023, 11, 1193424. [Google Scholar] [CrossRef]
- Huo, S.; Kong, M.; Zhu, F.; Qian, J.; Huang, D.; Chen, P.; Ruan, R. Co-culture of Chlorella and wastewater-borne bacteria in vinegar production wastewater: Enhancement of nutrients removal and influence of algal biomass generation. Algal Res. 2020, 45, 101744. [Google Scholar] [CrossRef]
- Sonowal, S.; Palani, N.P.; Ahmed, R.; Debbarma, J.; Chikkaputtaiah, C.; Basar, E.; Velmurugan, N. Himalayan bacterial endophytes enhance microalgal cell numbers and chlorophyll content in synthetic co-culture. J. Appl. Phycol. 2022, 34, 2383–2400. [Google Scholar] [CrossRef]
- da Silva Vaz, B.; Costa, J.A.; de Morais, M.G. CO2 Biofixation by the Cyanobacterium Spirulina sp. LEB 18 and the Green Alga Chlorella fusca LEB 111 Grown using gas effluents and solid residues of thermoelectric origin. Appl. Biochem. Biotechnol. 2016, 178, 418–429. [Google Scholar] [CrossRef]
- Harvath, P.; Reidy, S.; Byer, J. Identification of organic acids in used engine oil residues by pyrolysis-comprehensive 2D gas chromatography-time of flight mass spectrometry. SAE Mobilus 2016, 1, 2274. [Google Scholar]
- Wellburn, A.R. The spectral determination of Chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 1994, 144, 307–313. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, R.; Zhang, G.; Liu, Z.; Jiang, H.; Mao, X. Heterologous expression of the plant-derived astaxanthin biosynthesis pathway in Yarrowia lipolytica for glycosylated astaxanthin production. J. Agric. Food Chem. 2023, 71, 2943–2951. [Google Scholar] [CrossRef]
- ISO 8589; Sensory Analysis. General Guidance for the Design of Test Rooms. International Organization for Standardization: Geneva, Switzerland, 2007.
- Li, X.; Xie, W.; Bai, F.; Wang, J.; Zhou, X.; Gao, R.; Xu, X.; Zhao, Y. Influence of thermal processing on flavor and sensory profile of sturgeon meat. Food Chem. 2022, 374, 131689. [Google Scholar] [CrossRef]
- Qian, S.; Liu, K.; Wang, J.; Bai, F.; Gao, R.; Zeng, M.; Wu, J.; Zhao, Y.; Xu, X. Capturing the impact of oral processing behavior and bolus formation on the dynamic sensory perception and composition of steamed sturgeon meat. Food Chem. X 2023, 17, 100553. [Google Scholar] [CrossRef]
- ISO 5496:1984; Sensory Analysis—Methodology—Initiation and Training of Assessors in the Detection and Recognition of Odours. International Organization for Standardization (ISO): Geneva, Switzerland, 1984.
- ISO 8586:2012; Sensory Analysis—General Guidance for the Selection, Training and Monitoring of Selected Assessors and Expert Sensory Assessors. International Organization for Standardization (ISO): Geneva, Switzerland, 2012.
- GB/T 39625; Sensory Analysis—Methodology—General Guidance for Establishing a Sensory Profile. Standardization Administration of China: Beijing, China, 2020.
