Transcriptome Analysis of Sclerotium rolfsii: Unraveling Impact of Glycolytic Pathway on Substrate Utilization and Microbial Polysaccharide Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Culture Medium and Culture Conditions
2.3. Extraction of Extracellular Polysaccharides
2.4. Parameter Detection in Fermentation Process
- (1)
- Preparation of the Standard Curve
- (2)
- Sample Analysis
- (3)
- Blank Control
2.5. Transcriptome and Metabolome Analysis
2.6. RNA Extraction and cDNA Synthesis
2.7. RT-PCR Validation
2.8. Precursor Addition Validation Experiment
2.9. Mycelium Morphology
2.10. Statistical Analysis
3. Results
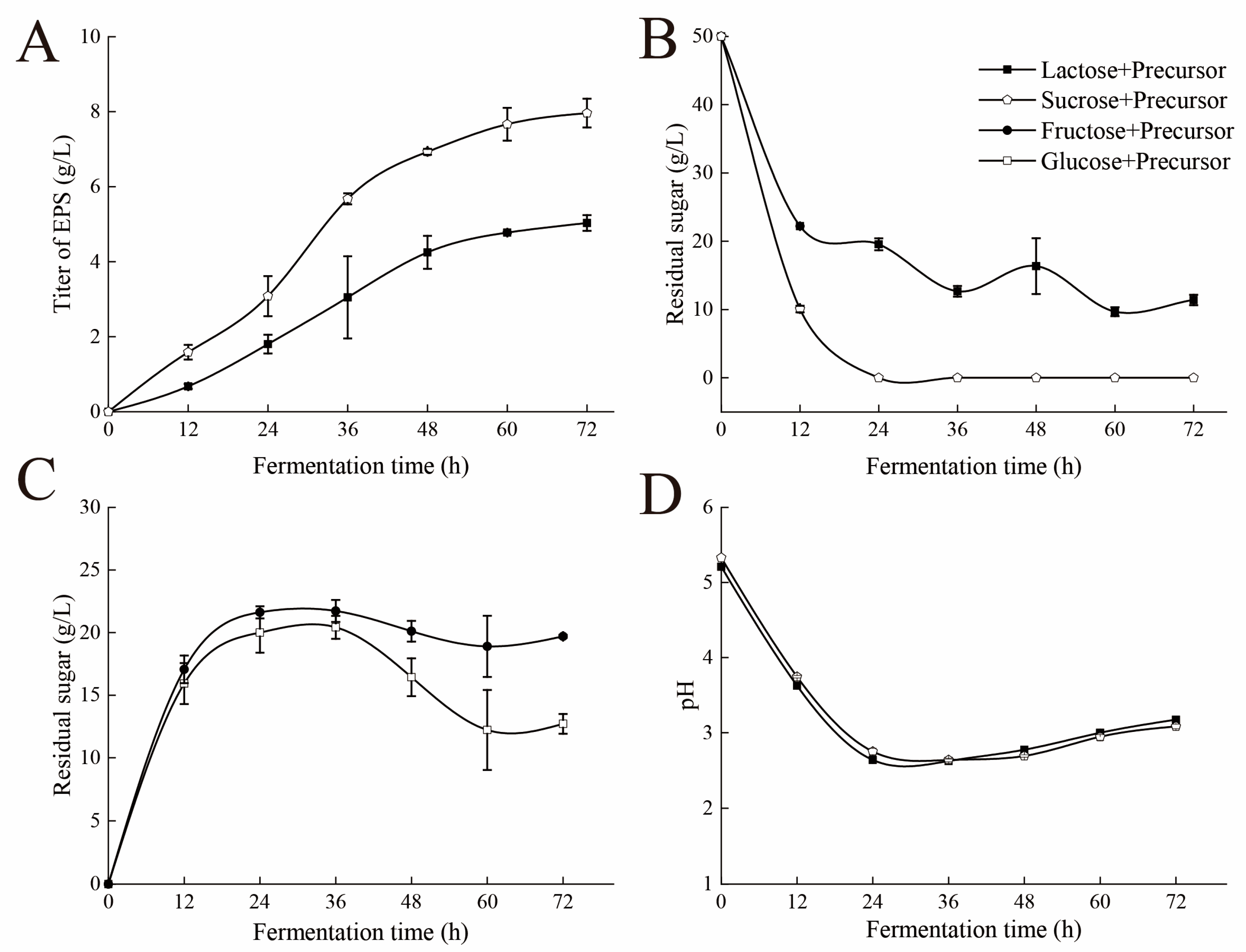
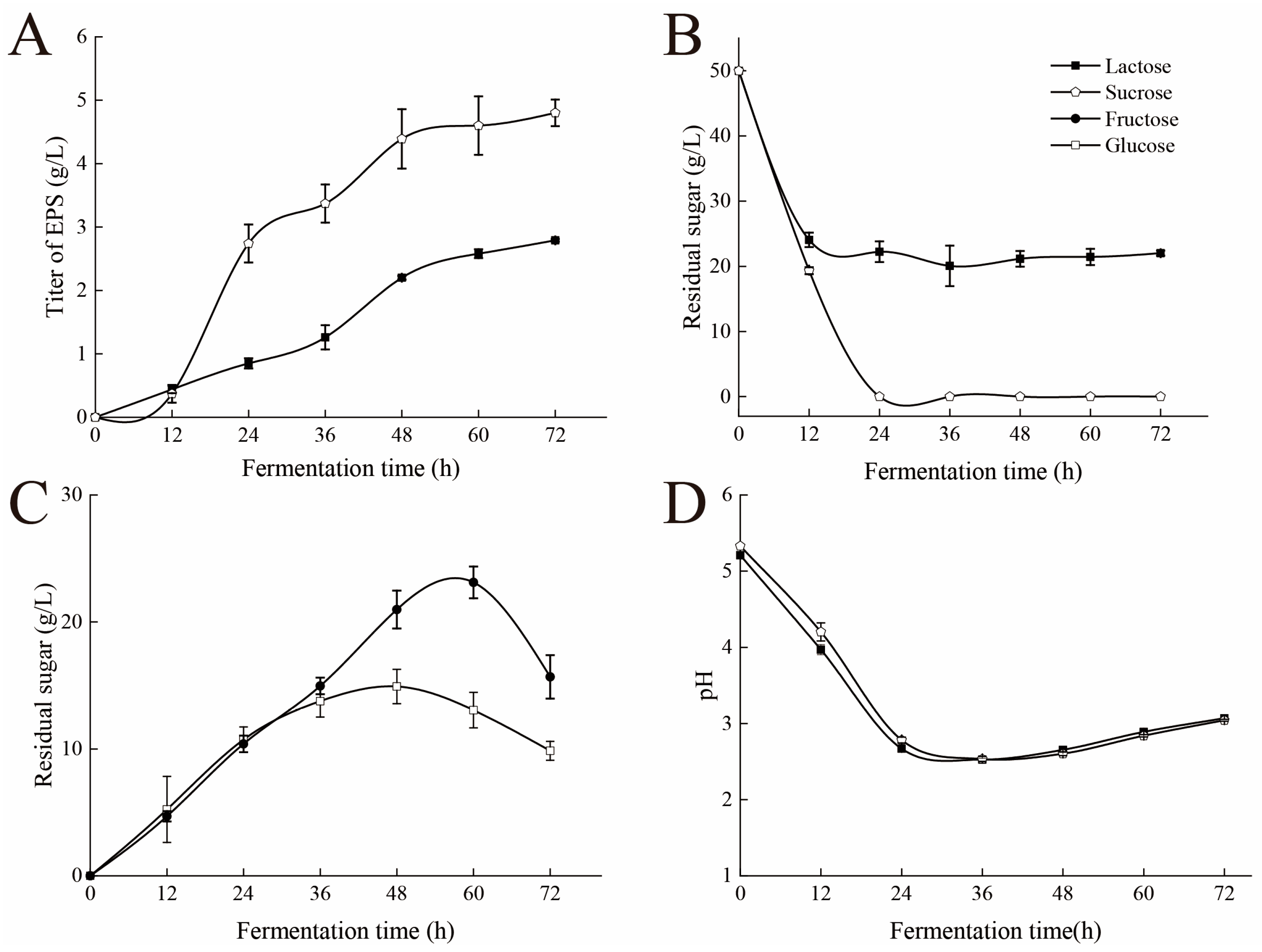
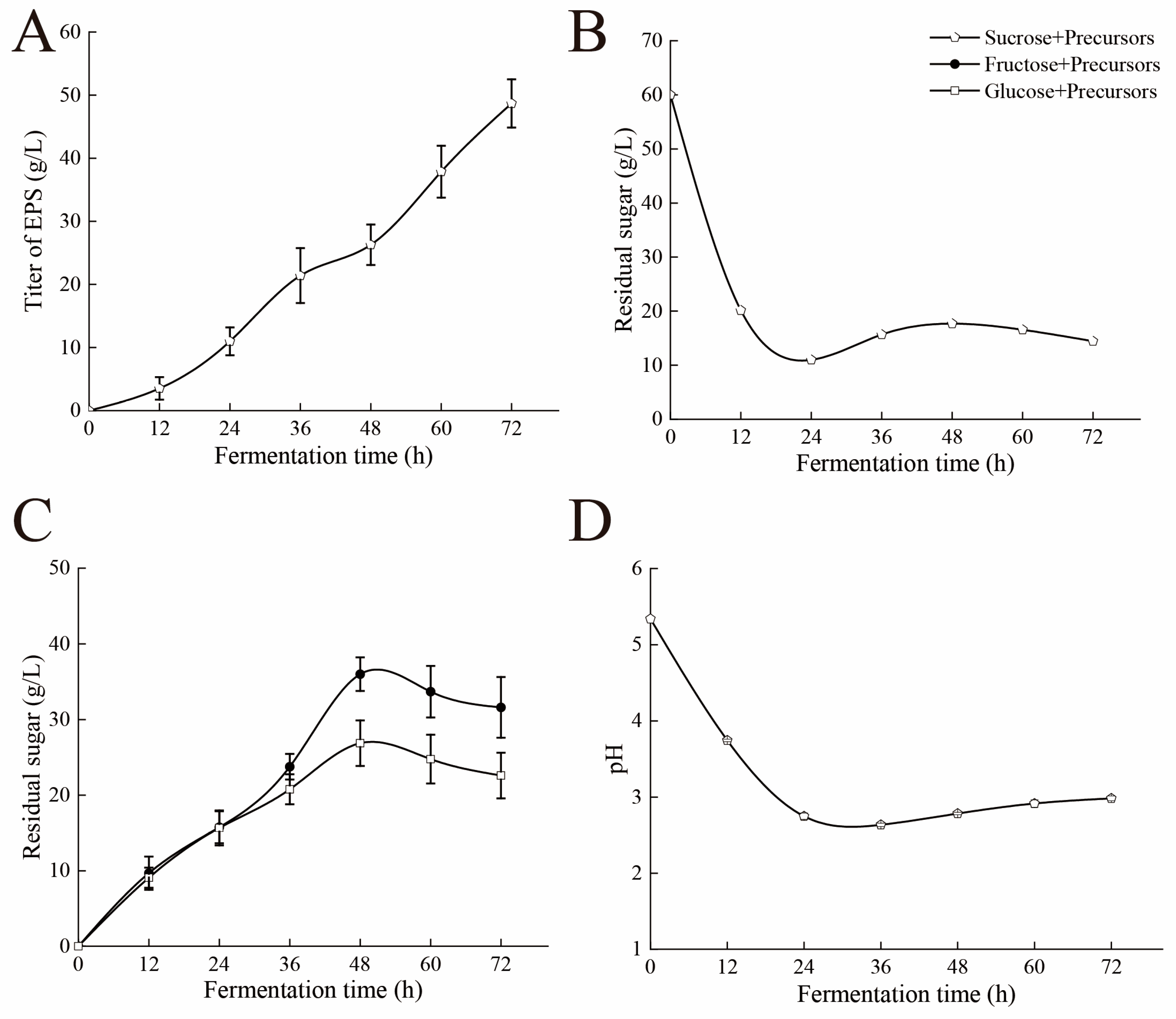
3.1. Dynamic Analysis of the Fermentation Process
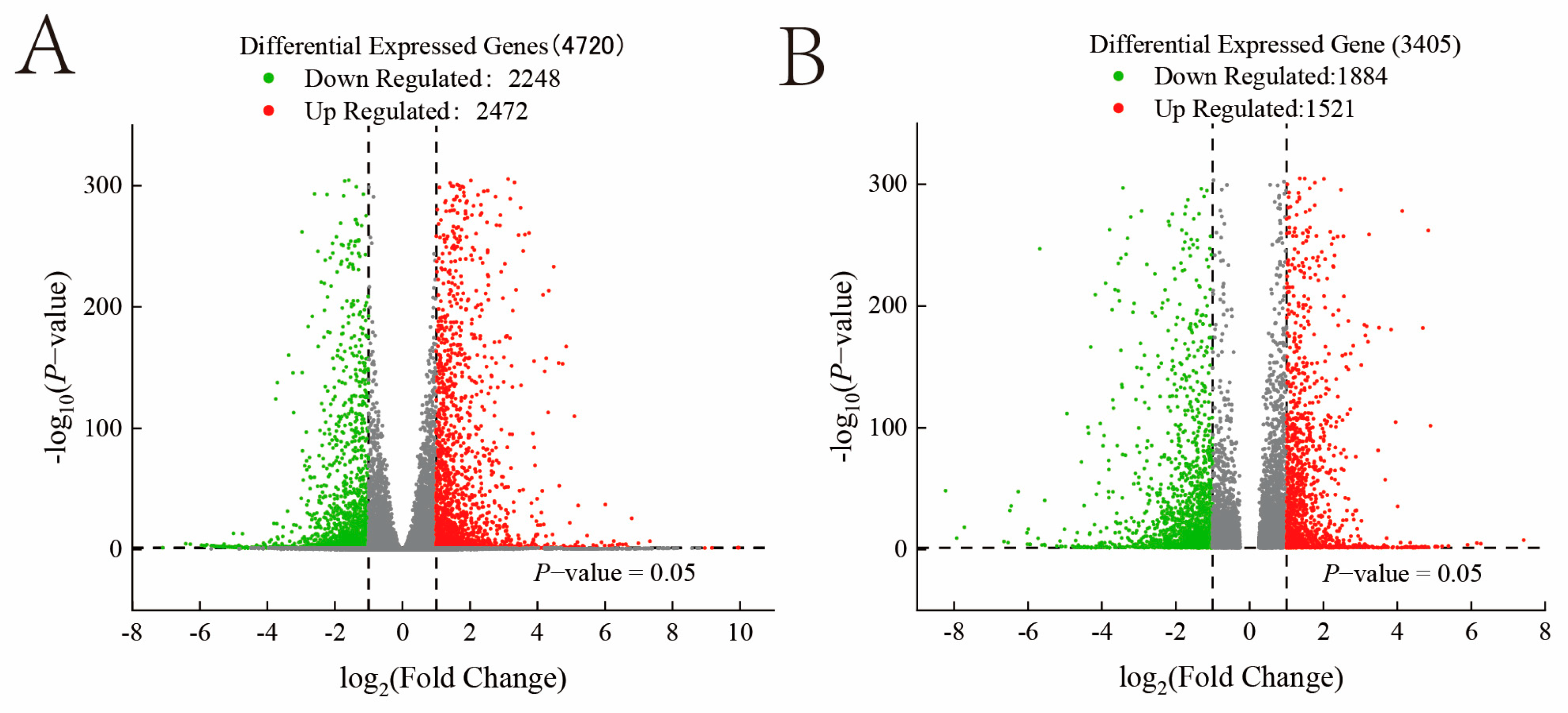
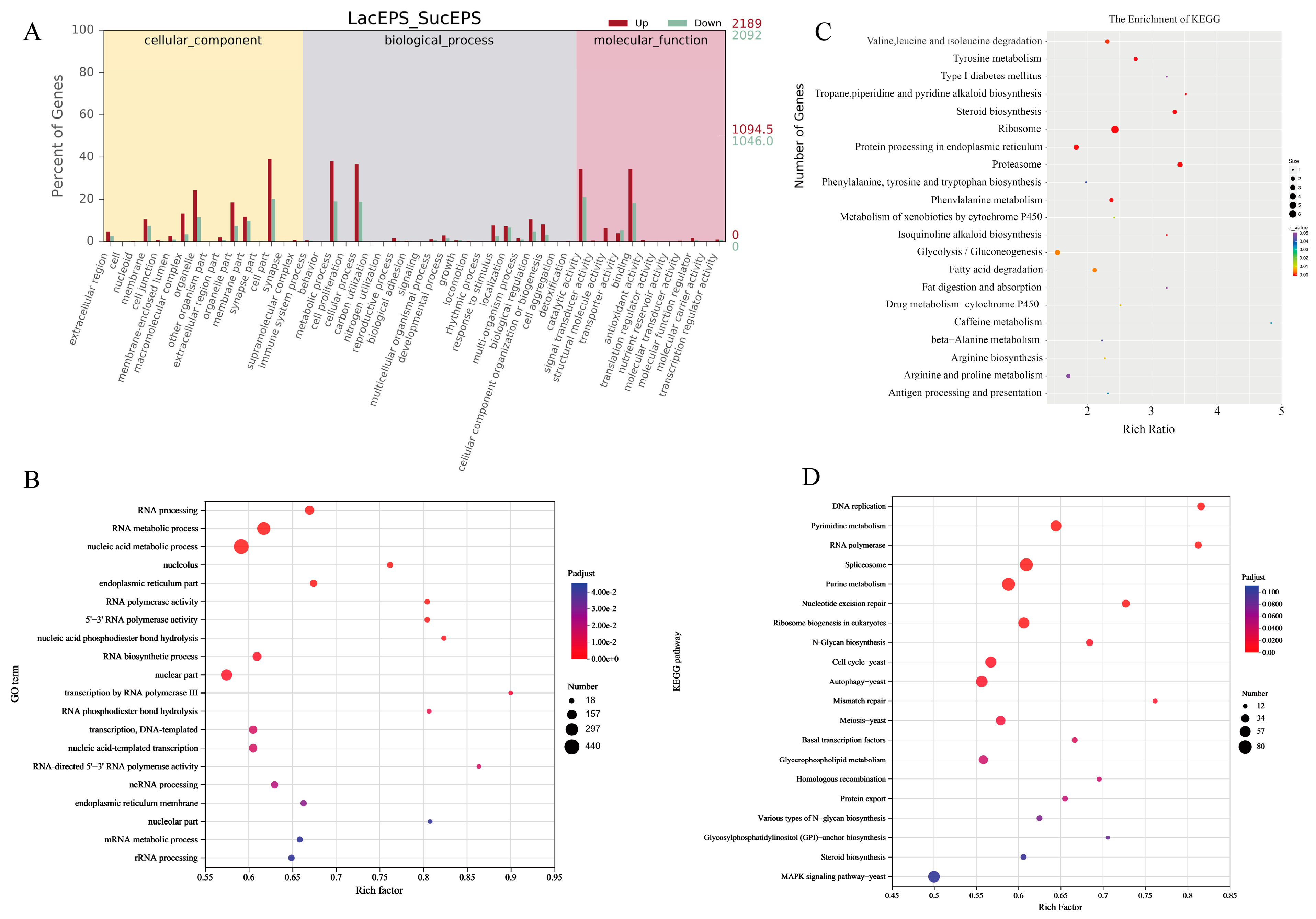
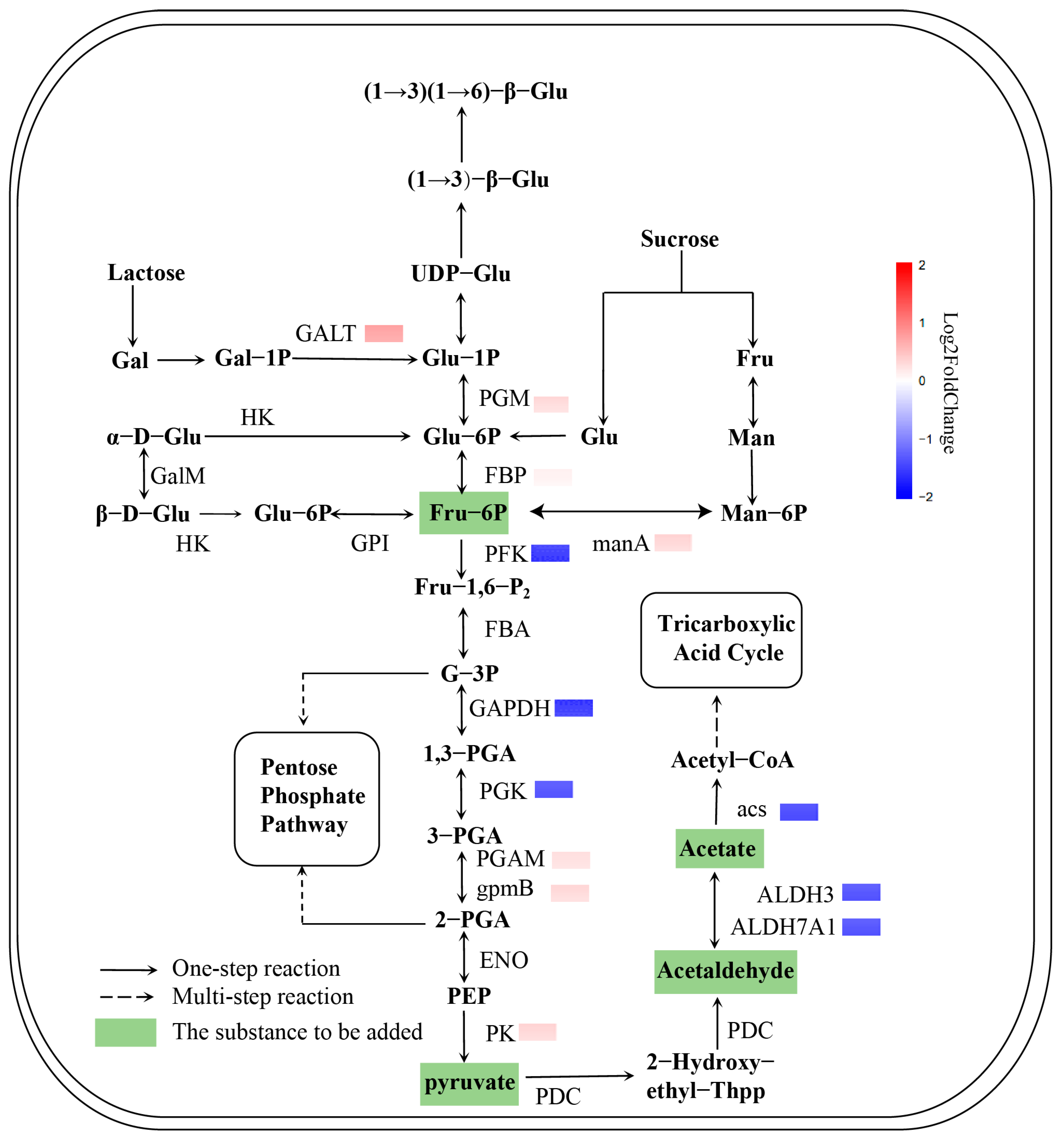
3.2. Differences in Gene Expression Levels Under Different Carbon Sources
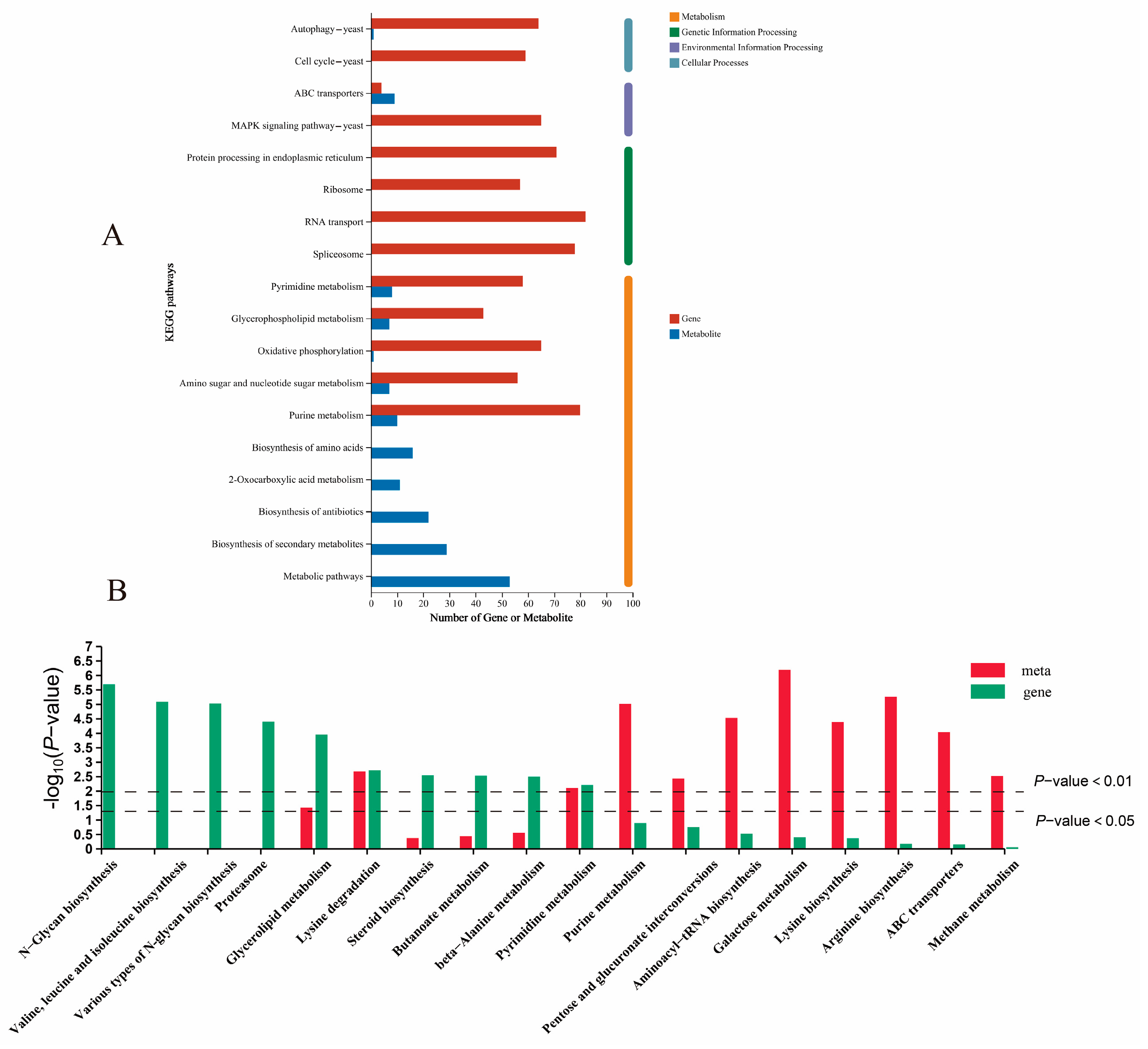
3.3. Functional Enrichment Analysis
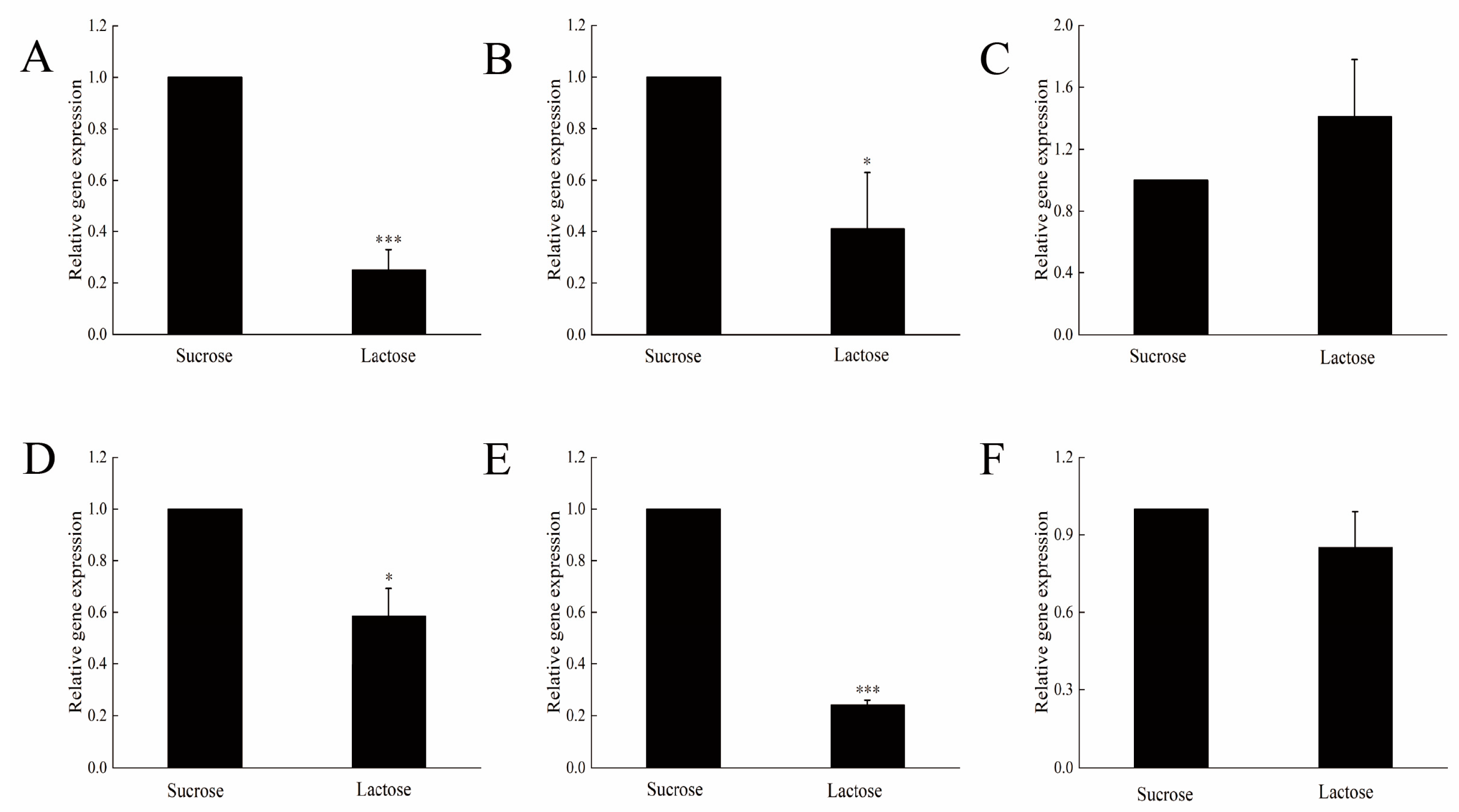
3.4. RT-PCR Analysis
3.5. Single-Factor Experiment
3.6. Response Surface Experiment
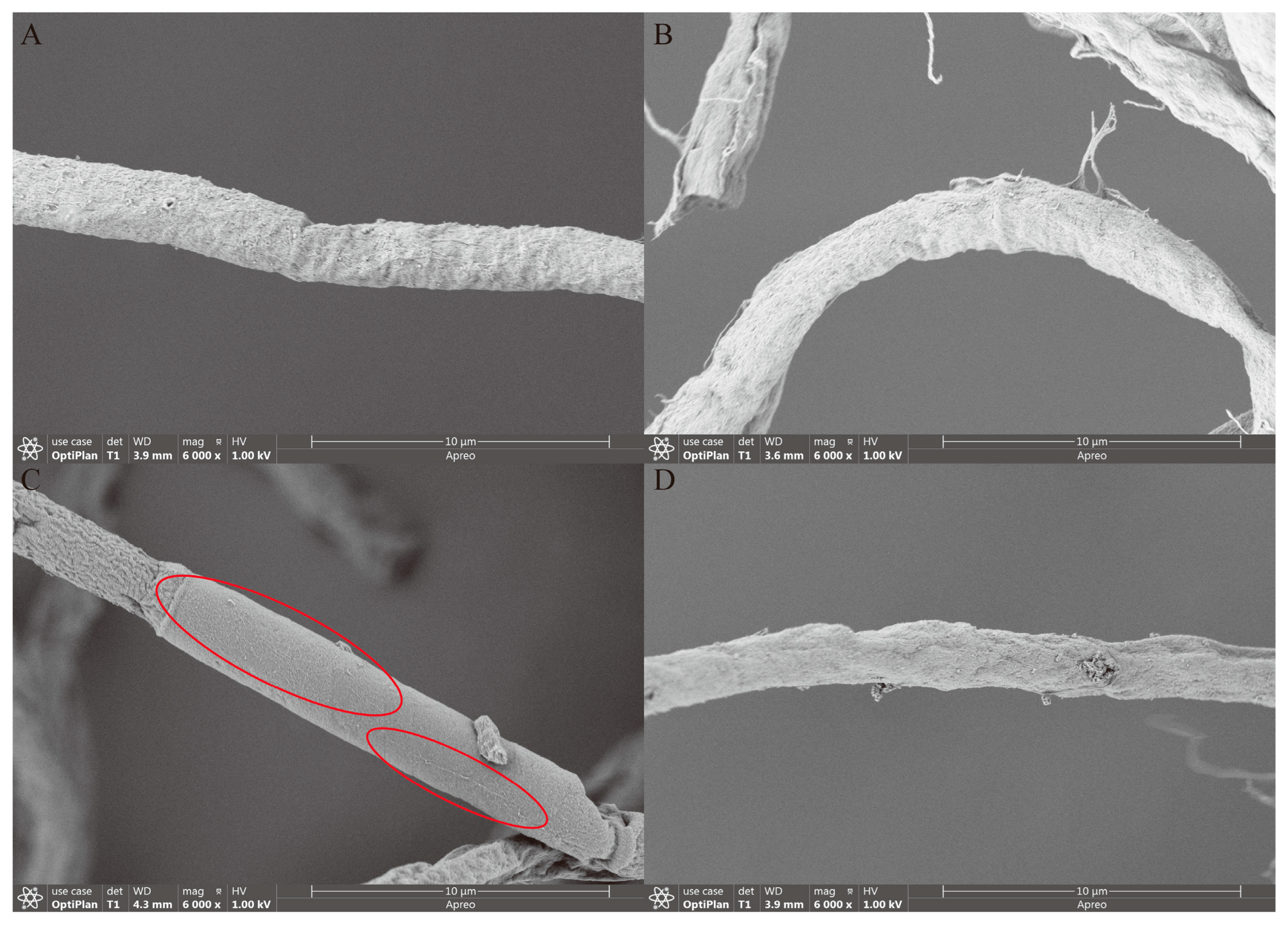
3.7. Growth Characteristics and Morphological Changes
3.8. Dynamic Analysis of Fermentation Process After Adding Precursors
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Number | A Acetic Acid (g/L) | B Aldehyde (g/L) | C Pyruvic Acid (g/L) | D Fructose 6-Phosphate (g/L) | Y Polysaccharide Titer (g/L) |
|---|---|---|---|---|---|
| 1 | 0.01 | 1.01 | 0.51 | 0.51 | 4.39 ± 0.12 |
| 2 | 0.51 | 0.51 | 1.01 | 0.01 | 3.19 ± 0.04 |
| 3 | 0.01 | 0.01 | 0.51 | 0.51 | 3.68 ± 0.07 |
| 4 | 0.51 | 0.01 | 1.01 | 0.51 | 3.82 ± 0.11 |
| 5 | 0.51 | 0.51 | 0.51 | 0.51 | 4.28 ± 0.17 |
| 6 | 0.51 | 0.51 | 0.51 | 0.51 | 4.31 ± 0.09 |
| 7 | 0.51 | 1.01 | 0.51 | 0.01 | 3.16 ± 0.08 |
