From Lebanese Soil to Antimicrobials: A Novel Streptomyces Species with Antimicrobial Potential
Abstract
1. Introduction
1.1. Sample Processing and Bacterial Isolation
1.1.1. Sample Collection
1.1.2. Sample Processing and Bacterial Isolation
1.2. Morphological, Biochemical, and Physiological Characterization
1.3. Scanning Electron Microscopy (SEM)
1.4. Genomic Characterization
1.4.1. DNA Extraction and WGS
1.4.2. Phylogenetic Classification
1.4.3. Functional Annotation
1.5. Small-Scale Secondary Metabolite Production and Extraction
1.6. Antibacterial Bioactivity Screening
1.6.1. Bacterial Strains
1.6.2. Broth Microdilution (BMD) Assay
2. Results
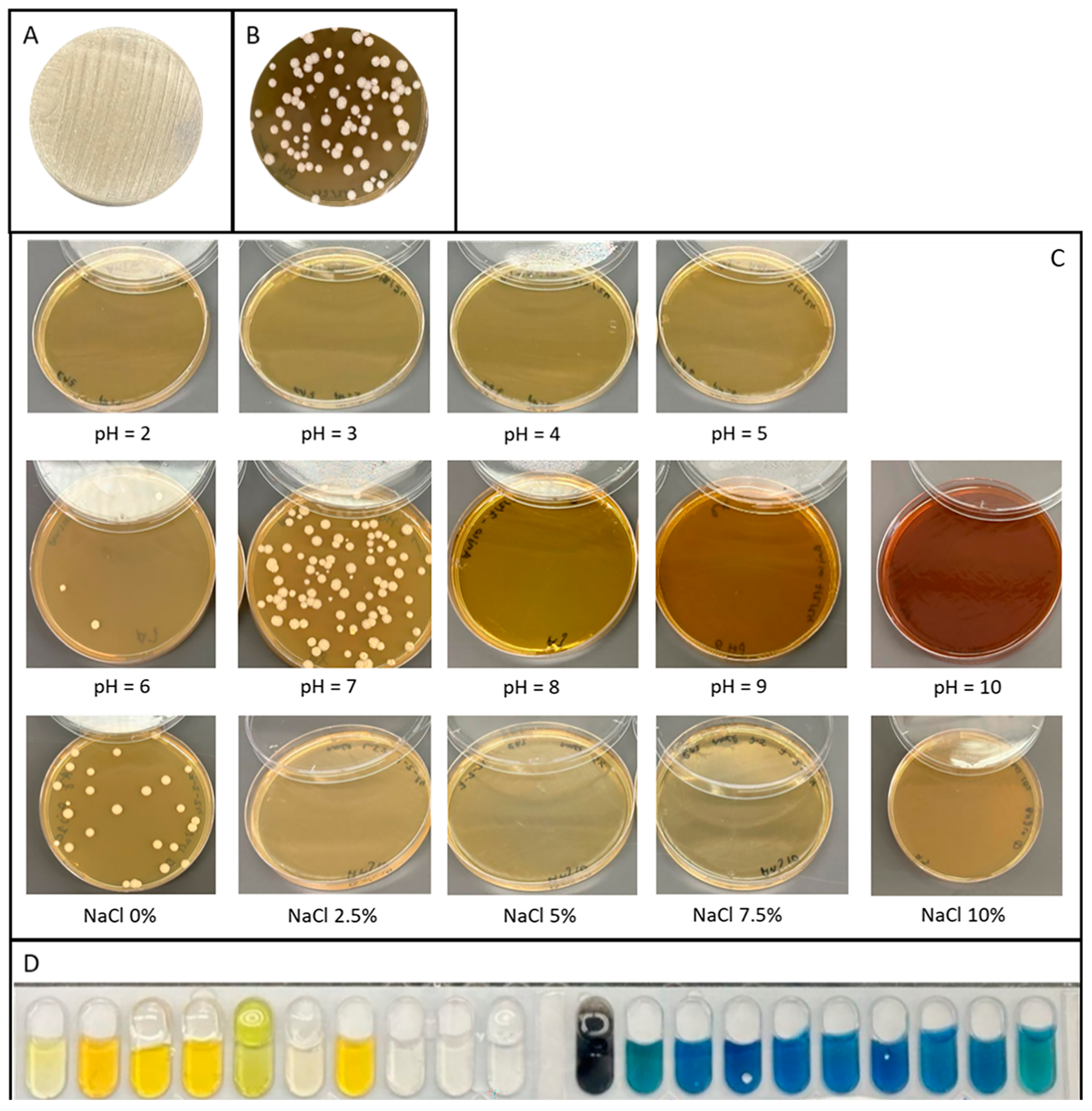
2.1. Morphological, Physiological, and Biochemical Characterization
2.1.1. Morphology
2.1.2. Gram Staining
2.1.3. Salt Tolerance
2.1.4. pH Tolerance
2.1.5. Biochemical Properties
2.2. Genomic Characterization
2.2.1. Genome Features
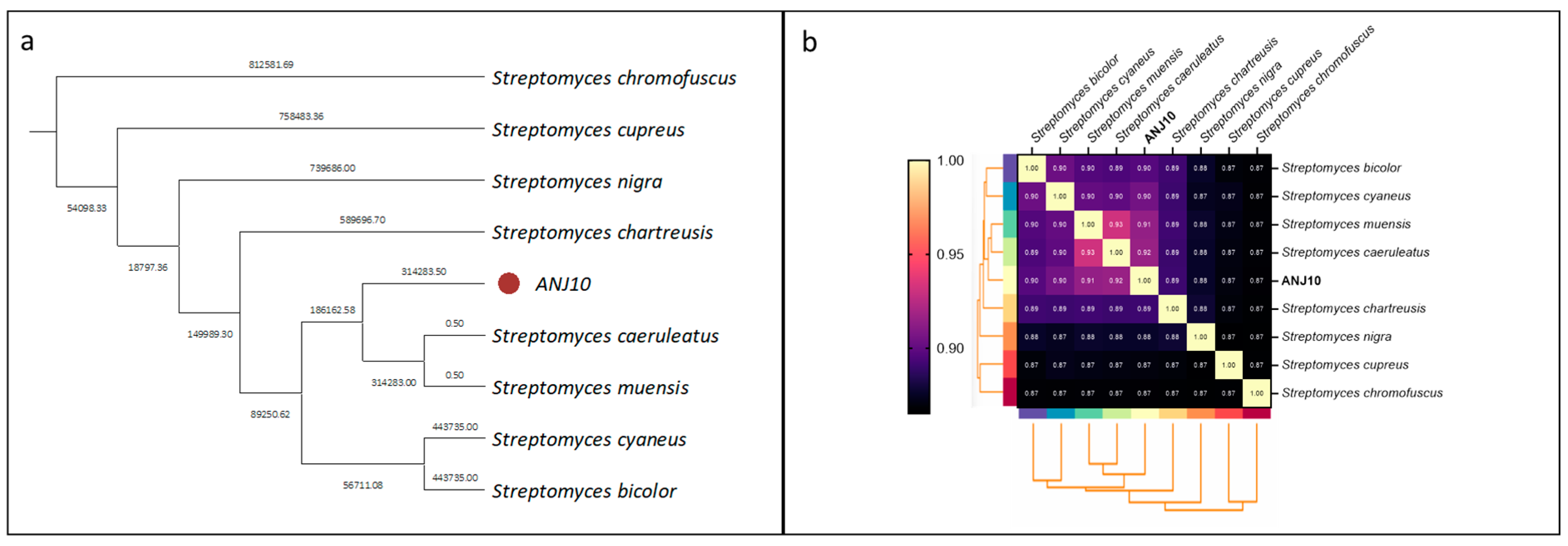
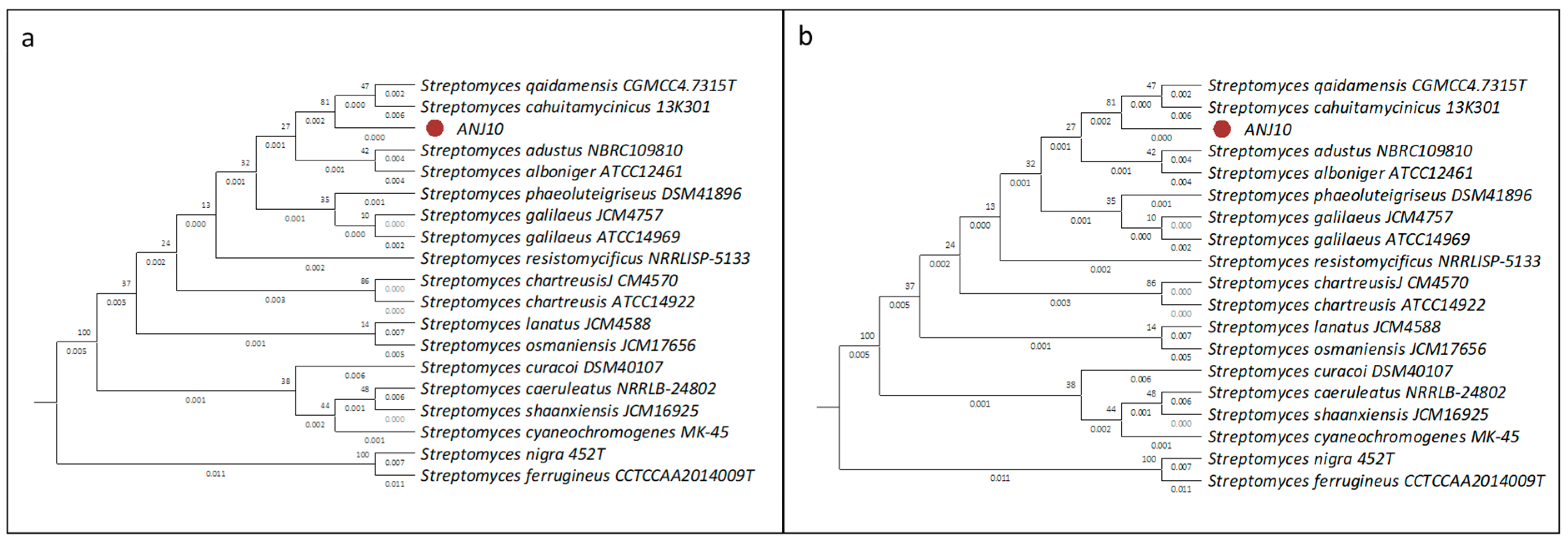
2.2.2. Phylogenetic Classification
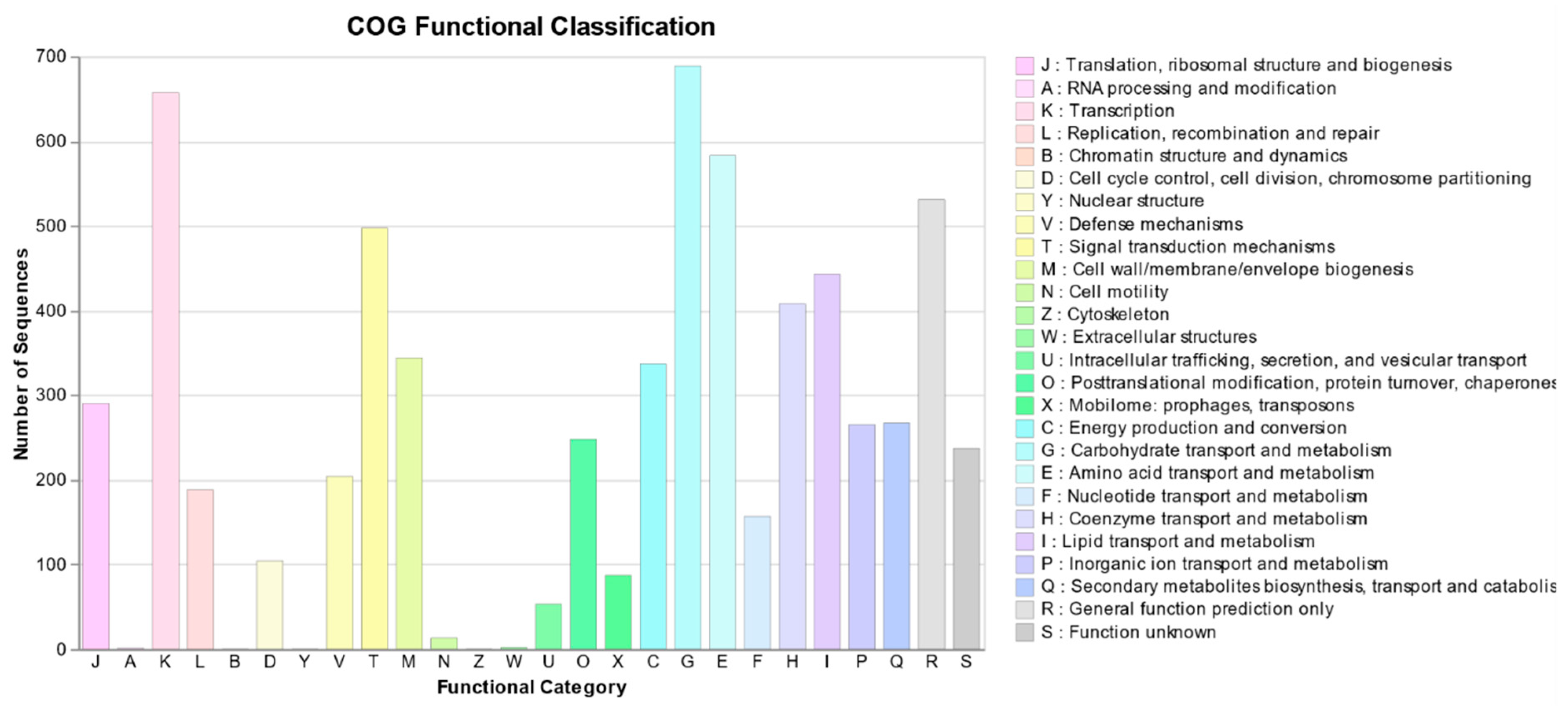
2.2.3. Functional Annotation
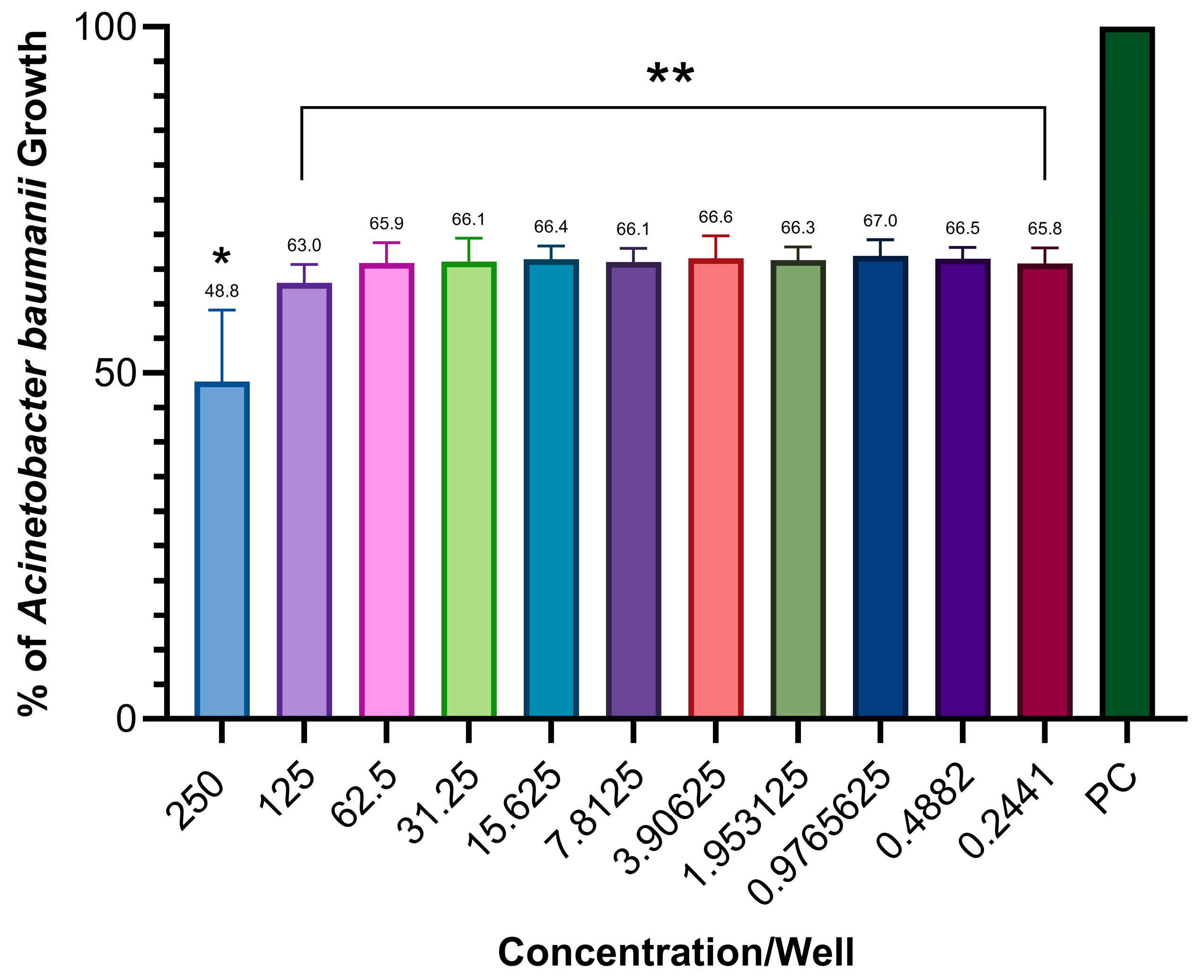
2.3. Antimicrobial Activity Testing
3. Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BGC | Biosynthetic Gene Cluster |
| WGS | Whole-Genome Sequencing |
| BMD | Broth Microdilution |
| NaCl | Sodium Chloride |
| SEM | Scanning Electron Microscopy |
| PBS | Phosphate-Buffered Saline |
| FA | Formaldehyde |
| GA | Glutaraldehyde |
| TSB | Tryptic Soy Broth |
| gDNA | Genomic DNA |
| CDS | Coding Sequences |
| rRNA | Ribosomal RNA |
| tRNA | Transfer RNA |
| tmRNA | Transfer-messenger RNA |
| ANI | Average Nucleotide Identity |
| dDDH | Digital DNA–DNA Hybridization |
| TYGS | Type Strain Genome Server |
| GBDP | Genome BLAST Distance Phylogeny |
| PC | Positive Control |
| NC | Negative Control |
| OD | Optical Density |
| MHCAB | Mueller Hinton Cation-Adjusted Broth |
| SMs | Secondary Metabolites |
