Improvement of L-Tryptophan Production in Escherichia coli Using Biosensor-Based, High-Throughput Screening and Metabolic Engineering
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Plasmids
2.2. Plasmid and Homologous Fragment Construction
2.3. Media and Growth Conditions
2.4. ARTP Mutagenesis
2.5. Flow Cytometric Cell Sorting (FACS)
2.6. High-Throughput Pre-Treatment and Analysis Using the UTPA Platform
2.7. Genome-Wide Mutation Analysis
2.8. Analysis of OD600 and L-Trp Accumulation
3. Results and Discussion
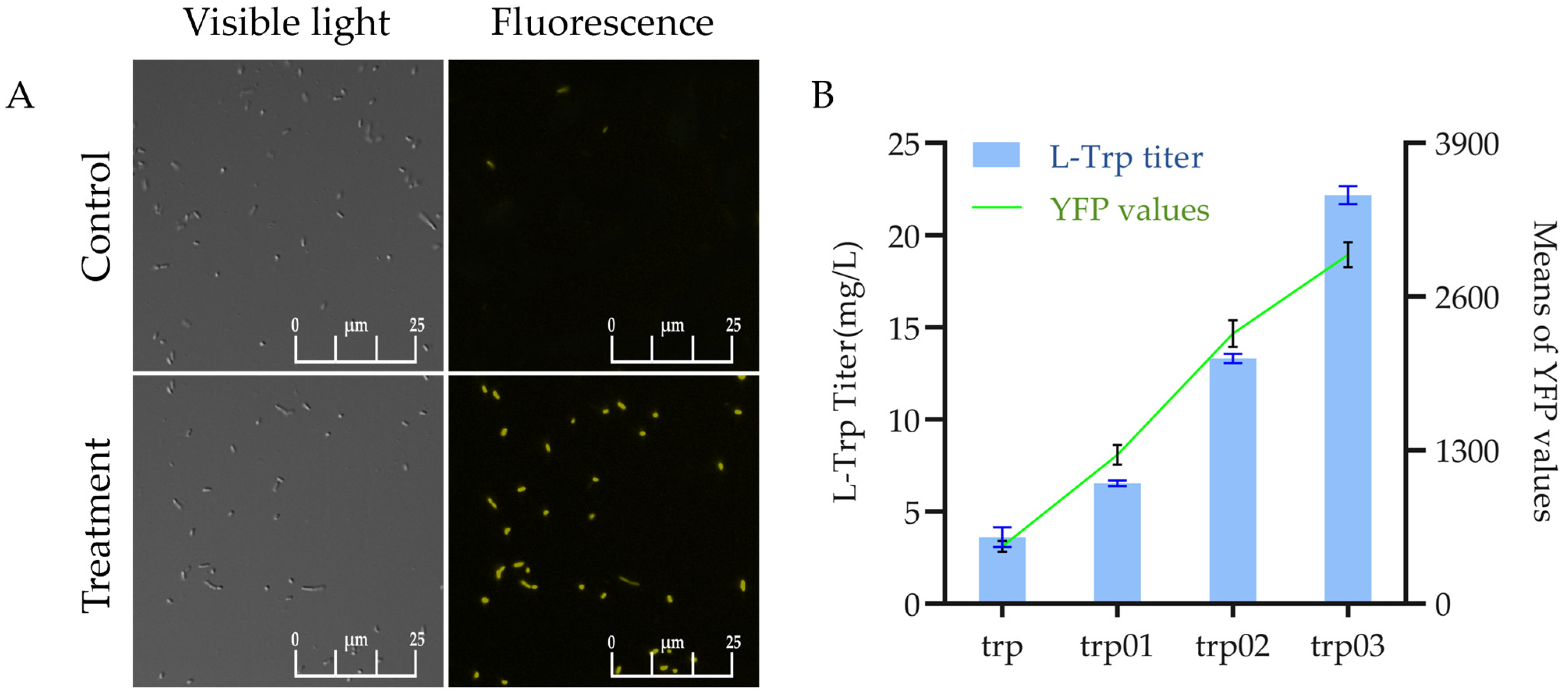
3.1. Construction and Validation of the L-Trp Biosensor
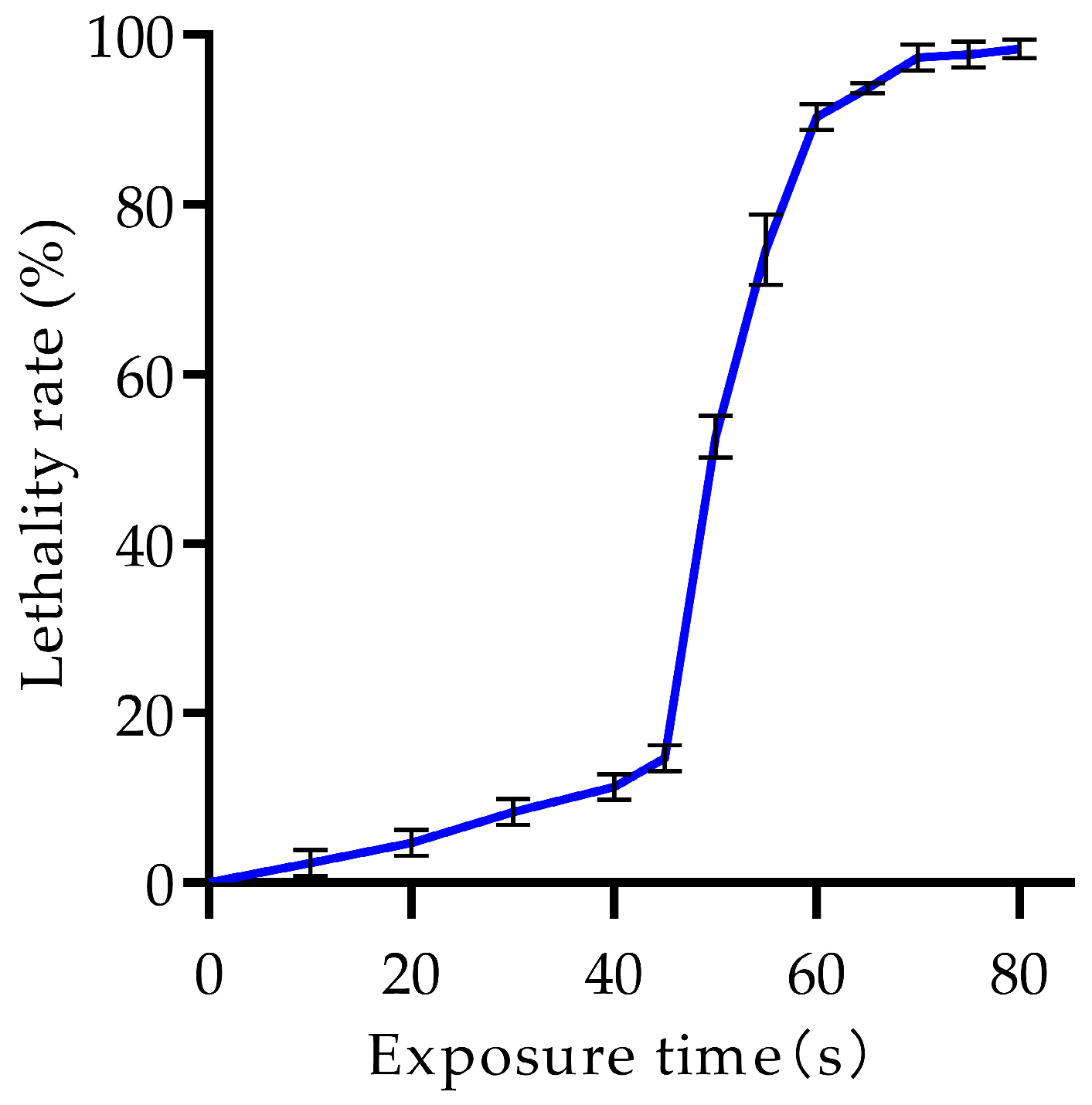
3.2. Determination and Optimization of ARTP Mutagenesis Conditions
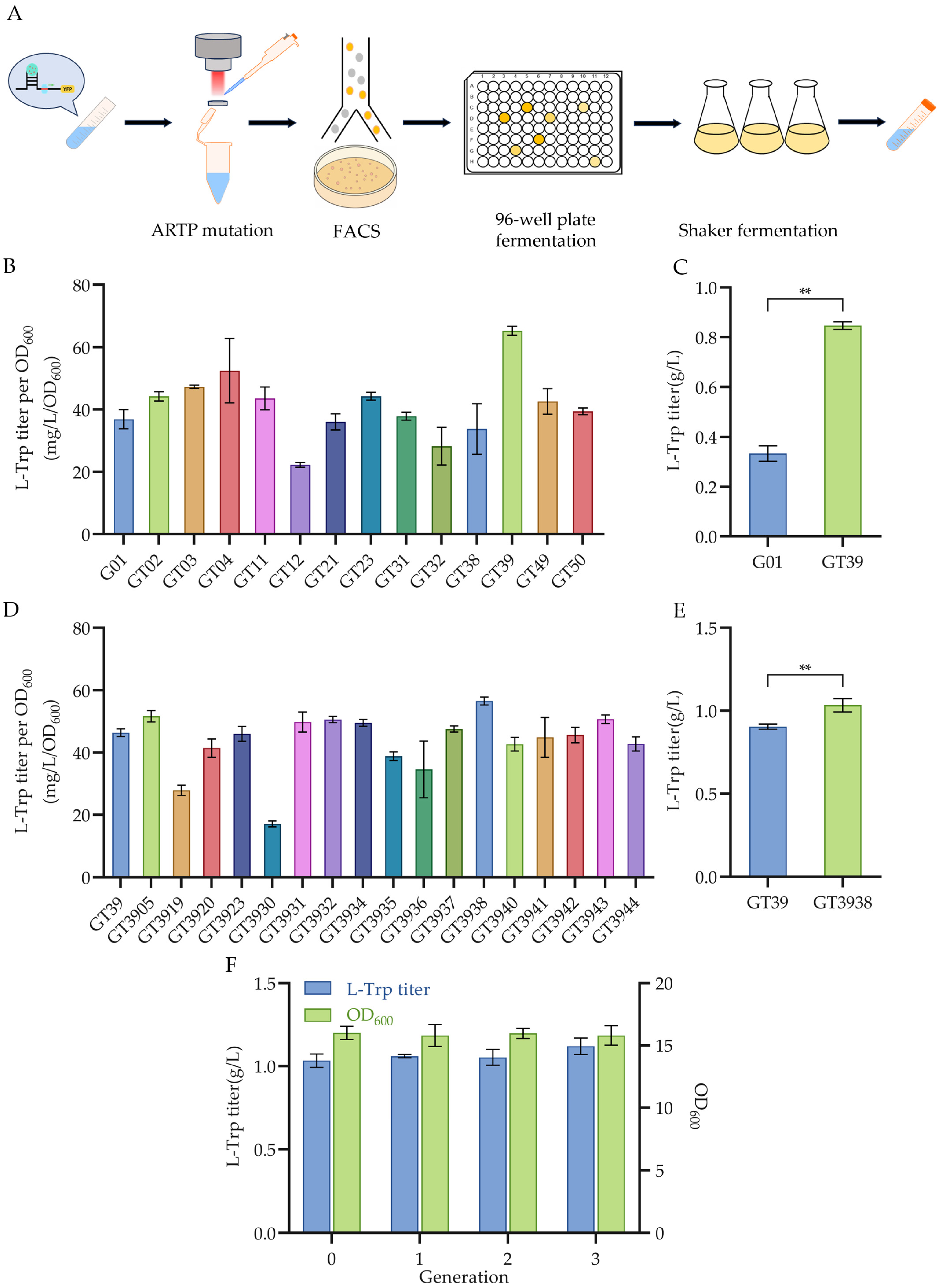
3.3. High-Throughput Screening of Mutant Strains with Enhanced L-Trp Production
3.4. Sequencing Analysis and Validation of the L-Trp Mutant GT3938
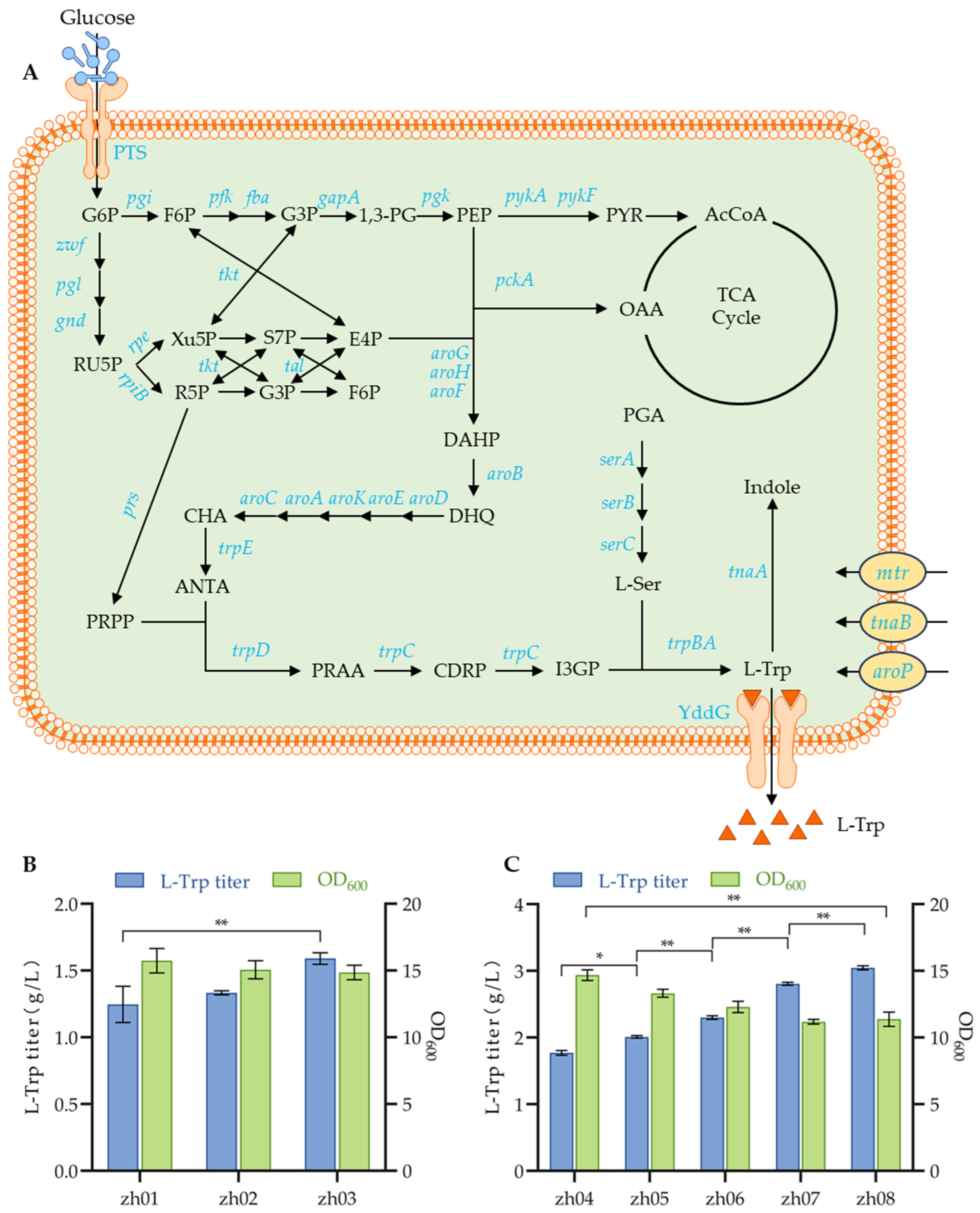
3.5. Enhancing L-Trp Efflux to Improve Production
3.6. Improvement of L-Trp Production by Increasing Introcellular PRPP and L-Ser Supplies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Richard, D.M.; Dawes, M.A.; Mathias, C.W.; Acheson, A.; Hill-Kapturczak, N.; Dougherty, D.M. L-Tryptophan: Basic Metabolic Functions, Behavioral Research and Therapeutic Indications. Int. J. Tryptophan Res. 2009, 2, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Li, G.; Zheng, Q.; Gu, X.; Shi, Q.; Su, Y.; Chu, Q.; Yuan, X.; Bao, Z.; Lu, J.; et al. Tryptophan metabolism in health and disease. Cell Metab. 2023, 35, 1304–1326. [Google Scholar] [CrossRef]
- Chan, E.-C.; Tsai, H.-L.; Chen, S.-L.; Mou, D.-G. Amplification of the tryptophan operon gene in Escherichia coli chromosome to increase l-tryptophan biosynthesis. Appl. Microbiol. Biotechnol. 1993, 40, 301–305. [Google Scholar] [CrossRef]