| 8 | 0.51 | 1.01 | 0.51 | 1.01 | 4.12 ± 0.14 |
| 9 | 0.51 | 0.51 | 1.01 | 1.01 | 4.45 ± 0.12 |
| 10 | 0.51 | 0.51 | 0.01 | 0.01 | 2.58 ± 0.05 |
| 11 | 0.51 | 0.01 | 0.51 | 1.01 | 4.35 ± 0.14 |
| 12 | 0.51 | 0.51 | 0.51 | 0.51 | 4.42 ± 0.10 |
| 13 | 1.01 | 0.51 | 0.51 | 1.01 | 3.31 ± 0.07 |
| 14 | 0.01 | 0.51 | 0.01 | 0.51 | 3.84 ± 0.07 |
| 15 | 0.51 | 0.51 | 0.51 | 0.51 | 4.19 ± 0.09 |
| 16 | 0.51 | 0.01 | 0.51 | 0.01 | 3.12 ± 0.06 |
| 17 | 0.51 | 0.51 | 0.01 | 1.01 | 4.4 ± 0.13 |
| 18 | 0.51 | 1.01 | 0.01 | 0.51 | 3.54 ± 0.10 |
| 19 | 1.01 | 0.51 | 0.01 | 0.51 | 3.11 ± 0.06 |
| 20 | 1.01 | 1.01 | 0.51 | 0.51 | 3.07 ± 0.06 |
| 21 | 0.51 | 0.51 | 0.51 | 0.51 | 4.28 ± 0.12 |
| 22 | 0.01 | 0.51 | 1.01 | 0.51 | 4.05 ± 0.13 |
| 23 | 0.01 | 0.51 | 0.51 | 1.01 | 5.01 ± 0.17 |
| 24 | 0.51 | 0.01 | 0.01 | 0.51 | 3.6 ± 0.12 |
| 25 | 0.01 | 0.51 | 0.51 | 1.01 | 3.2 ± 0.06 |
| 26 | 1.01 | 0.51 | 1.01 | 0.51 | 2.86 ± 0.05 |
| 27 | 1.01 | 0.51 | 0.51 | 0.01 | 2.64 ± 0.04 |
| Sum of | Mean | F | p-Value | Significant | ||
|---|---|---|---|---|---|---|
| Source | Squares | df | Square | Value | Prob > F | * |
| Model | 29.2151 | 9 | 3.2461 | 7.4799 | 0.0073 | * |
| A-A | 3.0454 | 1 | 3.0454 | 7.0175 | 0.0330 | |
| B-B | 2.6134 | 1 | 2.6134 | 6.0220 | 0.0439 | |
| C-C | 3.2917 | 1 | 3.2917 | 7.5850 | 0.0283 | |
| AB | 1.8957 | 1 | 1.8957 | 4.3682 | 0.0750 | |
| AC | 0.0666 | 1 | 0.0666 | 0.1535 | 0.7069 | |
| BC | 8.6500 | 1 | 8.6499 | 19.9318 | 0.0029 | |
| A2 | 1.6454 | 1 | 1.6454 | 3.7915 | 0.0926 | |
| B2 | 7.02560 | 1 | 7.02560 | 16.1888 | 0.0050 | |
| C2 | 0.3055 | 1 | 0.3055 | 0.7039 | 0.4292 | |
| Residual | 3.0378 | 7 | 0.4340 | |||
| Lack of Fit | 2.4523 | 3 | 0.8174 | 5.5841 | 0.0650 | |
| Pure Error | 0.5855 | 4 | 0.1464 | |||
| Cor Total | 32.2529 | 16 |
Appendix B
References