| NRPS | Nonribosomal Peptide Synthetase |
| PKS | Polyketide Synthase |
| T1PKS | Type I Polyketide Synthase |
| T2PKS | Type II Polyketide Synthase |
| RiPPs | Ribosomally synthesized and post-translationally modified Peptides |
| NRP | Nonribosomal Peptide |
| COG | Cluster of Orthologous Genes |
| ARTS | Antibiotic Resistant Target Seeker |
| MRSA | Methicillin-Resistant Staphylococcus aureus |
| MSSA | Methicillin-Sensitive Staphylococcus aureus |
| WHO | World Health Organization |
| MDR | Multi-Drug Resistant |
| miBIG | Minimum Information about a Biosynthetic Gene cluster |
References
- Wade, W. Unculturable bacteria—The uncharacterized organisms that cause oral infections. J. R. Soc. Med. 2002, 95, 81–83. [Google Scholar] [PubMed]
- Hug, L.A. Sizing Up the Uncultured Microbial Majority. mSystems 2018, 3, e00185-18. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, K.G.; Steen, A.D.; Ladau, J.; Yin, J.; Crosby, L. Phylogenetically Novel Uncultured Microbial Cells Dominate Earth Microbiomes. mSystems 2018, 3, e00055-18. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R.H. Marcel Faber Roundtable: Is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration? J. Ind. Microbiol. Biotechnol. 2006, 33, 507–513. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef]
- Fair, R.J.; Tor, Y. Antibiotics and bacterial resistance in the 21st century. Perspect. Med. Chem. 2014, 6, 25–64. [Google Scholar] [CrossRef]
- Genilloud, O. Actinomycetes: Still a source of novel antibiotics. Nat. Prod. Rep. 2017, 34, 1203–1232. [Google Scholar] [CrossRef]
- Bérdy, J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 2012, 65, 385–395. [Google Scholar] [CrossRef]
- Demain, A.L.; Sanchez, S. Microbial drug discovery: 80 years of progress. J. Antibiot. 2009, 62, 5–16. [Google Scholar] [CrossRef]
- Kämpfer, P.; Glaeser, S.P.; Parkes, L.; van Keulen, G.; Dyson, P. The Family Streptomycetaceae. In The Prokaryotes: Actinobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 889–1010. [Google Scholar]