- Liu, X.; Niu, H.; Huang, Z.; Li, Q.; Gu, P. Construction of a switchable synthetic Escherichia coli for aromatic amino acids by a tunable switch. J. Ind. Microbiol. Biotechnol. 2020, 47, 233–242. [Google Scholar] [CrossRef]
- Gu, P.; Yang, F.; Li, F.; Liang, Q.; Qi, Q. Knocking out analysis of tryptophan permeases in Escherichia coli for improving L-tryptophan production. Appl. Microbiol. Biotechnol. 2013, 97, 6677–6683. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.; Martínez, J.A.; Flores, N.; Escalante, A.; Gosset, G.; Bolivar, F. Engineering Escherichia coli to overproduce aromatic amino acids and derived compounds. Microb. Cell Fact. 2014, 13, 126. [Google Scholar] [CrossRef]
- Cruz-Casas, D.E.; Aguilar, C.N.; Ascacio-Valdés, J.A.; Rodríguez-Herrera, R.; Chávez-González, M.L.; Flores-Gallegos, A.C. Enzymatic hydrolysis and microbial fermentation: The most favorable biotechnological methods for the release of bioactive peptides. Food Chem. 2021, 3, 100047. [Google Scholar] [CrossRef]
- Begum, P.S.; Rajagopal, S.; Razak, M.A. Chapter 11—Emerging trends in microbial fermentation technologies. In Recent Developments in Applied Microbiology and Biochemistry; Viswanath, B., Ed.; Academic Press: Cambridge, MA, USA, 2021; pp. 113–119. [Google Scholar]
- Shen, G.; Liu, Y.; Ji, N.; Zhang, Y.; Wang, Q. Advances in fermentative production of L-tryptophan: A review. Sheng Wu Gong Cheng Xue Bao 2024, 40, 621–643. [Google Scholar] [CrossRef]
- Yu, J.; Wang, J.; Li, J.; Guo, C.; Huang, Y.; Xu, Q. Regulation of key enzymes in tryptophan biosynthesis pathway in Escherichia coli. Sheng Wu Gong Cheng Xue Bao 2008, 24, 844–850. [Google Scholar]
