Microbial Genome Editing with CRISPR–Cas9: Recent Advances and Emerging Applications Across Sectors
Abstract
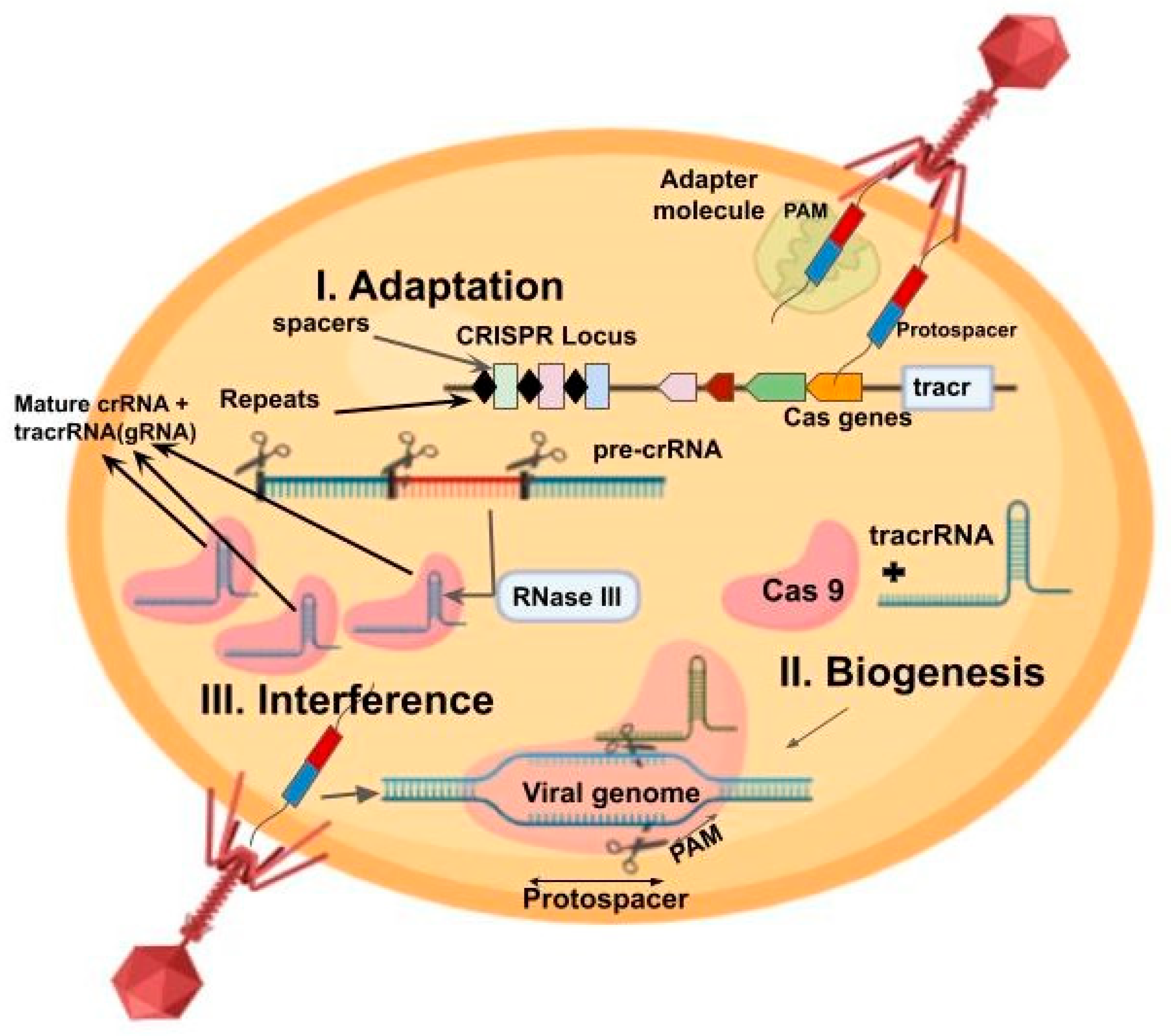
1. Introduction
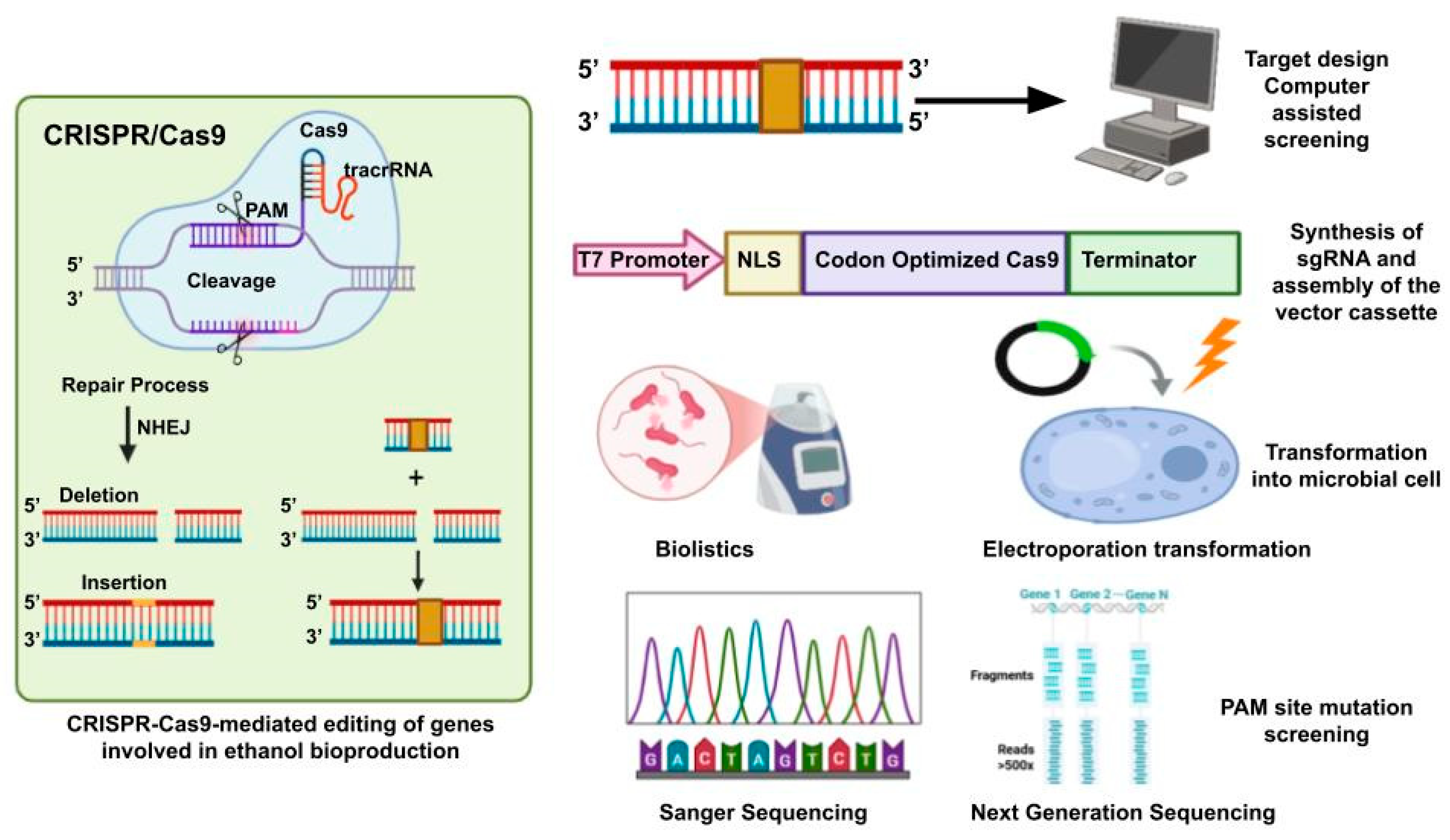
2. CRISPR–Cas9 as a Genome Editing Tool in Microbes
3. Applications of CRISPR–Cas9-Based Microbial Genome Editing
3.1. CRISPR–Cas9 in Industrial Fermentation Microbes
3.1.1. Escherichia coli and Its Use in Platform Chemical Production
3.1.2. Saccharomyces Cerevisiae and Non-Conventional Yeasts
3.1.3. Corynebacterium, Bacillus, Clostridium Species
3.1.4. Lactic Acid Bacteria and Acetic Acid Bacteria
| Serial Number | Microbial Strain | Industrial Product(s) | Key Genetic Targets | Enhancement Achieved | Reference |
|---|---|---|---|---|---|
| 1 | Escherichia coli | Succinic acid | ldhA, adhE, pta, pflB | Redirected flux to TCA cycle; increased succinate yield (80 g/L) | [40] |
| 2 | Saccharomyces cerevisiae | Isoprenoids (e.g., β-carotene) | tHMG1, ERG20, crtE/I/YB | Increased Isoprenoid flux via MVA pathway | [41] |
| 3 | Yarrowia lipolytica | Fatty acids, polyketides | β-oxidation genes, POX, LIP family | High lipid & polyketide productivity | [42] |
| 4 | Corynebacterium glutamicum | Lysine, Glutamate | NADPH supply genes, promoter regions | Cofactor balancing; increased amino acid titers | [43] |
| 5 | Bacillus subtilis | Enzymes, vitamins | Secretion genes (aprE, nprE) | Improved extracellular protein secretion | [44] |
| 6 | Clostridium beijerinckii | n-Butanol | adhE1, adhE2 (solvent genes) | Boosted solventogenesis; stable yields | [45] |
| 7 | Clostridium acetobutylicum | Acetone-butanol-ethanol (ABE) | Acid formation genes | Reduced acid byproducts; increased butanol:acetate ratio | [46] |
| 8 | Lactococcus lactis | Lactic acid, aroma compounds | ldh, adhE, sugar transporters | Optimized flavor compound production | [47] |
| 9 | Gluconobacter oxydans | Sorbitol, acetic acid | Respiratory dehydrogenases | Improved oxidative biotransformation | [48] |
| 10 | Synechocystis sp. | Isobutanol | alsS, ilvC, kivD | Cyanobacterial redirection of carbon flux | [49] |
3.2. Biofuels and Biochemicals Production
3.2.1. Metabolic Pathway Engineering
3.2.2. CRISPR for Tolerance Engineering
| S. No | Organisms | Product | Pathway | Substrate | CRISPR Tool Used | References |
|---|---|---|---|---|---|---|
| 1 | Corynebacterium glutamicum | 3-Hydroxypropionic acid | Glycerol Pathway | Glucose, xylose | CRISPR–Cas9 | [62] |
| 2 | Clostridium autoethanogenum | Ethanol | Ferredoxin oxidoreductase | Synthetic medium | CRISPR–Cas12a | [63] |
| 3 | Synechocystis sp. | Isobutanol | Ehrlich pathway | Glucose | CRISPR interference (CRISPRi) | [64] |
| 4 | S. elongates | 1,3-Propanediol | Synthetic metabolic pathway | Synthetic medium | Multiplex CRISPR editing | [65] |
| 5 | E. coli | Fatty alcohol | Fatty acyl-ACP reductase-dependent | Synthetic medium | CRISPR–Cas9 | [66] |
| 6 | Saccharomyces cerevisiae | 2,3-Butanediol | Butanediol biosynthetic | Glucose, galactose | CRISPRi | [67] |
| 7 | Klebsiella pneumoniae | 2-Butanol | Meso-2,3-butanediol synthesis | Glucose | CRISPRa | [68] |
| 8 | C. cellulolyticum | n-Butanol | CoA-dependent | Cellulose | CRISPR–Cas9 | [69] |
| 9 | Thermoanaerobacterium saccharolyticum | n-Butanol | n-butanol | Xylose | CRISPR–Cas12a | [70] |
| 10 | S. cerevisiae | Isobutanol | Embden-Meyerhof | Synthetic medium | Base Editing | [71] |
| 11 | Clostridium Tyrobutyricum | n-Butanol | Xylose metabolic pathway | Glucose and Xylose | CRISPRi | [72] |
| 12 | C. cellulovorans | Ethanol, n-Butanol | Fatty acyl-ACP reductase-dependent | Cellulose | CRISPR–Cas9 | [73] |
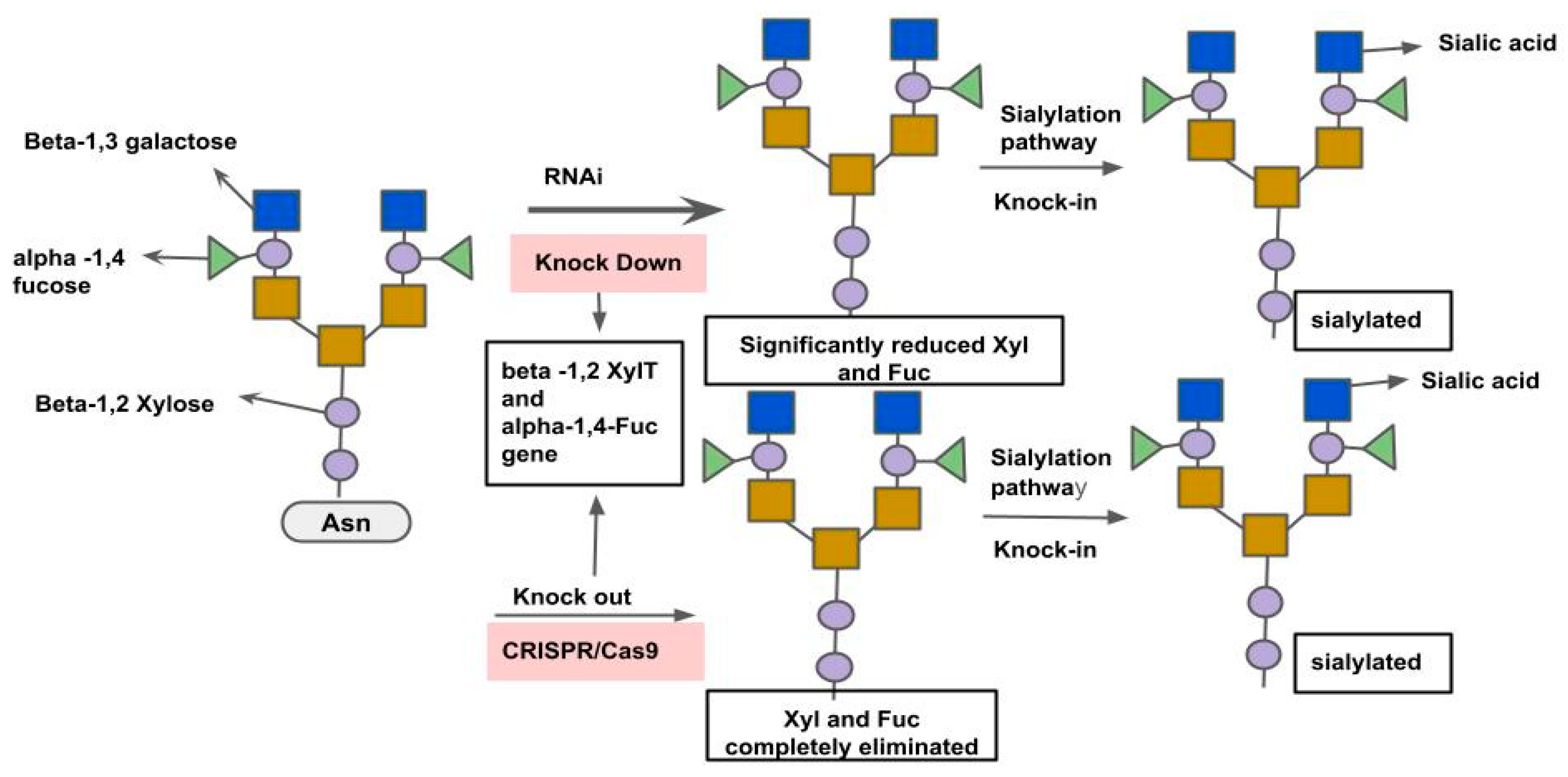
3.3. Microbial Production of Biopharmaceuticals and Enzymes
3.3.1. Recombinant Therapeutic Proteins in Bacteria and Yeast
3.3.2. Industrial Enzyme Engineering and Optimization
3.3.3. CRISPR for Pathway Balancing and Yield Improvement
3.4. CRISPR-Powered Advances in the Fermentation of Food
3.4.1. Improvement in Dairy and Brewing Starters
3.4.2. CRISPR in Probiotic and Functional Microorganism Engineering
4. Computational Tools and Predictive Modeling for CRISPR Design in Microorganisms