- Bashir, K.M.I.; Choi, J.S. Clinical and Physiological Perspectives of beta-Glucans: The Past, Present, and Future. Int. J. Mol. Sci. 2017, 18, 1906. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Mendieta, S.; Guillen, D.; Hernandez-Pando, R.; Sanchez, S.; Rodriguez-Sanoja, R. Potential of glucans as vaccine adjuvants: A review of the alpha-glucans case. Carbohydr. Polym. 2017, 165, 103–114. [Google Scholar] [CrossRef]
- Zlotko, K.; Wiater, A.; Wasko, A.; Pleszczynska, M.; Paduch, R.; Jaroszuk-Scisel, J.; Bieganowski, A. A Report on Fungal (1-->3)-alpha-d-glucans: Properties, Functions and Application. Molecules 2019, 24, 3972. [Google Scholar] [CrossRef] [PubMed]
- Castillo, N.A.; Valdez, A.L.; Farina, J.I. Microbial production of scleroglucan and downstream processing. Front. Microbiol. 2015, 6, 1106. [Google Scholar] [CrossRef]
- Wang, F.; Wang, X.; Tang, T.; Duan, Y.; Mao, T.; Guo, X.; Wang, Q.; You, J. De Novo RNA Sequencing and Transcriptome Analysis of Sclerotium rolfsii Gene Expression during Sclerotium Development. Genes 2023, 14, 2170. [Google Scholar] [CrossRef]
- Song, J.; Jia, Y.X.; Su, Y.; Zhang, X.Y.; Tu, L.N.; Nie, Z.Q.; Zheng, Y.; Wang, M. Initial Analysis on the Characteristics and Synthesis of Exopolysaccharides from Sclerotium rolfsii with Different Sugars as Carbon Sources. Polymers 2020, 12, 348. [Google Scholar] [CrossRef]
- Tan, R.Q.; Lyu, Y.B.; Zeng, W.Z.; Zhou, J.W. Enhancing scleroglucan production by Sclerotium rolfsii WSH-G01 through a pH-shift strategy based on kinetic analysis. Bioresour. Technol. 2019, 293, 122098. [Google Scholar] [CrossRef]
- Sen, I.K.; Mandal, A.K.; Chakraborty, R.; Behera, B.; Yadav, K.K.; Maiti, T.K.; Islam, S.S. Structural and immunological studies of an exopolysaccharide from Acinetobacter junii BB1A. Carbohydr. Polym. 2014, 101, 188–195. [Google Scholar] [CrossRef]
- Hernandez, L.L. ADSA Foundation Scholar Award: A role for serotonin in lactation physiology-Where do we go from here? J. Dairy. Sci. 2018, 101, 5671–5678. [Google Scholar] [CrossRef]