- Zhao, Y.; Wang, Y.; Li, C.; Li, L.; Yang, X.; Wu, Y.; Chen, S.; Zhao, Y. Novel insight into physicochemical and flavor formation in naturally fermented tilapia sausage based on microbial metabolic network. Food Res. Int. 2021, 141, 110122. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Lavoie, M.; Fan, X.; Tan, H.; Liu, G.; Xu, P.; Fu, Z.; Paerl, H.W.; Qian, H. Allelopathic interactions of linoleic acid and nitric oxide increase the competitive ability of Microcystis aeruginosa. ISME J. 2017, 11, 1865–1876. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2016, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. circlize implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef]
- Sui, Y.; Jiang, Y.; Moretti, M.; Vlaeminck, S.E. Harvesting time and biomass composition affect the economics of microalgae production. J. Clean. Prod. 2020, 259, 120782. [Google Scholar] [CrossRef]
- Strohm, J.; Dale, R. Dissolved Oxygen Measurement in Yeast Propagation. Ind. Eng. Chem. 1961, 53, 760–764. [Google Scholar] [CrossRef]
- Schlintl, C.; Schienle, A. Effects of coloring food images on the propensity to eat: A placebo approach with color suggestions. Front. Psychol. 2020, 11, 589826. [Google Scholar] [CrossRef]
- Van der Stricht, H.; Profeta, A.; Hung, Y.; Verbeke, W. Consumers’ willingness-to-buy pasta with microalgae proteins–Which label can promote sales? Food Qual. Prefer. 2023, 110, 104948. [Google Scholar] [CrossRef]
- Hutchings, J.B. Food Colour and Appearance in Perspective. Food Colour and Appearance; Springer: Boston, MA, USA, 1994; pp. 1–29. [Google Scholar]
- Xu, Z.; Theodoropoulos, C.; Pittman, J.K. Optimization of a Chlorella–Saccharomyces co–culture system for enhanced metabolite productivity. Algal Res. 2024, 79, 103455. [Google Scholar] [CrossRef]
- da Silva Souza, M.A.; Peres, L.E.; Peres, L.E.; Freschi, J.R.; Purgatto, E.; Lajolo, F.M.; Hassimotto, N.M. Changes in flavonoid and carotenoid profiles alter volatile organic compounds in purple and orange cherry tomatoes obtained by allele introgression. J. Sci. Food Agric. 2020, 100, 1662–1670. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Tian, Z.; Zheng, X.; Tang, X.; Li, W.; Huang, X. Characterization of flavour components and identification of lipid flavour precursors in different cuts of pork by phospholipidomics. Food Chem. 2024, 458, 139422. [Google Scholar] [CrossRef]
- Li, X.; Qi, L.; Zang, N.; Zhao, L.; Sun, Y.; Huang, X.; Wang, H.; Yin, Z.; Wang, A. Integrated metabolome and transcriptome analysis of the regulatory network of volatile ester formation during fruit ripening in pear. Plant Physiol. Biochem. 2022, 185, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Colonia, B.S.O.; de Melo Pereira, G.V.; Carvalho, J.C.d.; Karp, S.G.; Rodrigues, C.; Soccol, V.T.; Fanka, L.S.; Soccol, C.R. Deodorization of algae biomass to overcome off-flavors and odor issues for developing new food products: Innovations, trends, and applications. Food Chem. Adv. 2023, 2, 100270. [Google Scholar] [CrossRef]