4.1. Improved sgRNA Design Tools
4.2. Integrating Deep Learning and Artificial Intelligence
4.3. Challenges and Future Directions
5. Challenges, Limitations, and Ethical Considerations
5.1. Off-Target Effects
5.2. Efficiently Editing Microbial Genome
5.3. Host Immune Responses
5.4. Delivery Challenges in CRISPR–Cas9 Gene Therapy
6. Future Directions
6.1. Integration with AI, Automation, and HTS
6.2. CRISPR 3.0 Tools for Microbial Cell Factory Design
6.3. Next-Generation Fermentation System Vision
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Stahl, D.A.; Tiedje, J.M. Microbial Ecology and Genomics: A Crossroads of Opportunity; American Society of Microbiology: Washington, DC, USA, 2001. [Google Scholar]
- Tokuda, M.; Shintani, M. Microbial Evolution through Horizontal Gene Transfer by Mobile Genetic Elements. Microb. Biotechnol. 2024, 17, e14408. [Google Scholar] [CrossRef] [PubMed]
- Haudiquet, M.; de Sousa, J.M.; Touchon, M.; Rocha, E.P.C. Selfish, Promiscuous and Sometimes Useful: How Mobile Genetic Elements Drive Horizontal Gene Transfer in Microbial Populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2022, 377, 20210234. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; He, Z.; Yang, Y.; Deng, Y.; Tringe, S.G.; Alvarez-Cohen, L. High-Throughput Metagenomic Technologies for Complex Microbial Community Analysis: Open and Closed Formats. mBio 2015, 6, e02288-14. [Google Scholar] [CrossRef] [PubMed]
- Nazir, A.; Hussain, F.H.N.; Raza, A. Advancing Microbiota Therapeutics: The Role of Synthetic Biology in Engineering Microbial Communities for Precision Medicine. Front. Bioeng. Biotechnol. 2024, 12, 1511149. [Google Scholar] [CrossRef] [PubMed]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. ZFN, TALEN, and CRISPR/Cas-Based Methods for Genome Engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Zhang, Y.; Shi, S. Development and Application of CRISPR/Cas in Microbial Biotechnology. Front. Bioeng. Biotechnol. 2020, 8, 711. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.R.; Lee, S.J. Multiplex CRISPR-Cas Genome Editing: Next-Generation Microbial Strain Engineering. J. Agric. Food Chem. 2024, 72, 11871–11884. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Li, Y. CRISPR-Based Gene Editing Technology and Its Application in Microbial Engineering. Eng. Microbiol. 2023, 3, 100101. [Google Scholar] [CrossRef] [PubMed]
- Adli, M. The CRISPR Tool Kit for Genome Editing and Beyond. Nat. Commun. 2018, 9, 1911. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Lin, Q.; Jin, S.; Gao, C. The CRISPR-Cas Toolbox and Gene Editing Technologies. Mol. Cell 2022, 82, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Pattali, R.K.; Ornelas, I.J.; Nguyen, C.D.; Xu, D.; Divekar, N.S.; Nuñez, J.K. CRISPRoff Epigenetic Editing for Programmable Gene Silencing in Human Cells without DNA Breaks. bioRxiv 2024. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A. CRISPR-Cas9: A Fascinating Journey from Bacterial Immune System to Human Gene Editing. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2021; Volume 178, pp. 63–83. ISBN 978-0-12-821590-6. [Google Scholar]
- Filippova, J.; Matveeva, A.; Zhuravlev, E.; Stepanov, G. Guide RNA Modification as a Way to Improve CRISPR/Cas9-Based Genome-Editing Systems. Biochimie 2019, 167, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Beisel, C.L. The tracrRNA in CRISPR Biology and Technologies. Annu. Rev. Genet. 2021, 55, 161–181. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Gupta, D.; Ahmed, K.T.; Dey, D.; Singh, R.; Swarnakar, S.; Ravichandiran, V.; Roy, S.; Ghosh, D. Modification of Cas9, gRNA and PAM: Key to Further Regulate Genome Editing and Its Applications. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2021; Volume 178, pp. 85–98. ISBN 978-0-12-821590-6. [Google Scholar]