- Sathya, A.; Vijayabharathi, R.; Gopalakrishnan, S. Plant growth-promoting actinobacteria: A new strategy for enhancing sustainable production and protection of grain legumes. 3 Biotech 2017, 7, 102. [Google Scholar] [CrossRef]
- Cimermancic, P.; Medema, M.H.; Claesen, J.; Kurita, K.; Wieland Brown, L.C.; Mavrommatis, K.; Pati, A.; Godfrey, P.A.; Koehrsen, M.; Clardy, J.; et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 2014, 158, 412–421. [Google Scholar] [CrossRef]
- Doroghazi, J.R.; Albright, J.C.; Goering, A.W.; Ju, K.S.; Haines, R.R.; Tchalukov, K.A.; Labeda, D.P.; Kelleher, N.L.; Metcalf, W.W. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 2014, 10, 963–968. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.-H.; Kim, H.; Ji, C.-H.; Je, H.-W.; Lee, D.; Shim, S.H.; Joo, H.-S.; Kang, H.-S.; Gutierrez, M. Comparative Genomics Reveals a Remarkable Biosynthetic Potential of the Streptomyces Phylogenetic Lineage Associated with Rugose-Ornamented Spores. mSystems 2021, 6, e0048921. [Google Scholar] [CrossRef] [PubMed]
- Awada, B.; Hamie, M.; El Hajj, R.; Derbaj, G.; Najm, R.; Makhoul, P.; Ali, D.H.; Fayad, A.G.A.; El Hajj, H. HAS 1: A natural product from soil-isolated Streptomyces species with potent activity against cutaneous leishmaniasis caused by Leishmania tropica. Front. Pharmacol. 2022, 13, 1023114. [Google Scholar] [CrossRef]
- Ewels, P.A.; Peltzer, A.; Fillinger, S.; Patel, H.; Alneberg, J.; Wilm, A.; Garcia, M.U.; Di Tommaso, P.; Nahnsen, S. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020, 38, 276–278. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 2011, 28, 464–469. [Google Scholar] [CrossRef]
- Page, A.J.; Hunt, M.; Seemann, T.; Keane, J.A. SaffronTree: Fast, reference-free pseudo-phylogenomic trees from reads or contigs. J. Open Source Softw. 2017, 2, 243. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2021, 50, D801–D807. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genom. Sci. 2014, 9, 2. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef]