- Mele, M.A.; Kang, H.-M.; Lee, Y.T.; Islam, M.Z. Grape terpenoids: Flavor importance, genetic regulation, and future potential. Crit. Rev. Food Sci. Nutr. 2020, 61, 1429–1447. [Google Scholar] [CrossRef]
- Milovanović, I.; Mišan, A.; Simeunović, J.; Kovač, D.; Jambrec, D.; Mandić, A. Determination of volatile organic compounds in selected strains of cyanobacteria. J. Chem. 2015, 2015, 969542. [Google Scholar] [CrossRef]
- Ahrazem, O.; Gómez-Gómez, L.; Rodrigo, M.J.; Avalos, J.; Limón, M.C. Carotenoid cleavage oxygenases from microbes and photosynthetic organisms: Features and functions. Int. J. Mol. Sci. 2016, 17, 1781. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Oliveira, P.; Jimenez-Lopez, C.; Lourenço-Lopes, C.; Chamorro, F.; Pereira, A.G.; Carrera-Casais, A.; Fraga-Corral, M.; Carpena, M.; Simal-Gandara, J.; Prieto, M.A. Evolution of flavors in extra virgin olive oil shelf-life. Antioxidants 2021, 10, 368. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Qian, R.; Wang, D.; Liu, L.; Sun, C.; Lin, X. Lipid-Derived Aldehydes: New Key Mediators of plant growth and stress responses. Biology 2022, 11, 1590. [Google Scholar] [CrossRef] [PubMed]
- Achyuthan, K.E.; Harper, J.C.; Manginell, R.P.; Moorman, M.W. Volatile metabolites emission by in vivo microalgae—An overlooked opportunity? Metabolites 2017, 7, 39. [Google Scholar] [CrossRef]
- Karitani, Y.; Yamada, R.; Matsumoto, T.; Ogino, H. Improvement of cell growth in green algae Chlamydomonas reinhardtii through co-cultivation with yeast Saccharomyces cerevisiae. Biotechnol. Lett. 2024, 46, 431–441. [Google Scholar] [CrossRef]
- La, A.; Perré, P.; Taidi, B. Process for symbiotic culture of Saccharomyces cerevisiae and Chlorella vulgaris for in situ CO2 mitigation. Appl. Microbiol. Biotechnol. 2019, 103, 731–745. [Google Scholar] [CrossRef]
- Peña, A.; Sánchez, N.S.; Álvarez, H.; Calahorra, M.; Ramírez, J. Effects of high medium pH on growth, metabolism and transport in Saccharomyces cerevisiae. FEMS Yeast Res. 2015, 15, 25673753. [Google Scholar] [CrossRef]
- Arora, N.; Patel, A.; Mehtani, J.; Pruthi, P.A.; Pruthi, V.; Poluri, K.M. Co-culturing of oleaginous microalgae and yeast: Paradigm shift towards enhanced lipid productivity. Environ. Sci. Pollut. Res. 2019, 26, 16952–16973. [Google Scholar] [CrossRef]
- Proietti Tocca, G.; Agostino, V.; Menin, B.; Tommasi, T.; Fino, D.; Di Caprio, F. Mixotrophic and heterotrophic growth of microalgae using acetate from different production processes. Rev. Environ. Sci. Bio/Technol. 2024, 23, 93–132. [Google Scholar] [CrossRef]
- Liaqat, F.; Khazi, M.I.; Bahadar, A.; He, L.; Aslam, A.; Liaquat, R.; Agathos, S.N.; Li, J. Mixotrophic cultivation of microalgae for carotenoid production. Rev. Aquac. 2023, 15, 35–61. [Google Scholar] [CrossRef]