- Li, Z.; Wang, X.; Hu, G.; Li, X.; Song, W.; Wei, W.; Liu, L.; Gao, C. Engineering metabolic flux for the microbial synthesis of aromatic compounds. Metab. Eng. 2025, 88, 94–112. [Google Scholar] [CrossRef]
- Xu, X.; Lv, X.; Bi, X.; Chen, J.; Liu, L. Genetic circuits for metabolic flux optimization. Trends Microbiol. 2024, 32, 791–806. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.; Gao, S.; Wu, J.; Chen, S.; Zhang, K. Metabolic engineering of Escherichia coli to enhance L-tryptophan biosynthesis. Syst. Microbiol. Biomanuf. 2025, 5, 622–634. [Google Scholar] [CrossRef]
- Cheng, L.-K.; Wang, J.; Xu, Q.-Y.; Xie, X.-X.; Zhang, Y.-J.; Zhao, C.-G.; Chen, N. Effect of feeding strategy on l-tryptophan production by recombinant Escherichia coli. Ann. Microbiol. 2012, 62, 1625–1634. [Google Scholar] [CrossRef]
- Della Corte, D.; van Beek, H.L.; Syberg, F.; Schallmey, M.; Tobola, F.; Cormann, K.U.; Schlicker, C.; Baumann, P.T.; Krumbach, K.; Sokolowsky, S.; et al. Engineering and application of a biosensor with focused ligand specificity. Nat. Commun. 2020, 11, 4851. [Google Scholar] [CrossRef]
- Seok, J.Y.; Yang, J.; Choi, S.J.; Lim, H.G.; Choi, U.J.; Kim, K.-J.; Park, S.; Yoo, T.H.; Jung, G.Y. Directed evolution of the 3-hydroxypropionic acid production pathway by engineering aldehyde dehydrogenase using a synthetic selection device. Metab. Eng. 2018, 47, 113–120. [Google Scholar] [CrossRef]
- Jiang, S.; Wang, R.; Wang, D.; Zhao, C.; Ma, Q.; Wu, H.; Xie, X. Metabolic reprogramming and biosensor-assisted mutagenesis screening for high-level production of L-arginine in Escherichia coli. Metab. Eng. 2023, 76, 146–157. [Google Scholar] [CrossRef]