- Cenci-Goga, B.T.; Sechi, P.; Iulietto, M.F.; Amirjalali, S.; Barbera, S.; Karama, M.; Aly, S.S.; Grispoldi, L. Characterization and Growth under Different Storage Temperatures of Ropy Slime-Producing Leuconostoc mesenteroides Isolated from Cooked Meat Products. J. Food Prot. 2020, 83, 1043–1049. [Google Scholar] [CrossRef]
- Deepak, V.; Ramachandran, S.; Balahmar, R.M.; Pandian, S.R.; Sivasubramaniam, S.D.; Nellaiah, H.; Sundar, K. In vitro evaluation of anticancer properties of exopolysaccharides from Lactobacillus acidophilus in colon cancer cell lines. In Vitro Cell. Dev. Biol. Anim. 2016, 52, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Ali, K.; Mehmood, M.H.; Iqbal, M.A.; Masud, T.; Qazalbash, M.; Saleem, S.; Ahmed, S.; Tariq, M.R.; Safdar, W.; Nasir, M.A.; et al. Isolation and characterization of exopolysaccharide-producing strains of Lactobacillus bulgaricus from curd. Food Sci. Nutr. 2019, 7, 1207–1213. [Google Scholar] [CrossRef]
- Morona, J.K.; Morona, R.; Paton, J.C. Characterization of the locus encoding the Streptococcus pneumoniae type 19F capsular polysaccharide biosynthetic pathway. Mol. Microbiol. 1997, 23, 751–763. [Google Scholar] [CrossRef]
- De Vuyst, L.; Degeest, B. Heteropolysaccharides from lactic acid bacteria. FEMS Microbiol. Rev. 1999, 23, 153–177. [Google Scholar] [CrossRef]
- Sutherland, I.W. Microbial polysaccharides—A comparison with eukaryotic polymers. Symp. Soc. Exp. Biol. 1989, 43, 389–402. [Google Scholar] [PubMed]
- Schmid, J.; Müller-Hagen, D.; Bekel, T.; Funk, L.; Stahl, U.; Sieber, V.; Meyer, V. Transcriptome sequencing and comparative transcriptome analysis of the scleroglucan producer Sclerotium rolfsii. BMC Genomics 2010, 11, 329. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Y.; Wu, L.T.; Chen, Q.Y.; Li, S.; Zhu, Y.B.; Wu, J.N.; Chu, J.L.; Wu, S.S. Development of sugarcane resource for efficient fermentation of exopolysaccharide by using a novel strain of Kosakonia cowanii LT-1. Bioresour. Technol. 2019, 280, 247–254. [Google Scholar] [CrossRef]