- Kreft, Ł.; Botzki, A.; Coppens, F.; Vandepoele, K.; Van Bel, M. PhyD3: A phylogenetic tree viewer with extended phyloXML support for functional genomics data visualization. Bioinformatics 2017, 33, 2946–2947. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Farris, J.S. Estimating Phylogenetic Trees from Distance Matrices. Am. Nat. 1972, 106, 645–668. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Mungan, M.D.; Alanjary, M.; Blin, K.; Weber, T.; Medema, M.H.; Ziemert, N. ARTS 2.0: Feature updates and expansion of the Antibiotic Resistant Target Seeker for comparative genome mining. Nucleic Acids Res. 2020, 48, W546–W552. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Muñoz, J.C.; Selem-Mojica, N.; Mullowney, M.W.; Kautsar, S.A.; Tryon, J.H.; Parkinson, E.I.; De Los Santos, E.L.C.; Yeong, M.; Cruz-Morales, P.; Abubucker, S.; et al. A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 2020, 16, 60–68. [Google Scholar] [CrossRef]
- Shimoyama, Y. COGclassifier: A Tool for Classifying Prokaryote Protein Sequences into COG Functional Category 2022. Available online: https://github.com/moshi4/COGclassifier (accessed on 15 April 2024).
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Donald, L.; Pipite, A.; Subramani, R.; Owen, J.; Keyzers, R.A.; Taufa, T. Streptomyces: Still the Biggest Producer of New Natural Secondary Metabolites, a Current Perspective. Microbiol. Res. 2022, 13, 418–465. [Google Scholar] [CrossRef]
- Tribe, H.T. The discovery and development of cyclosporin. Mycologist 1998, 12, 20–22. [Google Scholar] [CrossRef]
- Seto, B. Rapamycin and mTOR: A serendipitous discovery and implications for breast cancer. Clin. Transl. Med. 2012, 1, 29. [Google Scholar] [CrossRef]