- Zhen, Z.; Xiang, L.; Li, S.; Li, H.; Lei, Y.; Chen, W.; Jin, J.-M.; Liang, C.; Tang, S.-Y. Designing a whole-cell biosensor applicable for S-adenosyl-l-methionine-dependent methyltransferases. Biosens. Bioelectron. 2025, 268, 116904. [Google Scholar] [CrossRef]
- Gong, X.; Zhang, R.; Wang, J.; Yan, Y. Engineering of a TrpR-Based Biosensor for Altered Dynamic Range and Ligand Preference. ACS Synth. Biol. 2022, 11, 2175–2183. [Google Scholar] [CrossRef]
- Mukdasai, S.; Poosittisak, S.; Ngeontae, W.; Srijaranai, S. A highly sensitive electrochemical determination of l-tryptophan in the presence of ascorbic acid and uric acid using in situ addition of tetrabutylammonium bromide on the ß-cyclodextrin incorporated multi-walled carbon nanotubes modified electrode. Sens. Actuators B Chem. 2018, 272, 518–525. [Google Scholar] [CrossRef]
- Liu, Y.; Yuan, H.; Ding, D.; Dong, H.; Wang, Q.; Zhang, D. Establishment of a Biosensor-based High-Throughput Screening Platform for Tryptophan Overproduction. ACS Synth. Biol. 2021, 10, 1373–1383. [Google Scholar] [CrossRef]
- Weidemüller, P.; Kholmatov, M.; Petsalaki, E.; Zaugg, J.B. Transcription factors: Bridge between cell signaling and gene regulation. Proteomics 2021, 21, e2000034. [Google Scholar] [CrossRef]
- Garst, A.D.; Edwards, A.L.; Batey, R.T. Riboswitches: Structures and mechanisms. Cold Spring Harb. Perspect. Biol. 2011, 3, a003533. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, B.; Duan, C.; Sun, B.; Yang, J.; Yang, S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015, 81, 2506–2514. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Tang, J.; Liu, Y.; Zhu, X.; Zhang, T.; Zhang, X. Combinatorial modulation of galP and glk gene expression for improved alternative glucose utilization. Appl. Microbiol. Biotechnol. 2012, 93, 2455–2462. [Google Scholar] [CrossRef]
