From Raw to Fermented: Uncovering the Microbial Wealth of Dairy
Abstract
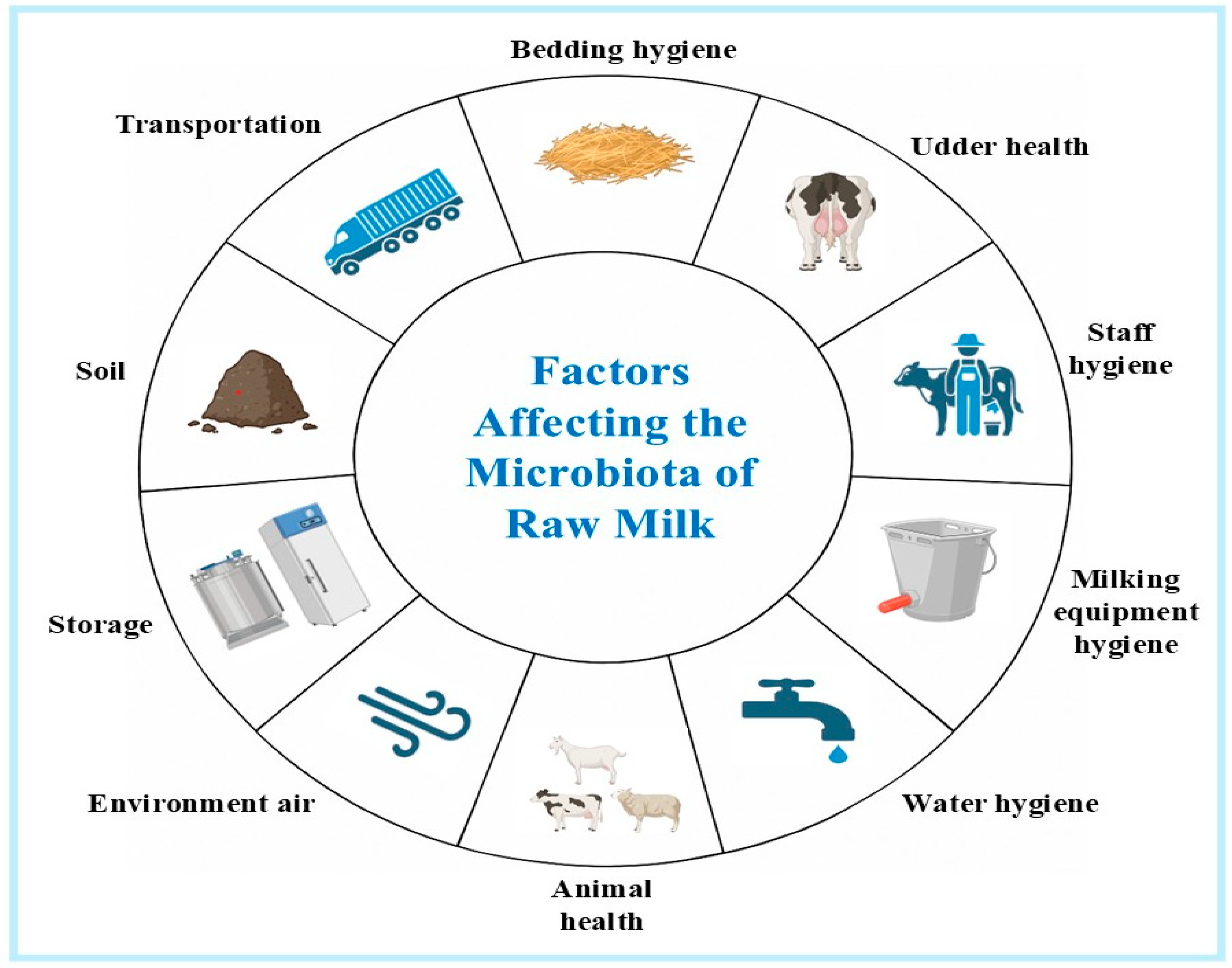
1. Introduction
2. Microbial Composition of Raw Milk
3. Evolution of Microbial Diversity During Fermentation
3.1. Natural (Spontaneous) vs. Starter Culture Fermentation
3.2. Traditional Fermented Dairy Products: Hidden Microbial Treasures
3.2.1. Kefir
3.2.2. Cheese
3.2.3. Comparison of Raw Milk, Kefir, and Cheese Microbiota
4. Techniques for Identifying Microbial Diversity in Dairy
5. Effects of the Microbiome on Flavor and Texture
5.1. Flavor and Aroma Development
5.2. Texture and Rheology
6. Conclusions and Recommendations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yap, M.; O’Sullivan, O.; Cotter, P.D. Microbiota of raw milk and raw milk cheeses. In Cheese, 5th ed.; McSweeney, P.L.H., Fox, P.F., Cotter, P.D., Everett, D.W., Eds.; Academic Press: Cambridge, MA, USA, 2025; pp. 335–347. [Google Scholar] [CrossRef]
- Gori, K.; Sørensen, L.M.; Petersen, M.A.; Jespersen, L.; Arneborg, N. Debaryomyces hansenii strains differ in their production of flavor compounds in a cheese-surface model. Microbiol. Open 2012, 1, 161–168. [Google Scholar] [CrossRef]
- Martin, N.; Berger, C.; Le Du, C.; Spinnler, H.E. Aroma compound production in cheese curd by Kluyveromyces marxianus. Appl. Environ. Microbiol. 2001, 67, 884–890. [Google Scholar] [CrossRef]
- Celano, G.; Calasso, M.; Costantino, G.; Vacca, M.; Ressa, A.; Nikoloudaki, O.; De Palo, P.; Calabrese, F.M.; Gobbetti, M.; De Angelis, M. Effect of seasonality on microbiological variability of raw cow milk from Apulian dairy farms in Italy. Microbiol. Spectr. 2022, 10, e00514-22. [Google Scholar] [CrossRef]
- Nalepa, B.; Olszewska, M.A.; Markiewicz, L.H. Seasonal variances in bacterial microbiota and volatile organic compounds in raw milk. Int. J. Food Microbiol. 2018, 267, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Yap, M.; O’Sullivan, O.; O’Toole, P.W.; Sheehan, J.J.; Fenelon, M.A.; Cotter, P.D. Seasonal and geographical impact on the Irish raw milk microbiota correlates with chemical composition and climatic variables. mSystems 2024, 9, e01290-23. [Google Scholar] [CrossRef]