- Fu, L.; Jiang, B.; Wei, J.; Liu, J.; Hu, X.; Zhang, L. Transcriptome analysis of polysaccharide-based microbial flocculant MBFA9 biosynthesis regulated by nitrogen source. Sci. Rep. 2020, 10, 2918. [Google Scholar] [CrossRef] [PubMed]
- Fariña, J.I.; Santos, V.E.; Perotti, N.I.; Casas, J.A.; Molina, O.E.; García-Ochoa, F. Influence of the nitrogen source on the production and rheological properties of scleroglucan produced. World J. Microb. Biotechnol. 1999, 15, 309–316. [Google Scholar] [CrossRef]
- Lin, S.; Liu, Z.Q.; Baker, P.J.; Yi, M.; Wu, H.; Xu, F.; Teng, Y.; Zheng, Y.G. Enhancement of cordyceps polysaccharide production via biosynthetic pathway analysis in Hirsutella sinensis. Int. J. Biol. Macromol. 2016, 92, 872–880. [Google Scholar] [CrossRef]
- Song, J.; Qiu, Y.; Zhao, R.; Hou, J.; Tu, L.; Nie, Z.; Wang, J.; Zheng, Y.; Wang, M. Transcriptomics and Metabolomics Analysis of Sclerotium rolfsii Fermented with Differential Carbon Sources. Foods 2022, 11, 3706. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2009, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Shi, C.; Liu, L.; Han, J.; Yang, Q.; Wang, Y.; Li, X.; Fu, W.; Gao, H.; Huang, H.; et al. Majorbio Cloud 2024: Update single-cell and multiomics workflows. iMeta 2024, 3, e217. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Guo, W.; Zou, L.F.; Ji, Z.Y.; Cai, L.L.; Chen, G.Y. Glucose 6-phosphate isomerase (Pgi) is required for extracellular polysaccharide biosynthesis, DSF signals production and full virulence of Xanthomonas oryzae pv. oryzicola in rice. Physiol. Mol. 2017, 100, 209–219. [Google Scholar] [CrossRef]
- West, T.P.; Strohfus, B. Short Communication: Effect of manganese on polysaccharide production and cellular pigmentation in the fungus Aureobasidium pullulans. World J. Microbiol. Biotechnol. 1997, 13, 233–235. [Google Scholar] [CrossRef]