- Jiang, G.; Yang, Z.; Wang, Y.; Yao, M.; Chen, Y.; Xiao, W.; Yuan, Y. Enhanced astaxanthin production in yeast via combined mutagenesis and evolution. Biochem. Eng. J. 2020, 156, 107519. [Google Scholar] [CrossRef]
- Liu, J.; Lu, Y. Fast Colorimetric Sensing of Adenosine and Cocaine Based on a General Sensor Design Involving Aptamers and Nanoparticles. Angew. Chem. Int. Ed. 2006, 45, 90–94. [Google Scholar] [CrossRef]
- Carpenter, A.C.; Paulsen, I.T.; Williams, T.C. Blueprints for Biosensors: Design, Limitations, and Applications. Genes 2018, 9, 375. [Google Scholar] [CrossRef] [PubMed]
- Qian, S.; Li, Y.; Cirino, P.C. Biosensor-guided improvements in salicylate production by recombinant Escherichia coli. Microb. Cell Factories 2019, 18, 18. [Google Scholar] [CrossRef]
- Han, Y.; Zhou, M.; Han, J.; Xiang, H. Research progress and application of L-tryptophan biosensors in synthetic biology. Chin. J. Biotechnol. 2024, 1–18. [Google Scholar] [CrossRef]
- Pu, W.; Chen, J.; Liu, P.; Shen, J.; Cai, N.; Liu, B.; Lei, Y.; Wang, L.; Ni, X.; Zhang, J.; et al. Directed evolution of linker helix as an efficient strategy for engineering LysR-type transcriptional regulators as whole-cell biosensors. Biosens. Bioelectron. 2023, 222, 115004. [Google Scholar] [CrossRef]
- Camacho, D.M.; Collins, K.M.; Powers, R.K.; Costello, J.C.; Collins, J.J. Next-Generation Machine Learning for Biological Networks. Cell 2018, 173, 1581–1592. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Chung, M.T.; Yao, Y.; Nidetz, R.; Lee, L.M.; Liu, A.P.; Feng, Y.; Kurabayashi, K.; Yang, G.-Y. Efficient molecular evolution to generate enantioselective enzymes using a dual-channel microfluidic droplet screening platform. Nat. Commun. 