- Babu, K.; Kathiresan, V.; Kumari, P.; Newsom, S.; Parameshwaran, H.P.; Chen, X.; Liu, J.; Qin, P.Z.; Rajan, R. Coordinated Actions of Cas9 HNH and RuvC Nuclease Domains Are Regulated by the Bridge Helix and the Target DNA Sequence. Biochemistry 2021, 60, 3783–3800. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Arantes, P.R.; Ahsan, M.; Sinha, S.; Kyro, G.W.; Maschietto, F.; Allen, B.; Skeens, E.; Lisi, G.P.; Batista, V.S.; et al. Twisting and Swiveling Domain Motions in Cas9 to Recognize Target DNA Duplexes, Make Double-Strand Breaks, and Release Cleaved Duplexes. Front. Mol. Biosci. 2023, 9, 1072733. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M.R. The Mechanism of Double-Strand DNA Break Repair by the Nonhomologous DNA End-Joining Pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Zhang, M.; Wang, X.; Ying, W.; Hu, X.; Dai, P.; Meng, F.; Shi, L.; Sun, Y.; Yao, N.; et al. Tild-CRISPR Allows for Efficient and Precise Gene Knockin in Mouse and Human Cells. Dev. Cell 2018, 45, 526–536.e5. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Zheng, P.; Sun, J.; Wang, M. Microbial Base Editing: A Powerful Emerging Technology for Microbial Genome Engineering. Trends Biotechnol. 2021, 39, 165–180. [Google Scholar] [CrossRef] [PubMed]
- Calero, P.; Nikel, P.I. Chasing Bacterial Chassis for Metabolic Engineering: A Perspective Review from Classical to Non-traditional Microorganisms. Microb. Biotechnol. 2019, 12, 98–124. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Kumar, P.; Maity, S.K.; Agrawal, D.; Narisetty, V.; Jacob, S.; Kumar, G.; Bhatia, S.K.; Kumar, D.; Vivekanand, V. Recent Advances in Bio-Based Production of Top Platform Chemical, Succinic Acid: An Alternative to Conventional Chemistry. Biotechnol. Biofuels 2024, 17, 72. [Google Scholar] [CrossRef] [PubMed]
- Skorokhodova, A.Y.; Stasenko, A.A.; Krasilnikova, N.V.; Gulevich, A.Y.; Debabov, V.G. Engineering Escherichia coli for Efficient Aerobic Conversion of Glucose to Malic Acid through the Modified Oxidative TCA Cycle. Fermentation 2022, 8, 738. [Google Scholar] [CrossRef]
- Yin, L.; Zhou, Y.; Ding, N.; Fang, Y. Recent Advances in Metabolic Engineering for the Biosynthesis of Phosphoenol Pyruvate-Oxaloacetate-Pyruvate-Derived Amino Acids. Molecules 2024, 29, 2893. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Jiang, T.; Li, M.; Zou, Y.; Yan, Y. Fine-Tuning Gene Expression for Improved Biosynthesis of Natural Products: From Transcriptional to Post-Translational Regulation. Biotechnol. Adv. 2022, 54, 107853. [Google Scholar] [CrossRef] [PubMed]
- Christensen, T.S.; Oliveira, A.P.; Nielsen, J. Reconstruction and Logical Modeling of Glucose Repression Signaling Pathways in Saccharomyces Cerevisiae. BMC Syst. Biol. 2009, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Lyu, X.; Lyu, Y.; Yu, H.; Chen, W.; Ye, L.; Yang, R. Biotechnological Advances for Improving Natural Pigment Production: A State-of-the-Art Review. Bioresour. Bioprocess. 2022, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Cao, J. Improving and Streamlining Gene Editing in Yarrowia Lipolytica via Integration of Engineered Cas9 Protein. J. Fungi 2024, 10, 63. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.-J.; Zhao, J.; Bhadauria, V.; Peng, Y.-L. Efficient Gene Editing with an Arg-tRNA Promoter-Driven CRISPR/Cas9 in the Rice Blast Fungus Pyricularia Oryzae. Phytopathol. Res. 2024, 6, 56. [Google Scholar] [CrossRef]
- Cho, S.; Shin, J.; Cho, B.-K. Applications of CRISPR/Cas System to Bacterial Metabolic Engineering. Int. J. Mol. Sci. 2018, 19, 1089. [Google Scholar] [CrossRef] [PubMed]
- Zahoor, A.; Lindner, S.N.; Wendisch, V.F. Metabolic Engineering of Corynebacterium Glutamicum Aimed at Alternative Carbon Sources and New Products. Comput. Struct. Biotechnol. J. 2012, 3, e201210004. [Google Scholar] [CrossRef] [PubMed]