- Russell, A.H.; Truman, A.W. Genome Mining Strategies for Ribosomally Synthesised and Post-Translationally Modified Peptides. Comput. Struct. Biotechnol. J. 2020, 18, 1838–1851. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Mining and unearthing hidden biosynthetic potential. Nat. Commun. 2021, 12, 3864. [Google Scholar] [CrossRef]
- Liu, W.; Sun, F.; Hu, Y. Genome Mining-Mediated Discovery of a New Avermipeptin Analogue in Streptomyces Actuosus ATCC 25421. ChemistryOpen 2018, 7, 558–561. [Google Scholar] [CrossRef]
- Zheng, D.; Ding, N.; Jiang, Y.; Zhang, J.; Ma, J.; Chen, X.; Liu, J.; Han, L.; Huang, X. Albaflavenoid, a New Tricyclic Sesquiterpenoid from Streptomyces Violascens. J. Antibiot. 2016, 69, 773–775. [Google Scholar] [CrossRef]
- Hur, J.; Jang, J.; Sim, J. A Review of the Pharmacological Activities and Recent Synthetic Advances of γ-Butyrolactones. IJMS 2021, 22, 2769. [Google Scholar] [CrossRef] [PubMed]
- Purev, E.; Kondo, T.; Takemoto, D.; Niones, J.T.; Ojika, M. Identification of ε-Poly-L-Lysine as an Antimicrobial Product from an Epichloë Endophyte and Isolation of Fungal ε-PL Synthetase Gene. Molecules 2020, 25, 1032. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, Y.; Sun, J.; Huang, W.-C.; Xue, C.; Mao, X. A Novel Soluble Squalene-Hopene Cyclase and Its Application in Efficient Synthesis of Hopene. Front. Bioeng. Biotechnol. 2020, 8, 426. [Google Scholar] [CrossRef] [PubMed]
- Chater, K.F. Recent advances in understanding Streptomyces. F1000Res 2016, 5, 2795. [Google Scholar] [CrossRef]
- Awada, B.; Chahine, D.; Derbaj, G.; Abdel Khalek, P.; Kallassy, M.; Abou Fayad, A. Antimicrobial Natural Products Derived from Microorganisms Inhabiting the MENA Region. Nat. Prod. Commun. 2023, 18, 1–27. [Google Scholar] [CrossRef]
- Liu, Z.; Zhao, Y.; Huang, C.; Luo, Y. Recent Advances in Silent Gene Cluster Activation in Streptomyces. Front. Bioeng. Biotechnol. 2021, 9, 632230. [Google Scholar] [CrossRef]



| Region | Type | From | To | Most Similar Known Cluster | Type | Similarity |
|---|---|---|---|---|---|---|
| Region 1.1 | lanthipeptide-class-i | 270,498 | 296,733 | |||