- Judge, A.; Dodd, M.S. Metabolism. Essays Biochem. 2020, 64, 607–647. [Google Scholar] [CrossRef]
- Zhang, Z.W.; Luo, S.; Zhang, G.C.; Feng, L.Y.; Zheng, C.; Zhou, Y.H.; Du, J.B.; Yuan, M.; Chen, Y.E.; Wang, C.Q.; et al. Nitric oxide induces monosaccharide accumulation through enzyme S-nitrosylation. Plant Cell Environ. 2017, 40, 1834–1848. [Google Scholar] [CrossRef]
- Woo, J.E.; Seong, H.J.; Lee, S.Y.; Jang, Y.S. Metabolic Engineering of Escherichia coli for the Production of Hyaluronic Acid From Glucose and Galactose. Front. Bioeng. Biotechnol. 2019, 7, 351. [Google Scholar] [CrossRef]
- Kong, X.; Zhang, B.; Hua, Y.; Zhu, Y.; Li, W.; Wang, D.; Hong, J. Efficient l-lactic acid production from corncob residue using metabolically engineered thermo-tolerant yeast. Bioresour. Technol. 2019, 273, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhang, X. Construction of pyruvate producing strain with intact pyruvate dehydrogenase and genome-wide transcription analysis. World J. Microbiol. Biotechnol. 2017, 33, 59. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.K.; Uppada, V.; Swaminathan, A.G.; Noronha, S.B. Engineering of yeast pyruvate decarboxylase for enhanced selectivity towards carboligation. Bioresour. Technol. 2015, 192, 90–96. [Google Scholar] [CrossRef]
- Wood, A.J.; Duff, R.J. The aldehyde dehydrogenase (ALDH) gene superfamily of the moss Physcomitrella patens and the algae Chlamydomonas reinhardtii and Ostreococcus tauri. Bryologist 2009, 112, 1–11. [Google Scholar] [CrossRef]
- Rodrigues, S.M.; Andrade, M.O.; Gomes, A.P.; Damatta, F.M.; Baracat-Pereira, M.C.; Fontes, E.P. Arabidopsis and tobacco plants ectopically expressing the soybean antiquitin-like ALDH7 gene display enhanced tolerance to drought, salinity, and oxidative stress. J. Exp. Bot. 2006, 57, 1909–1918. [Google Scholar] [CrossRef]