2018, 9, 1030. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, B.B.; Xu, J.Z.; Zhang, W.G. Engineering of Shikimate Pathway and Terminal Branch for Efficient Production of L-Tryptophan in Escherichia coli. Int. J. Mol. Sci. 2023, 24, 11866. [Google Scholar] [CrossRef]
- Minliang, C.; Chengwei, M.; Lin, C.; Zeng, A.-P. Integrated laboratory evolution and rational engineering of GalP/Glk-dependent Escherichia coli for higher yield and productivity of L-tryptophan biosynthesis. Metab. Eng. Commun. 2021, 12, e00167. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Y.; Ding, D.; Cong, L.; Zhang, D. Rational design and analysis of an Escherichia coli strain for high-efficiency tryptophan production. J. Ind. Microbiol. Biotechnol. 2018, 45, 357–367. [Google Scholar] [CrossRef]
- Pflüger-Grau, K.; Chavarría, M.; de Lorenzo, V. The interplay of the EIIA(Ntr) component of the nitrogen-related phosphotransferase system (PTS(Ntr)) of Pseudomonas putida with pyruvate dehydrogenase. Biochim. Biophys. Acta 2011, 1810, 995–1005. [Google Scholar] [CrossRef]
- Gravina, F.; Degaut, F.L.; Gerhardt, E.C.M.; Pedrosa, F.O.; Souza, E.M.; Antônio de Souza, G.; Huergo, L.F. The protein–protein interaction network of the Escherichia coli EIIANtr regulatory protein reveals a role in cell motility and metabolic control. Res. Microbiol. 2021, 172, 103882. [Google Scholar] [CrossRef]
- Alper, H.; Stephanopoulos, G. Global transcription machinery engineering: A new approach for improving cellular phenotype. Metab. Eng. 2007, 9, 258–267. [Google Scholar] [CrossRef]
- Lüttmann, D.; Göpel, Y.; Görke, B. The phosphotransferase protein EIIA(Ntr) modulates the phosphate starvation response through interaction with histidine kinase PhoR in Escherichia coli. Mol. Microbiol. 2012, 86, 96–110. [Google Scholar] [CrossRef]