- Adiego-Pérez, B.; Randazzo, P.; Daran, J.M.; Verwaal, R.; Roubos, J.A.; Daran-Lapujade, P.; van der Oost, J. Multiplex Genome Editing of Microorganisms Using CRISPR-Cas. FEMS Microbiol. Lett. 2019, 366, fnz086. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.-K.; Hong, Y.; Wu, Y.-R. Emerging Technologies for Genetic Modification of Solventogenic Clostridia: From Tool to Strategy Development. Bioresour. Technol. 2021, 334, 125222. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Wang, H.; Fu, S.; Zhang, D. CRISPR/Cas Tools for Enhancing the Biopreservation Ability of Lactic Acid Bacteria in Aquatic Products. Front. Bioeng. Biotechnol. 2022, 10, 1114588. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Zhang, Y.; Guo, D.; Qin, L.; Ashraf, M.; Ahmad, N. Recent Advances in Yeast Genome Evolution with Stress Tolerance for Green Biological Manufacturing. Biotechnol. Bioeng. 2022, 119, 2689–2697. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Olarte, R.D.; Bravo Rodríguez, R.; Morales-Ríos, E. Genome Editing in Bacteria: CRISPR-Cas and Beyond. Microorganisms 2021, 9, 844. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.-C.; Hsiao, W.-C.; Hsu, Y.-C.; Lin, M.-C.; Hsu, C.-C.; Zhang, M.M. Highly Efficient CRISPR-Cas9 Base Editing in Bifidobacterium with Bypass of Restriction Modification Systems. Appl. Environ. Microbiol. 2025, 91, e0198524. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.-K.; Wang, G.-Y.; Zeng, J.; Zhang, J.-A. Improved Succinate Production by Metabolic Engineering. Biomed. Res. Int. 2013, 2013, 538790. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.-Y.; Liu, Y.; Wang, D.; Lu, F.-P.; Huang, L.-Q.; Dai, Z.-B.; Zhang, X.-L. [Construction of Saccharomyces cerevisiae cell factories for lycopene production]. Zhongguo Zhong Yao Za Zhi 2014, 39, 3978–3985. [Google Scholar] [PubMed]
- Dong, T.; Shu, Y.; Wang, Y.; Yao, M.; Xiao, W. An Engineered Yarrowia Lipolytica with Rapid Growth and Efficient Lipid Utilization. Synth. Syst. Biotechnol. 2025, 10, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Ma, Z.; Cao, Y.; Zhang, H.; Qi, Y.; Wei, L.; Jiang, J.; Xu, N.; Liu, J. Metabolic Engineering of Corynebacterium Glutamicum for Efficient l -Homoserine Production from Lignocellulose-Derived Sugars. ACS Sustain. Chem. Eng. 2025, 13, 3441–3451. [Google Scholar] [CrossRef]
- Put, H.; Gerstmans, H.; Vande Capelle, H.; Fauvart, M.; Michiels, J.; Masschelein, J. Bacillus Subtilis as a Host for Natural Product Discovery and Engineering of Biosynthetic Gene Clusters. Nat. Prod. Rep. 2024, 41, 1113–1151. [Google Scholar] [CrossRef] [PubMed]
- Vamsi Krishna, K.; Bharathi, N.; George Shiju, S.; Alagesan Paari, K.; Malaviya, A. An Updated Review on Advancement in Fermentative Production Strategies for Biobutanol Using Clostridium spp. Environ. Sci. Pollut. Res. 2022, 29, 47988–48019. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Jadhav, S.M.; Moholkar, V.S. Acetone-Butanol-Ethanol (ABE) Fermentation with Clostridial Co-Cultures for Enhanced Biobutanol Production. Process Saf. Environ. Prot. 2024, 185, 277–285. [Google Scholar] [CrossRef]
- Chen, L.; Liu, R.; He, C.; Wu, M.; Ge, Q.; Yu, H. Biosynthesis Pathway of Flavor Compound 3-Methylbutanal Derived from Leucine by Lactococcus Lactis 517: From a Transcriptomics Perspective. Food Biosci. 2024, 61, 104740. [Google Scholar] [CrossRef]
- Da Silva, G.A.R.; Oliveira, S.S.D.S.; Lima, S.F.; Do Nascimento, R.P.; Baptista, A.R.D.S.; Fiaux, S.B. The Industrial Versatility of Gluconobacter Oxydans: Current Applications and Future Perspectives. World J. Microbiol. Biotechnol. 2022, 38, 134. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Lindblad, P. Expressing 2-Keto Acid Pathway Enzymes Significantly Increases Photosynthetic Isobutanol Production. Microb. Cell Fact. 2022, 21, 17. [Google Scholar] [CrossRef] [PubMed]
- Xing, M.-N.; Zhang, X.-Z.; Huang, H. Application of Metagenomic Techniques in Mining Enzymes from Microbial Communities for Biofuel Synthesis. Biotechnol. Adv. 2012, 30, 920–929. [Google Scholar] [CrossRef] [PubMed]
- Javed, M.R.; Noman, M.; Shahid, M.; Ahmed, T.; Khurshid, M.; Rashid, M.H.; Ismail, M.; Sadaf, M.; Khan, F. Current Situation of Biofuel Production and Its Enhancement by CRISPR/Cas9-Mediated Genome Engineering of Microbial Cells. Microbiol. Res. 2019, 219, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Periyasamy, S.; Beula Isabel, J.; Kavitha, S.; Karthik, V.; Mohamed, B.A.; Gizaw, D.G.; Sivashanmugam, P.; Aminabhavi, T.M. Recent Advances in Consolidated Bioprocessing for Conversion of Lignocellulosic Biomass into Bioethanol—A Review. Chem. Eng. J. 2023, 453, 139783. [Google Scholar] [CrossRef]