- von Neubeck, M.; Baur, C.; Krewinkel, M.; Stoeckel, M.; Kranz, B.; Stressler, T.; Wenning, M. Biodiversity of refrigerated raw milk microbiota and their enzymatic spoilage potential. Int. J. Food Microbiol. 2015, 211, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Hou, Y.; Wang, Y.; Gao, Y.; Chen, Y.; Qi, J.; Ju, N. Relationship between microorganisms and milk metabolites during quality changes in refrigerated raw milk: A metagenomic and metabolomic exploration. Int. J. Food Microbiol. 2024, 425, 110891. [Google Scholar] [CrossRef]
- Lu, H.; Dang, C.; Liu, R.; Zhang, S.; Xue, Y.; Feng, L.; Wang, S. The effects of sampling sites, collection time, refrigerated storage duration on microbiota of raw milk from a Chinese dairy farm: An exploratory study. J. Food Prot. 2025, 88, 100504. [Google Scholar] [CrossRef]
- Espi-Malillos, A.; López-Almela, I.; Ruiz-Garcia, P.; López-Mendoza, M.C.; Carrón, N.; Gonzalez-Torres, P.; Quereda, J.J. Raw milk at refrigeration temperature displays an independent microbiota dynamic regardless of Listeria monocytogenes contamination. Food Res. Int. 2025, 202, 115637. [Google Scholar] [CrossRef]
- Ban, G.H.; Lee, J.I.; Kang, D.H. Effects of storage temperature on microbiota shifts in raw milk biofilm developed on stainless steel. Food Microbiol. 2023, 110, 104163. [Google Scholar] [CrossRef]
- Duarte, R.V.; Pinto, C.A.; Gomes, A.M.; Delgadillo, I.; Saraiva, J.A. A microbiological perspective of raw milk preserved at room temperature using hyperbaric storage compared to refrigerated storage. Innov. Food Sci. Emerg. Technol. 2022, 78, 103019. [Google Scholar] [CrossRef]
- McInnis, E.A.; Kalanetra, K.M.; Mills, D.A.; Maga, E.A. Analysis of raw goat milk microbiota: Impact of stage of lactation and lysozyme on microbial diversity. Food Microbiol. 2015, 46, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Doyle, C.J.; Gleeson, D.; O’Toole, P.W.; Cotter, P.D. High-throughput metataxonomic characterization of the raw milk microbiota identifies changes reflecting lactation stage and storage conditions. Int. J. Food Microbiol. 2017, 255, 1–6. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, L.; Jiang, X.; Song, Y.; Wang, D.; Liu, H.; Yao, J. Multiomics analysis revealed that the metabolite profile of raw milk is associated with the lactation stage of dairy cows and could be affected by variations in the ruminal microbiota. J. Dairy Sci. 2024, 107, 8709–8721. [Google Scholar] [CrossRef]