- Commichau, F.M.; Forchhammer, K.; Stülke, J. Regulatory links between carbon and nitrogen metabolism. Curr. Opin. Microbiol. 2006, 9, 167–172. [Google Scholar] [CrossRef]
- Wu, Z.; Wang, Y.; Li, L.; Zhen, S.; Du, H.; Wang, Z.; Xiao, S.; Wu, J.; Zhu, L.; Shen, J.; et al. New insights into the antimicrobial action and protective therapeutic effect of tirapazamine towards Escherichia coli-infected mice. Int. J. Antimicrob. Agents 2023, 62, 106923. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Z.; Wu, F.; Zhang, Z.; Zhang, X.; Hu, Y.; Wang, Q.; Zhang, S. Improvement of L-Tryptophan Production in Escherichia coli Using Biosensor-Based, High-Throughput Screening and Metabolic Engineering. Fermentation 2025, 11, 267. https://doi.org/10.3390/fermentation11050267
Gao Z, Wu F, Zhang Z, Zhang X, Hu Y, Wang Q, Zhang S. Improvement of L-Tryptophan Production in Escherichia coli Using Biosensor-Based, High-Throughput Screening and Metabolic Engineering. Fermentation. 2025; 11(5):267. https://doi.org/10.3390/fermentation11050267
Chicago/Turabian StyleGao, Zhenghao, Fengli Wu, Zhidan Zhang, Xu Zhang, Yuansen Hu, Qinhong Wang, and Shuaibing Zhang. 2025. "Improvement of L-Tryptophan Production in Escherichia coli Using Biosensor-Based, High-Throughput Screening and Metabolic Engineering" Fermentation 11, no. 5: 267. https://doi.org/10.3390/fermentation11050267
APA StyleGao, Z., Wu, F., Zhang, Z., Zhang, X., Hu, Y., Wang, Q., & Zhang, S. (2025). Improvement of L-Tryptophan Production in Escherichia coli Using Biosensor-Based, High-Throughput Screening and Metabolic Engineering. Fermentation, 11(5), 267. https://doi.org/10.3390/fermentation11050267