- Adegboye, M.F.; Ojuederie, O.B.; Talia, P.M.; Babalola, O.O. Bioprospecting of Microbial Strains for Biofuel Production: Metabolic Engineering, Applications, and Challenges. Biotechnol. Biofuels 2021, 14, 5. [Google Scholar] [CrossRef] [PubMed]
- Abdelaal, A.S.; Jawed, K.; Yazdani, S.S. CRISPR/Cas9-Mediated Engineering of Escherichia coli for n-Butanol Production from Xylose in Defined Medium. J. Ind. Microbiol. Biotechnol. 2019, 46, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Dong, S.; Wang, P.; Tao, Y.; Wang, Y. Genome Editing in Clostridium Saccharoperbutylacetonicum N1-4 with the CRISPR-Cas9 System. Appl. Environ. Microbiol. 2017, 83, e00233-17. [Google Scholar] [CrossRef] [PubMed]
- Wasels, F.; Jean-Marie, J.; Collas, F.; López-Contreras, A.M.; Lopes Ferreira, N. A Two-Plasmid Inducible CRISPR/Cas9 Genome Editing Tool for Clostridium Acetobutylicum. J. Microbiol. Methods 2017, 140, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, L.; Zhang, W. Proteomic and Metabolomic Analyses Reveal Metabolic Responses to 3-Hydroxypropionic Acid Synthesized Internally in Cyanobacterium Synechocystis sp. PCC 6803. Biotechnol. Biofuels 2016, 9, 209. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Wang, Y.; Yang, Q.; Zhang, Y.; Wu, Y.; Yang, Y.; Mei, M.; He, M.; Wang, X.; Yang, S. Molecular Mechanism of Enhanced Ethanol Tolerance Associated with Hfq Overexpression in Zymomonas Mobilis. Front. Bioeng. Biotechnol. 2022, 10, 1098021. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Z.; Yan, X.; Yuan, Y.; Wang, R.; Zhu, H.; Wang, Y.; Li, J.; Ye, J.; Yue, H.; Yang, X. Advances in Stress-Tolerance Elements for Microbial Cell Factories. Synth. Syst. Biotechnol. 2024, 9, 793–808. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Jiang, J.; Zhang, H.; Liu, J.; Ruan, H. Transcending Membrane Barriers: Advances in Membrane Engineering to Enhance the Production Capacity of Microbial Cell Factories. Microb. Cell Fact. 2024, 23, 154. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, V.; Lind, U.; St. Onge, R.P.; Blomberg, A.; Nygård, Y. A CRISPR Interference Screen of Essential Genes Reveals That Proteasome Regulation Dictates Acetic Acid Tolerance in Saccharomyces cerevisiae. mSystems 2021, 6, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Huang, J.; Wu, Y.; Wu, W.; Zhang, Y.; Liu, D. Metabolic Engineering of Corynebacterium Glutamicum for the Production of 3-Hydroxypropionic Acid from Glucose and Xylose. Metab. Eng. 2017, 39, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Liew, F.; Henstra, A.M.; Kӧpke, M.; Winzer, K.; Simpson, S.D.; Minton, N.P. Metabolic Engineering of Clostridium Autoethanogenum for Selective Alcohol Production. Metab. Eng. 2017, 40, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Varman, A.M.; Xiao, Y.; Pakrasi, H.B.; Tang, Y.J. Metabolic Engineering of Synechocystis Sp. Strain PCC 6803 for Isobutanol Production. Appl. Environ. Microbiol. 2013, 79, 908–914. [Google Scholar] [CrossRef] [PubMed]
- Hirokawa, Y.; Maki, Y.; Hanai, T. Improvement of 1,3-Propanediol Production Using an Engineered Cyanobacterium, Synechococcus Elongatus by Optimization of the Gene Expression Level of a Synthetic Metabolic Pathway and Production Conditions. Metab. Eng. 2017, 39, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Zhu, F.; Lu, L.; Fu, A.; Lu, J.; Deng, Z.; Liu, T. Metabolic Engineering of Fatty Acyl-ACP Reductase-Dependent Pathway to Improve Fatty Alcohol Production in Escherichia coli. Metab. Eng. 2014, 22, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Lian, J.; Chao, R.; Zhao, H. Metabolic Engineering of a Saccharomyces Cerevisiae Strain Capable of Simultaneously Utilizing Glucose and Galactose to Produce Enantiopure (2R,3R)-Butanediol. Metab. Eng. 2014, 23, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wu, Y.; Huang, J.; Liu, D. Metabolic Engineering of Klebsiella Pneumoniae for the de Novo Production of 2-Butanol as a Potential Biofuel. Bioresour. Technol. 2015, 197, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Gaida, S.M.; Liedtke, A.; Jentges, A.H.W.; Engels, B.; Jennewein, S. Metabolic Engineering of Clostridium cellulolyticum for the Production of N-Butanol from Crystalline Cellulose. Microb. Cell Fact. 2016, 15, 6. [Google Scholar] [CrossRef] [PubMed]