- Ma, X.; Mi, Y.; Zhao, C.; Wei, Q. A comprehensive review on carbon source effect of microalgae lipid accumulation for biofuel production. Sci. Total Environ. 2022, 806, 151387. [Google Scholar] [CrossRef]
- De Bellis, L.; Luvisi, A.; Alpi, A. Aconitase: To be or not to be inside plant glyoxysomes, that is the question. Biology 2020, 9, 162. [Google Scholar] [CrossRef]
- Kong, F.; Romero, I.T.; Warakanont, J.; Li-Beisson, Y. Lipid catabolism in microalgae. New Phytol. 2018, 218, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Raita, S.; Feldmane, L.; Kusnere, Z.; Spalvins, K.; Kuzmika, I.; Berzina, I.; Mika, T. Microbial carotenoids production: Strains, conditions, and yield affecting factors. Environ. Clim. Technol. 2023, 27, 1027–1048. [Google Scholar] [CrossRef]
- Nabeta, K.; Saitoh, T.; Adachi, K.; Komuro, K. Biosynthesis of phytyl side-chain of Chlorophyll a: Apparent reutilization of carbon dioxide evolved during acetate assimilation in biosynthesis of chloroplastidic isoprenoid. Chem. Commun. 1998, 29, 671–672. [Google Scholar] [CrossRef]

| Classification | Volatile Compounds | CAS# | LRI | Concentration (μg/kg) | |
|---|---|---|---|---|---|
| Monoculture | Co-Culture | ||||
| Ketones | |||||
| A1 | 2-Butanone | 78–93–3 | 894 | 0.305 ± 0.031 | 0.061 ± 0.002 *** |
| A2 | 2,3-Butanedione | 431–03–8 | 971 | 0.286 ± 0.016 | 0.352 ± 0.015 * |
| A3 | 2-Methyl-3-Heptanone | 13,019–20–0 | 1161 | 0.489 ± 0.009 | 10.25 ± 0.204 *** |
| A4 | 2-Heptanone | 110–43–0 | 1177 | 1.82 ± 0.024 | 2.851 ± 0.042 *** |
| A5 | 3-Octanone | 106–68–3 | 1249 | 0.896 ± 0.005 | 0.167 ± 0.003 *** |
| A6 | 6-Methyl-2-Heptanone | 928–68–7 | 1232 | 0.285 ± 0.012 | 0.166 ± 0.003 *** |
| A7 | 2-Octanone | 111–13–7 | 1281 | 1.155 ± 0.028 | 0.365 ± 0.037 *** |
| A8 | 1-Hydroxy-2-Propanone | 116–09–6 | 1292 | 0.283 ± 0.006 | ND |
| A9 | 2,2,6-Trimethyl-Cyclohexanone | 2408–37–9 | 1314 | 0.964 ± 0.008 | 1.126 ± 0.06 * |
| A10 | 6-Methyl-5-Hepten-2-one | 110–93–0 | 1332 | 1.525 ± 0.029 | 0.761 ± 0.068 *** |
| A11 | 2-Methyl-2-Hepten-4-one | 22,319–24–0 | 1357 | 0.901 ± 0.015 | 0.305 ± 0.004 *** |
| A12 | 2-Nonanone | 821–55–6 | 1384 | 2.039 ± 0.008 | 0.119 ± 0.001 *** |
| A13 | 3-Octen-2-one | 1669–44–9 | 1402 | ND | 0.87 ± 0 |
| A14 | 2-Decanone | 693–54–9 | 1489 | 0.591 ± 0.009 | 0.591 ± 0.001 |
| A15 | 1-(2-Methyl-1-Cyclopenten-1-yl)-Ethanone | 3168–90–9 | 1589 | 0.299 ± 0.001 | 0.241 ± 0.007 *** |
| A16 | 4-Hydroxy-4-Methyl-Cyclohexanone | 17,429–02–6 | 1601 | 25.174 ± 1.775 | 50.598 ± 0.329 *** |
| A17 | Acetophenone | 98–86–2 | 1648 | 0.21 ± 0.015 | 0.167 ± 0.022 |
| A18 | 3-Acetyl-2-Octanone | 27,970–50–9 | 1663 | 1.067 ± 0.017 | 1.744 ± 0.019 *** |
| A19 | 2,6,6-Trimethyl-2-Cyclohexene-1,4-Dione | 1125–21–9 | 1691 | 0.999 ± 0.001 | 0.576 ± 0.005 *** |