| Region 1.2 | NRPS, NRPS-like, arylpolyene | 320,092 | 418,525 | o-dialkylbenzene 1/o-dialkylbenzene 2 | Polyketide+NRP | 96% |
| Region 1.3 | T2PKS | 443,846 | 516,361 | spore pigment | Polyketide | 75% |
| Region 1.4 | NRPS | 537,820 | 594,302 | arginomycin | Other | 20% |
| Region 1.5 | terpene | 662,834 | 683,847 | albaflavenone | Terpene | 100% |
| Region 1.6 | CDPS | 1,675,437 | 1,696,183 | nocardiopsistin A/nocardiopsistin B/nocardiopsistin C | Polyketide | 9% |
| Region 1.7 | furan | 1,848,929 | 1,869,942 | methylenomycin A | Other | 9% |
| Region 1.8 | butyrolactone | 1,891,210 | 1,902,202 | triacsin C | Other | 6% |
| Region 1.9 | blactam | 2,025,385 | 2,046,998 | clavulanic acid | Other:Non-NRP beta-lactam | 16% |
| Region 1.10 | NI-siderophore | 3,395,933 | 3,425,705 | desferrioxamin B/desferrioxamine E | Other | 100% |
| Region 1.11 | melanin | 3,522,101 | 3,532,601 | istamycin | Saccharide | 4% |
| Region 1.12 | ectoine | 4,844,671 | 4,855,075 | ectoine | Other | 100% |
| Region 1.13 | NAPAA | 5,081,439 | 5,115,026 | ε-Poly-L-lysine | NRP | 100% |
| Region 1.14 | NI-siderophore | 5,302,674 | 5,332,887 | kinamycin | Polyketide | 16% |
| Region 1.15 | betalactone | 5,567,365 | 5,593,487 | JBIR-34/JBIR-35 | NRP | 8% |
| Region 1.16 | RiPP-like | 5,681,751 | 5,693,103 | |||
| Region 1.17 | butyrolactone, terpene | 5,768,985 | 5,792,627 | γ-butyrolactone | Other | 100% |
| Region 1.18 | terpene | 5,918,195 | 5,939,997 | rubiginone A2/rubiginone J/rubiginone K/rubiginone L/rubiginone M/rubiginone N/ochromycinone/rubiginone B2 | Polyketide | 20% |
| Region 1.19 | NI-siderophore | 6,035,520 | 6,066,813 | paulomycin | Other | 13% |
| Region 1.20 | lassopeptide | 6,088,809 | 6,111,354 | citrulassin D | RiPP | 100% |
| Region 2.1 | RiPP-like | 116,451 | 128,376 | |||
| Region 2.2 | terpene | 187,177 | 208,133 | |||
| Region 2.3 | NRPS, T1PKS | 339,857 | 390,604 | Sch-47554/Sch-47555 | Polyketide | 3% |
| Region 2.4 | NI-siderophore | 424,258 | 456,845 | peucechelin | NRP | 30% |