- Bhandiwad, A.; Shaw, A.J.; Guss, A.; Guseva, A.; Bahl, H.; Lynd, L.R. Metabolic Engineering of Thermoanaerobacterium saccharolyticum for N-Butanol Production. Metab. Eng. 2014, 21, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Avalos, J.L.; Fink, G.R.; Stephanopoulos, G. Compartmentalization of Metabolic Pathways in Yeast Mitochondria Improves the Production of Branched-Chain Alcohols. Nat. Biotechnol. 2013, 31, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Xu, M.; Tang, I.; Yang, S. Metabolic Engineering of Clostridium tyrobutyricum for N-butanol Production through Co-utilization of Glucose and Xylose. Biotechnol. Bioeng. 2015, 112, 2134–2141. [Google Scholar] [CrossRef]
- Yang, X.; Xu, M.; Yang, S.-T. Metabolic and Process Engineering of Clostridium Cellulovorans for Biofuel Production from Cellulose. Metab. Eng. 2015, 32, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Fang, H.; Zhang, D. Expanding Application of CRISPR-Cas9 System in Microorganisms. Synth. Syst. Biotechnol. 2020, 5, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Jozala, A.F.; Geraldes, D.C.; Tundisi, L.L.; Feitosa, V.d.A.; Breyer, C.A.; Cardoso, S.L.; Mazzola, P.G.; Oliveira-Nascimento, L.d.; Rangel-Yagui, C.d.O.; Magalhães, P.d.O.; et al. Biopharmaceuticals from Microorganisms: From Production to Purification. Braz. J. Microbiol. 2016, 47 (Suppl. 1), 51–63. [Google Scholar] [CrossRef] [PubMed]
- He, Y.-Z.; Yan, J.-R.; He, B.; Ren, H.; Kuang, X.; Long, T.-F.; Chen, C.-P.; Liao, X.-P.; Liu, Y.-H.; Sun, J. A Transposon-Associated CRISPR/Cas9 System Specifically Eliminates Both Chromosomal and Plasmid-Borne Mcr-1 in Escherichia coli. Antimicrob. Agents Chemother. 2021, 65, e0105421. [Google Scholar] [CrossRef] [PubMed]
- Glass, Z.; Lee, M.; Li, Y.; Xu, Q. Engineering the Delivery System for CRISPR-Based Genome Editing. Trends Biotechnol. 2018, 36, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Raschmanová, H.; Weninger, A.; Glieder, A.; Kovar, K.; Vogl, T. Implementing CRISPR-Cas Technologies in Conventional and Non-Conventional Yeasts: Current State and Future Prospects. Biotechnol. Adv. 2018, 36, 641–665. [Google Scholar] [CrossRef]
- Pan, Y.; Yang, J.; Wu, J.; Yang, L.; Fang, H. Current Advances of Pichia Pastoris as Cell Factories for Production of Recombinant Proteins. Front. Microbiol. 2022, 13, 1059777. [Google Scholar] [CrossRef] [PubMed]
- Karagöz, G.E.; Acosta-Alvear, D.; Walter, P. The Unfolded Protein Response: Detecting and Responding to Fluctuations in the Protein-Folding Capacity of the Endoplasmic Reticulum. Cold Spring Harb. Perspect. Biol. 2019, 11, a033886. [Google Scholar] [CrossRef] [PubMed]
- Call, S.N.; Andrews, L.B. CRISPR-Based Approaches for Gene Regulation in Non-Model Bacteria. Front. Genome Ed. 2022, 4, 892304. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, Z. CRISPR-Cas Systems: Overview, Innovations and Applications in Human Disease Research and Gene Therapy. Comput. Struct. Biotechnol. J. 2020, 18, 2401–2415. [Google Scholar] [CrossRef] [PubMed]
- Boukid, F.; Ganeshan, S.; Wang, Y.; Tülbek, M.Ç.; Nickerson, M.T. Bioengineered Enzymes and Precision Fermentation in the Food Industry. Int. J. Mol. Sci. 2023, 24, 10156. [Google Scholar] [CrossRef] [PubMed]
- Hamburger, F.; Schlichting, N.; Eichenlaub, M.; Costea, P.I.; Sauer, C.; Jenewein, S.; Kabisch, J. Automation-Aided Construction and Characterization of Bacillus subtilis PrsA Strains for the Secretion of Amylases. Front. Bioeng. Biotechnol. 2024, 12, 1479626. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Li, X.; Guo, L.; Gao, C.; Song, W.; Wei, W.; Wu, J.; Liu, L.; Chen, X. Reprogramming Metabolic Flux in Escherichia coli to Enhance Chondroitin Production. Adv. Sci. 2024, 11, e2307351. [Google Scholar] [CrossRef] [PubMed]
- Ruan, S.; He, C.; Wang, A.; Lin, Y.; Liang, S. Establishment and Application of an RNAi System in Pichia Pastoris. Front. Bioeng. Biotechnol. 2025, 13, 1548187. [Google Scholar] [CrossRef] [PubMed]