- Kirch, H.H.; Bartels, D.; Wei, Y.; Schnable, P.S.; Wood, A.J. The ALDH gene superfamily of Arabidopsis. Trends Plant Sci. 2004, 9, 371–377. [Google Scholar] [CrossRef]
- Mori, Y.; Yamawaki, K.; Ishiguro, T.; Yoshihara, K.; Ueda, H.; Sato, A.; Ohata, H.; Yoshida, Y.; Minamino, T.; Okamoto, K.; et al. ALDH-Dependent Glycolytic Activation Mediates Stemness and Paclitaxel Resistance in Patient-Derived Spheroid Models of Uterine Endometrial Cancer. Stem Cell Rep. 2019, 13, 730–746. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Schnable, P.S. Functional specialization of maize mitochondrial aldehyde dehydrogenases. Plant. Physiol. 2002, 130, 1657–1674. [Google Scholar] [CrossRef][Green Version]
- Szekely, G.; Abraham, E.; Cseplo, A.; Rigo, G.; Zsigmond, L.; Csiszar, J.; Ayaydin, F.; Strizhov, N.; Jasik, J.; Schmelzer, E.; et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 2008, 53, 11–28. [Google Scholar] [CrossRef]
- Kumari, S.; Tishel, R.; Eisenbach, M.; Wolfe, A.J. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J. Bacteriol. 1995, 177, 2878–2886. [Google Scholar] [CrossRef] [PubMed]
- Makris, D.P.; Rossiter, J.T. Heat-induced, metal-catalyzed oxidative degradation of quercetin and rutin (Quercetin 3-O-rhamnosylglucoside) in aqueous model systems. J. Agric. Food Chem. 2000, 48, 3830–3838. [Google Scholar] [CrossRef] [PubMed]
- Gordon, J.I.; Duronio, R.J.; Rudnick, D.A.; Adams, S.P.; Gokel, G.W. Protein N-myristoylation. J. Biol. Chem. 1991, 266, 8647–8650. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lama, S.; Agrawal, D.; Kumar, V.; Park, S. Acetate as a potential feedstock for the production of value-added chemicals: Metabolism and applications. Biotechnol. Adv. 2021, 49, 107736. [Google Scholar] [CrossRef]
| Regulated Proteins | Added Precursor | Time (h) | Concentration (g/L) | Polysaccharide Titer (g/L) |
|---|---|---|---|---|
| Blank group | 2.79 ± 0.06 | |||