| Region 2.5 | lanthipeptide-class-iii, RiPP-like | 467,702 | 494,891 | informatipeptin | RiPP:Lanthipeptide | 100% |
| Region 2.6 | terpene | 527,615 | 548,646 | cyphomycin | Polyketide | 2% |
| Region 2.7 | T1PKS, NRPS-like, NRPS | 626,113 | 713,163 | antimycin | NRP+Polyketide | 93% |
| Region 2.8 | NRP-metallophore, NRPS | 840,099 | 907,141 | cahuitamycin A/cahuitamycin B/cahuitamycin C | NRP | 62% |
| Region 2.9 | NRPS-like | 997,843 | 1,040,518 | deoxyhangtaimycin | Polyketide+NRP | 11% |
| Region 2.10 | T1PKS | 1,080,440 | 1,125,278 | herbimycin A | Polyketide | 30% |
| Region 2.11 | linaridin, lanthipeptide-class-iv | 1,466,740 | 1,516,503 | Sch-47554/Sch-47555 | Polyketide | 3% |
| Region 3.1 | terpene | 12,267 | 33,244 | rimomycin A/rimomycin B/rimomycin C | NRP | 6% |
| Region 3.2 | terpene, T1PKS | 130,006 | 180,149 | malacidin A/malacidin B | NRP:Lipopeptide:Ca+-dependent lipopeptide | 5% |
| Region 3.3 | T1PKS, NRPS | 181,637 | 269,950 | foxicin A/foxicin B/foxicin C/foxicin | NRP+Polyketide | 14% |
| Region 3.4 | terpene | 331,138 | 352,196 | ebelactone | Polyketide | 5% |
| Region 3.5 | hydrogen-cyanide, betalactone, NRPS, T1PKS | 739,801 | 807,911 | rakicidin A/rakicidin B | NRP:Cyclic depsipeptide+Polyketide:Modular type I polyketide | 20% |
| Region 3.6 | T1PKS | 1,001,424 | 1,032,706 | niphimycins C-E | Polyketide | 29% |
| Region 4.1 | PKS-like | 40,613 | 81,626 | sanglifehrin A | NRP+Polyketide | 6% |
| Region 4.2 | terpene, PKS-like | 118,451 | 167,319 | sanglifehrin A | NRP+Polyketide | 4% |
| Region 4.3 | terpene | 182,354 | 209,122 | hopene | Terpene | 92% |
| Region 4.4 | NRPS, T1PKS | 512,984 | 576,593 | althiomycin | NRP+Polyketide:Modular type I polyketide | 100% |
| Region 4.5 | T1PKS | 760,445 | 800,778 | niphimycins C-E | Polyketide | 35% |
| Region 5.1 | T1PKS | 1 | 20,202 | griseochelin | Polyketide | 46% |
| Region 6.1 | T1PKS | 1 | 14,357 | ECO-0501 | Polyketide | 14% |