- Fourie, K.R.; Wilson, H.L. Understanding GroEL and DnaK Stress Response Proteins as Antigens for Bacterial Diseases. Vaccines 2020, 8, 773. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; Chen, H.; Li, N.; Liang, W. The Application of the CRISPR-Cas System in Antibiotic Resistance. Infect. Drug Resist. 2022, 15, 4155–4168. [Google Scholar] [CrossRef] [PubMed]
- Druzhinina, I.S.; Kubicek, C.P. Genetic Engineering of Trichoderma reesei Cellulases and Their Production. Microb. Biotechnol. 2017, 10, 1485–1499. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, C.; Lu, H.; Wu, Q.; Wu, Y.; Li, W.; Li, X. Improvement of Thermostability and Catalytic Efficiency of Xylanase from Myceliophthora thermophilar by N-Terminal and C-Terminal Truncation. Front. Microbiol. 2024, 15, 1385329. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Jiang, T.; Yan, Y. The Expanded CRISPR Toolbox for Constructing Microbial Cell Factories. Trends Biotechnol. 2024, 42, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Bendixen, L.; Jensen, T.I.; Bak, R.O. CRISPR-Cas-Mediated Transcriptional Modulation: The Therapeutic Promises of CRISPRa and CRISPRi. Mol. Ther. 2023, 31, 1920–1937. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Q.; Wu, X.-Y.; Xu, X.; Tan, X.; Li, Z.; Zhang, B. Production of L-Glutamate Family Amino Acids in Corynebacterium glutamicum: Physiological Mechanism, Genetic Modulation, and Prospects. Synth. Syst. Biotechnol. 2021, 6, 302–325. [Google Scholar] [CrossRef] [PubMed]
- Madzak, C. Yarrowia Lipolytica Strains and Their Biotechnological Applications: How Natural Biodiversity and Metabolic Engineering Could Contribute to Cell Factories Improvement. J. Fungi 2021, 7, 548. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.; Gao, C.; Wang, Q.; Liu, H.; Qi, Q. A Novel Strategy for Succinate and Polyhydroxybutyrate Co-Production in Escherichia coli. Bioresour. Technol. 2010, 101, 7675–7678. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Bao, T.; Yang, S.-T. Engineering Clostridium for Improved Solvent Production: Recent Progress and Perspective. Appl. Microbiol. Biotechnol. 2019, 103, 5549–5566. [Google Scholar] [CrossRef] [PubMed]
- Naseri, G.; Koffas, M.A.G. Application of Combinatorial Optimization Strategies in Synthetic Biology. Nat. Commun. 2020, 11, 2446. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, K.; Shen, W.; Xia, Y.; Li, Y.; Chen, X. Boosting Production of Cembratriene-ol in Saccharomyces cerevisiae via Systematic Optimization. Biotechnol. J. 2024, 19, 2300324. [Google Scholar] [CrossRef] [PubMed]
- Punjabi, M.; Bharadvaja, N.; Jolly, M.; Dahuja, A.; Sachdev, A. Development and Evaluation of Low Phytic Acid Soybean by siRNA Triggered Seed Specific Silencing of Inositol Polyphosphate 6-/3-/5-Kinase Gene. Front. Plant Sci. 2018, 9, 804. [Google Scholar] [CrossRef] [PubMed]
- Millen, A.M.; Horvath, P.; Boyaval, P.; Romero, D.A. Mobile CRISPR/Cas-Mediated Bacteriophage Resistance in Lactococcus Lactis. PLoS ONE 2012, 7, e51663. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jeong, G.-T. Enhancement of Ethanol Productivity with Saccharomyces cerevisiae by Overexpression of Lipid Elongation Gene Using CRISPR/CAS9. Kor. J. Microbiol. Biotechnol. 2021, 49, 210–216. [Google Scholar] [CrossRef]
- Liu, L.; Helal, S.E.; Peng, N. CRISPR-Cas-Based Engineering of Probiotics. BioDesign Res. 2023, 5, 0017. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Yu, H.; Feng, X.; Tang, H.; Xiong, Z.; Xia, Y.; Ai, L.; Song, X. Specific Bile Salt Hydrolase Genes in Lactobacillus Plantarum AR113 and Relationship with Bile Salt Resistance. LWT 2021, 145, 111208. [Google Scholar] [CrossRef]
- Balabanova, L.; Averianova, L.; Marchenok, M.; Son, O.; Tekutyeva, L. Microbial and Genetic Resources for Cobalamin (Vitamin B12) Biosynthesis: From Ecosystems to Industrial Biotechnology. Int. J. Mol. Sci. 2021, 22, 4522. [Google Scholar] [CrossRef] [PubMed]
- Byun, G.; Yang, J.; Seo, S.W. CRISPRi-Mediated Tunable Control of Gene Expression Level with Engineered Single-Guide RNA in Escherichia coli. Nucleic Acids Res. 2023, 51, 4650–4659. [Google Scholar] [CrossRef] [PubMed]