| A20 | 1-(4-Methylphenyl)-Ethanone | 122–00–9 | 1775 | ND | 0.216 ± 0.003 |
| A21 | Geranylacetone | 3796–70–1 | 1849 | 0.807 ± 0.137 | 1.701 ± 0.04 *** |
| A22 | trans-β-Ionone | 79–77–6 | 1939 | 11.847 ± 0.125 | 29.688 ± 0.255 *** |
| A23 | Epoxy-β-Ionone | 23,267–57–4 | 1993 | 4.212 ± 0.01 | 13.234 ± 0.191 *** |
| A24 | 3,4-Dehydro-β-Ionone | 1203–08–3 | 1999 | ND | 0.235 ± 0.004 |
| A25 | 6,10,14-Trimethyl- 2-Pentadecanone | 502–69–2 | 2121 | 1.017 ± 0.058 | 3.106 ± 0.077 *** |
| A26 | (E)-4-Oxo-β-Ionone | 27,185–77–9 | 2460 | 0.136 ± 0.004 | 0.296 ± 0.003 *** |
| A27 | 2-Pyrrolidinone | 616–45–5 | 2027 | 1.55 ± 0.008 | 0.467 ± 0.027 *** |
| A28 | Piperitenone Oxide | 35,178–55–3 | 2144 | ND | 0.647 ± 0.006 |
| A29 | 3-Ethyl-4-Methyl-1H-Pyrrole-2,5-Dione | 20,189–42–8 | 2251 | 9.652 ± 0.042 | 21.381 ± 0.066 *** |
| Aldehydes | |||||
| B1 | Acetaldehyde | 75–07–0 | 691 | 0.293 ± 0.011 | 0.272 ± 0.018 |
| B2 | 2-Methyl-Butanal | 96–17–3 | 907 | 0.812 ± 0.01 | 0.127 ± 0.005 *** |
| B3 | 3-Methyl-Butanal | 590–86–3 | 911 | 1.3 ± 0.081 | 0.293 ± 0.05 *** |
| B4 | Pentanal | 110–62–3 | 972 | 0.41 ± 0.008 | 1.256 ± 0.045 *** |
| B5 | Hexanal | 66–25–1 | 1075 | 7.415 ± 0.094 | 14.336 ± 0.275 *** |
| B6 | Heptanal | 111–71–7 | 1180 | 0.9 ± 0.008 | 1.416 ± 0.069 *** |
| B7 | Octanal | 124–13–0 | 1283 | ND | 0.531 ± 0.009 |
| B8 | (E)-2-Heptenal | 18,829–55–5 | 1320 | ND | 0.199 ± 0 |
| B9 | Nonanal | 124–19–6 | 1388 | 0.622 ± 0.001 | 3.47 ± 0.22 *** |
| B10 | (E)-2-Octenal | 2548–87–0 | 1425 | 0.319 ± 0.001 | 0.601 ± 0.002 *** |
| B11 | 3-Furaldehyde | 498–60–2 | 1454 | 0.117 ± 0.002 | 0.127 ± 0.002 ** |
| B12 | Benzaldehyde | 100–52–7 | 1519 | ND | 0.127 ± 0.002 |
| B13 | (E)-2-Nonenal | 18,829–56–6 | 1533 | 0.251 ± 0.007 | 1.04 ± 0.032 *** |
| B14 | β-Cyclocitral | 432–25–7 | 1621 | 5.31 ± 0.09 | 5.515 ± 0.004 * |
| B15 | Safranal | 116–26–7 | 1645 | 0.271 ± 0.009 | 0.259 ± 0.001 |
| B16 | 3-Ethyl-Benzaldehyde | 34,246–54–3 | 1707 | 0.054 ± 0.002 | ND |
| B17 | (E, Z)-2,4-Decadienal | 25,152–83–4 | 1810 | ND | 0.165 ± 0.012 |
| Alcohols | |||||
| C1 | Ethanol | 64–17–5 | 925 | 1.854 ± 0.082 | 0.376 ± 0.011 *** |
| C2 | 1-Butanol | 71–36–3 | 1137 | 12.617 ± 0.177 | 3.464 ± 0.216 *** |
| C3 | 1-Pentanol | 71–41–0 | 1241 | 1.573 ± 0.006 | ND |
| C4 | 1-Hexanol | 111–27–3 | 1341 | 0.763 ± 0.121 | 0.397 ± 0.064 * |
| C5 | 1-Octen-3-ol | 3391–86–4 | 1437 | 5.504 ± 0.085 | 4.424 ± 0.101 *** |
| C6 | 1-Heptanol | 111–70–6 | 1442 | 0.442 ± 0.011 | ND |
| C7 | 2-Ethyl-1-Hexanol | 104–76–7 | 1476 | 2.418 ± 0.043 | 4.569 ± 0.767 * |
| C8 | 1-Octanol | 111–87–5 | 1544 | 0.565 ± 0.012 | 0.147 ± 0.022 *** |
| C9 | 1-Nonanol | 143–08–8 | 1646 | 0.52 ± 0.008 | 0.164 ± 0.02 *** |
| C10 | Benzyl Alcohol | 100–51–6 | 1862 | 1.818 ± 0.08 | 1.357 ± 0.039 *** |
| C11 | Phenylethyl Alcohol | 60–12–8 | 1898 | 0.196 ± 0.003 | 0.217 ± 0.011 |