| ALHD3 | Acetaldehyde | 0 | 0.1 | 3.65 ± 0.12 * |
| 12 | 0.1 | 3.93 ± 0.07 * | ||
| 24 | 0.1 | 4.36 ± 0.12 ** | ||
| Alcohol Dehydrogenase (ADH) | Acetic acid | 0 | 0.1 | 4.03 ± 0.13 * |
| 12 | 0.1 | 3.82 ± 0.09 * | ||
| 24 | 0.1 | 2.82 ± 0.07 | ||
| PKF1 | D-Fructose 6-phosphate disodium | 0 | 0.1 | 3.14 ± 0.27 |
| 12 | 0.1 | 3.42 ± 0.08 | ||
| 24 | 0.1 | 4.55 ± 0.18 ** | ||
| PDC | Pyruvic acid | 0 | 0.1 | 4.00 ± 0.18 * |
| 12 | 0.1 | 3.77 ± 0.15 * | ||
| 24 | 0.1 | 3.59 ± 0.22 * | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, J.; Li, J.; Zhen, C.; Du, J.; Zhao, R.; Fan, B.; Hou, J.; Gao, B.; Zheng, Y.; Tu, L.; et al. Transcriptome Analysis of Sclerotium rolfsii: Unraveling Impact of Glycolytic Pathway on Substrate Utilization and Microbial Polysaccharide Production. Fermentation 2025, 11, 143. https://doi.org/10.3390/fermentation11030143
Song J, Li J, Zhen C, Du J, Zhao R, Fan B, Hou J, Gao B, Zheng Y, Tu L, et al. Transcriptome Analysis of Sclerotium rolfsii: Unraveling Impact of Glycolytic Pathway on Substrate Utilization and Microbial Polysaccharide Production. Fermentation. 2025; 11(3):143. https://doi.org/10.3390/fermentation11030143
Chicago/Turabian StyleSong, Jia, Junfeng Li, Chenrui Zhen, Juan Du, Rui Zhao, Bingqian Fan, Jiayi Hou, Bingning Gao, Yu Zheng, Linna Tu, and et al. 2025. "Transcriptome Analysis of Sclerotium rolfsii: Unraveling Impact of Glycolytic Pathway on Substrate Utilization and Microbial Polysaccharide Production" Fermentation 11, no. 3: 143. https://doi.org/10.3390/fermentation11030143
APA StyleSong, J., Li, J., Zhen, C., Du, J., Zhao, R., Fan, B., Hou, J., Gao, B., Zheng, Y., Tu, L., & Wang, M. (2025). Transcriptome Analysis of Sclerotium rolfsii: Unraveling Impact of Glycolytic Pathway on Substrate Utilization and Microbial Polysaccharide Production. Fermentation, 11(3), 143. https://doi.org/10.3390/fermentation11030143