- Lee, M. Deep Learning in CRISPR-Cas Systems: A Review of Recent Studies. Front. Bioeng. Biotechnol. 2023, 11, 1226182. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chu, W.; Gill, R.A.; Sang, S.; Shi, Y.; Hu, X.; Yang, Y.; Zaman, Q.U.; Zhang, B. Computational Tools and Resources for CRISPR/Cas Genome Editing. Genom. Proteom. Bioinform. 2023, 21, 108–126. [Google Scholar] [CrossRef] [PubMed]
- Konstantakos, V.; Nentidis, A.; Krithara, A.; Paliouras, G. CRISPR-Cas9 gRNA Efficiency Prediction: An Overview of Predictive Tools and the Role of Deep Learning. Nucleic Acids Res. 2022, 50, 3616–3637. [Google Scholar] [CrossRef] [PubMed]
- Xiang, T.; Feng, H.; Xing, X.; Zhang, C. GLiDe: A Web-Based Genome-Scale CRISPRi sgRNA Design Tool for Prokaryotes. BMC Bioinform. 2025, 26, 1. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lamson, A.R.; Shelley, M.; Troyanskaya, O. Interpretable Neural Architecture Search and Transfer Learning for Understanding CRISPR–Cas9 off-Target Enzymatic Reactions. Nat. Comput. Sci. 2023, 3, 1056–1066. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Qu, Y.; Cousins, H.; Johnson, W.A.; Yin, D.; Shah, M.; Zhou, D.; Altman, R.; Wang, M.; Cong, L. CRISPR-GPT: An LLM Agent for Automated Design of Gene-Editing Experiments. arXiv 2024, arXiv:2404.18021. [Google Scholar]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-Frequency off-Target Mutagenesis Induced by CRISPR-Cas Nucleases in Human Cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.W.; Kim, S.; Kim, Y.; Kweon, J.; Kim, H.S.; Bae, S.; Kim, J.-S. Analysis of Off-Target Effects of CRISPR/Cas-Derived RNA-Guided Endonucleases and Nickases. Genome Res. 2014, 24, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Bikard, D.; Cox, D.; Zhang, F.; Marraffini, L.A. RNA-Guided Editing of Bacterial Genomes Using CRISPR-Cas Systems. Nat. Biotechnol. 2013, 31, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, J.-S. A Guide to Genome Engineering with Programmable Nucleases. Nat. Rev. Genet. 2014, 15, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Shmakov, S.; Abudayyeh, O.O.; Makarova, K.S.; Wolf, Y.I.; Gootenberg, J.S.; Semenova, E.; Minakhin, L.; Joung, J.; Konermann, S.; Severinov, K.; et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cell 2015, 60, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, C.T.; Deshpande, P.S.; Dever, D.P.; Camarena, J.; Lemgart, V.T.; Cromer, M.K.; Vakulskas, C.A.; Collingwood, M.A.; Zhang, L.; Bode, N.M.; et al. Identification of Preexisting Adaptive Immunity to Cas9 Proteins in Humans. Nat. Med. 2019, 25, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Potter, J.; Kumar, S.; Zou, Y.; Quintanilla, R.; Sridharan, M.; Carte, J.; Chen, W.; Roark, N.; Ranganathan, S.; et al. Rapid and Highly Efficient Mammalian Cell Engineering via Cas9 Protein Transfection. J. Biotechnol. 2015, 208, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Guo, L.; Xu, S.; Chen, J.; Zhou, J. High-Throughput Screening Technology in Industrial Biotechnology. Trends Biotechnol. 2020, 38, 888–906. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome Editing with CRISPR-Cas Nucleases, Base Editors, Transposases and Prime Editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dudeja, C.; Mishra, A.; Ali, A.; Singh, P.P.; Jaiswal, A.K. Microbial Genome Editing with CRISPR–Cas9: Recent Advances and Emerging Applications Across Sectors. Fermentation 2025, 11, 410. https://doi.org/10.3390/fermentation11070410
Dudeja C, Mishra A, Ali A, Singh PP, Jaiswal AK. Microbial Genome Editing with CRISPR–Cas9: Recent Advances and Emerging Applications Across Sectors. Fermentation. 2025; 11(7):410. https://doi.org/10.3390/fermentation11070410
Chicago/Turabian StyleDudeja, Chhavi, Amish Mishra, Ansha Ali, Prem Pratap Singh, and Atul Kumar Jaiswal. 2025. "Microbial Genome Editing with CRISPR–Cas9: Recent Advances and Emerging Applications Across Sectors" Fermentation 11, no. 7: 410. https://doi.org/10.3390/fermentation11070410
APA StyleDudeja, C., Mishra, A., Ali, A., Singh, P. P., & Jaiswal, A. K. (2025). Microbial Genome Editing with CRISPR–Cas9: Recent Advances and Emerging Applications Across Sectors. Fermentation, 11(7), 410. https://doi.org/10.3390/fermentation11070410