| C12 | 1-Decanol | 112–30–1 | 1952 | ND | 0.156 ± 0.003 |
| C13 | Phytol | 150–86–7 | 2575 | ND | 0.259 ± 0.009 |
| Nitrogenous compounds | |||||
| D1 | N, N-Dimethyl-Methylamine | 75–50–3 | 642 | 2.41 ± 0.325 | 5.367 ± 0.165 *** |
| D2 | Pyrazine | 290–37–9 | 1203 | 0.09 ± 0.002 | ND |
| D3 | 2-Methyl-Pyrazine | 109–08–0 | 1259 | 1.181 ± 0.066 | 0.162 ± 0.007 *** |
| D4 | 2,5-Dimethyl-Pyrazine | 123–32–0 | 1316 | 1.784 ± 0.005 | 0.321 ± 0.001 *** |
| D5 | 2,6-Dimethyl-Pyrazine | 108–50–9 | 1322 | 0.298 ± 0.002 | ND |
| D6 | 2-Ethyl-6-Methyl-Pyrazine | 13,925–03–6 | 1379 | 0.117 ± 0.003 | ND |
| D7 | Pyrrole | 109–97–7 | 1501 | 0.072 ± 0.01 | ND |
| D8 | 1-(1H-Pyrrol-2-yl)-Ethanone | 1072–83–9 | 1958 | 0.078 ± 0.005 | 0.17 ± 0 *** |
| Phenols | |||||
| E1 | 3-Methyl-4-Isopropylphenol | 3228–02–2 | 1410 | 1.258 ± 0.002 | 0.736 ± 0.013 *** |
| E2 | Butylated Hydroxytoluene | 128–37–0 | 1903 | 0.057 ± 0.002 | 0.5 ± 0.016 *** |
| E3 | Phenol | 108–95–2 | 1986 | 0.204 ± 0.003 | 15.146 ± 0.012 *** |
| E4 | P-Tert-Butyl-Phenol | 98–54–4 | 2267 | 0.144 ± 0.001 | 1.358 ± 0.001 *** |
| E5 | 2,4-Di-Tert-Butylphenol | 96–76–4 | 2286 | 0.369 ± 0.017 | 1.104 ± 0.078 *** |
| Sulfur compounds | |||||
| F1 | Methanethiol | 74–93–1 | 677 | 0.069 ± 0.008 | ND |
| F2 | Dimethyl Disulfide | 624–92–0 | 1063 | 0.3 ± 0.021 | ND |
| F3 | Dimethyl Trisulfide | 3658–80–8 | 1375 | 0.114 ± 0.005 | ND |
| F4 | Dimethyl Sulfoxide | 67–68–5 | 1568 | 0.069 ± 0 | ND |
| Organic acids | |||||
| G1 | Acetic Acid | 64–19–7 | 1435 | 1.319 ± 0.001 | 0.589 ± 0.001 *** |
| G2 | Octanoic Acid | 124–07–2 | 2037 | 0.774 ± 0.032 | 0.408 ± 0.058 *** |
| G3 | n-Decanoic Acid | 334–48–5 | 2248 | ND | 0.264 ± 0.003 |
| Furans | |||||
| H1 | 3-Methyl-Furan | 930–27–8 | 857 | 0.102 ± 0.009 | 0.073 ± 0.005 * |
| H2 | 2-Pentyl-Furan | 3777–69–3 | 1224 | 0.898 ± 0.026 | 2.368 ± 0.169 *** |
| Ester | |||||
| I1 | Dihydroactinidiolide | 17,092–92–1 | 2358 | 4.982 ± 0.014 | 16.727 ± 0.223*** |
| Hydrocarbons | |||||
| J1 | Decane | 124–18–5 | 996 | 0.84 ± 0.041 | 0.279 ± 0.001 *** |
| J2 | Undecane | 1120–21–4 | 1089 | 4.533 ± 0.78 | 0.284 ± 0.003 *** |
| J3 | 1-(1-Cyclohexen-1-yl)-Ethanone | 932–66–1 | 1115 | ND | 0.221 ± 0.001 |
| J4 | 5-Ethyldecane | 17,302–36–2 | 1129 | 0.551 ± 0.025 | ND |
| J5 | D-Limonene | 5989–27–5 | 1191 | ND | 0.078 ± 0.006 |
| J6 | Dodecane | 112–40–3 | 1196 | 12.567 ± 0.079 | 1.954 ± 0.038 *** |
| J7 | Styrene | 100–42–5 | 1251 | 1.153 ± 0.027 | ND |
| J8 | Tridecane | 629–50–5 | 1297 | 6.323 ± 0.1 | 5.507 ± 0.087 *** |
| J9 | 2-Methyl-Tridecane | 1560–96–9 | 1353 | 0.401 ± 0.007 | 0.409 ± 0.001 |
| J10 | 3-Methyl-Tridecane | 6418–41–3 | 1363 | 1.939 ± 0.003 | 2.193 ± 0.005 *** |
| J11 | Tetradecane | 629–59–4 | 1395 | 2.979 ± 0.517 | 5.455 ± 0.118 *** |
| J12 | Pentadecane | 629–62–9 | 1495 | 49.193 ± 0.109 | 30.276 ± 5.551 ** |