| Name of Producing Strain | Media | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria | V | Veg | A | B | C | INA | RA3 | GPMY | V6 | AF/MS | GYM | M8 | COM | NL2 |
| S. aureus ATCC 29213 | - | - | - | - | 250 | - | - | 250 | - | - | - | - | - | - |
| S. aureus Newman | 125 | 125 | 125 | 125 | - | - | - | - | - | - | - | 250 | - | - |
| S. aureus N315 | - | - | - | - | 250 | - | - | - | - | - | - | - | - | - |
| E. feacalis ATCC 19433 | - | - | - | - | - | - | - | - | 125 | - | - | - | - | - |
| K. pneumonaie DSM | - | - | - | - | 250 | - | - | - | - | - | 250 | - | - | - |
| A. baumannii DSM 30008 | 125 | 250 | 250 | 250 | 62.5 | 250 | 250 | 250 | 125 | 250 | 250 | 250 | 250 | 125 |
| K. pneumonaie ATCC 13883 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| P. aeruginosa ATCC 27853 | 250 | - | - | - | 250 | - | - | - | - | 250 | 250 | 250 | 250 | 250 |
| P. aeruginosa mexAB | 250 | - | 250 | - | - | - | - | - | 250 | 250 | 250 | 125 | 250 | 250 |
| E. coli ATCC 25922 | 62.5 | 125 | 62.5 | 250 | - | - | - | - | - | 125 | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamiyeh, R.; Hanna, A.; Abou Fayad, A. From Lebanese Soil to Antimicrobials: A Novel Streptomyces Species with Antimicrobial Potential. Fermentation 2025, 11, 406. https://doi.org/10.3390/fermentation11070406
Hamiyeh R, Hanna A, Abou Fayad A. From Lebanese Soil to Antimicrobials: A Novel Streptomyces Species with Antimicrobial Potential. Fermentation. 2025; 11(7):406. https://doi.org/10.3390/fermentation11070406
Chicago/Turabian StyleHamiyeh, Razane, Aya Hanna, and Antoine Abou Fayad. 2025. "From Lebanese Soil to Antimicrobials: A Novel Streptomyces Species with Antimicrobial Potential" Fermentation 11, no. 7: 406. https://doi.org/10.3390/fermentation11070406
APA StyleHamiyeh, R., Hanna, A., & Abou Fayad, A. (2025). From Lebanese Soil to Antimicrobials: A Novel Streptomyces Species with Antimicrobial Potential. Fermentation, 11(7), 406. https://doi.org/10.3390/fermentation11070406