| J13 | Hexadecane | 544–76–3 | 1595 | 39.117 ± 1.442 | 24.063 ± 0.051 *** |
| J14 | Heptadecane | 629–78–7 | 1705 | 421.347 ± 19.469 | 260.721 ± 4.31 *** |
| J15 | Octadecane | 593–45–3 | 1797 | 1.331 ± 0.017 | ND |
| J16 | (Z)-3-Heptadecene | 1,000,141–67–3 | 1717 | 36.228 ± 1.801 | 12.014 ± 0.011 *** |
| Genes | Forward and Reverse Primers (5′ → 3′) | |
|---|---|---|
| 16S (internal control) | AP 16S-F | CGTAAACCTCTCCTCAGTTCAG |
| AP 16S-F | GAACGGATTCACCGCAGTAT | |
| Ribulose-1,5-bisphosphate carboxylase/oxygenase | RuBisCO-F | TTCTGCTTTGTTGCCTATCCG |
| RuBisCO-R | ATCCAAATACGTTACCCACGA | |
| Geranylgeranyl diphosphate synthase | GGPPS-F | ATCTGGAAGCTCAAAAGGCTAC |
| GGPPS-R | CGCAACCACCGTTTCCAAA | |
| Phytoene desaturase | PDS-F | AATTATATAGATCCGCTGCAT |
| PDS-R | TCTTCACTGACATTATGGGGAC | |
| 1-Deoxy-D-xylulose 5-phosphate reductase | DXR-F | AGGCTCATTTTCTCTTTGGTT |
| DXR-R | AACACAGAAGTATCCTGCAAC | |
| 1-Deoxy-D-xylulose 5-phosphate synthase | DXS-F | CTGTCTCCCCAATATGACCA |
| DXS-R | ATTAATACCAGTCACCAGCAT | |
| Phytoene synthase | PSY-F | GCTCTCGGTATTGCTAACCAG |
| PSY-R | CCAGCGTTCATCTACTATGCC | |
| Acetyl-CoA synthetase | ACS-F | TCGGTGATCTAATTCTAGCTG |
| ACS-R | ATCGCATCAATAAACCCAT | |
| Acetyl-CoA carboxylase | ACCase-F | GGAATGTTAAGCCTCATGCAA |
| ACCase-R | CATGGCAAAACTAGCCGTCA | |
| Fatty acid synthase | FAS-F | ATTCGGGGTATTGATCGCCC |
| FAS-R | CTCCCCCATGTTCAACGTGA | |
| Acyl-CoA desaturase | ACD-F | TATTTATGGCATTCCTCCACA |
| ACD-R | GTAATTCCTAAGCCACCAGT | |
| Delta (6)-fatty-acid desaturase | FAD6-F | GACACCGCTCATTTTCTCGGA |
| FAD6-R | ATTCTTCTTTACGCCAGGGTT | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Sui, J.; Cui, Y.; Zeng, M.; Wu, H.; Feng, G.; Lu, X. Enhancing Nutritional Value and Sensory Quality of Spirulina (Arthrospira platensis) Through Preharvest Co-Cultivation with Yeast Saccharomyces cerevisiae. Fermentation 2025, 11, 462. https://doi.org/10.3390/fermentation11080462
Zhao Y, Sui J, Cui Y, Zeng M, Wu H, Feng G, Lu X. Enhancing Nutritional Value and Sensory Quality of Spirulina (Arthrospira platensis) Through Preharvest Co-Cultivation with Yeast Saccharomyces cerevisiae. Fermentation. 2025; 11(8):462. https://doi.org/10.3390/fermentation11080462
Chicago/Turabian StyleZhao, Yue, Jikang Sui, Yuxuan Cui, Mingyong Zeng, Haohao Wu, Guangxin Feng, and Xiangning Lu. 2025. "Enhancing Nutritional Value and Sensory Quality of Spirulina (Arthrospira platensis) Through Preharvest Co-Cultivation with Yeast Saccharomyces cerevisiae" Fermentation 11, no. 8: 462. https://doi.org/10.3390/fermentation11080462
APA StyleZhao, Y., Sui, J., Cui, Y., Zeng, M., Wu, H., Feng, G., & Lu, X. (2025). Enhancing Nutritional Value and Sensory Quality of Spirulina (Arthrospira platensis) Through Preharvest Co-Cultivation with Yeast Saccharomyces cerevisiae. Fermentation, 11(8), 462. https://doi.org/10.3390/fermentation11080462