- Secchi, G.; Bisutti, V.; Toscano, A.; Pegolo, S.; Giannuzzi, D.; Cecchinato, A.; Franciosi, E. Changes in the milk and fecal microbiota in Holstein cows with subclinical intramammary infection. J. Dairy Sci. 2025, 108, 10220–10236. [Google Scholar] [CrossRef]
- Zhang, Y.; Bai, X.; Wang, X.; Lan, L.; Yang, D.; Zhang, B.; Wang, J. Characteristics of the milk microbiota of healthy goats and goats diagnosed with clinical mastitis in Western China. Microb. Pathog. 2025, 206, 107764. [Google Scholar] [CrossRef]
- Kable, M.E.; Srisengfa, Y.; Laird, M.; Zaragoza, J.; McLeod, J.; Heidenreich, J.; Marco, M.L. The core and seasonal microbiota of raw bovine milk in tanker trucks and the impact of transfer to a milk processing facility. mBio 2016, 7, e00836-16. [Google Scholar] [CrossRef]
- Skeie, S.B.; Håland, M.; Thorsen, I.M.; Narvhus, J.; Porcellato, D. Bulk tank raw milk microbiota differs within and between farms: A moving goalpost challenging quality control. J. Dairy Sci. 2019, 102, 1959–1971. [Google Scholar] [CrossRef]
- Ouamba, A.J.; Gagnon, M.; LaPointe, G.; Chouinard, P.Y.; Roy, D. Graduate student literature review: Farm management practices: Potential microbial sources that determine the microbiota of raw bovine milk. J. Dairy Sci. 2022, 105, 7276–7287. [Google Scholar] [CrossRef] [PubMed]
- Feligini, M.; Panelli, S.; Sacchi, R.; Ghitti, M.; Capelli, E. Tracing the origin of raw milk from farm by using automated ribosomal intergenic spacer analysis (ARISA) fingerprinting of microbiota. Food Control 2015, 50, 51–56. [Google Scholar] [CrossRef]
- Tarrah, A.; Callegaro, S.; Pakroo, S.; Finocchiaro, R.; Giacomini, A.; Corich, V.; Cassandro, M. New insights into the raw milk microbiota diversity from animals with a different genetic predisposition for feed efficiency and resilience to mastitis. Sci. Rep. 2022, 12, 13498. [Google Scholar] [CrossRef]
- Steinberg, R.S.; Silva e Silva, L.C.; de Souza, M.R.; Reis, R.B.; da Silva, P.C.L.; Lacorte, G.A.; Nicoli, J.R.; Neumann, E.; Nunes, Á.C. Changes in bovine milk bacterial microbiome from healthy and subclinical mastitis affected animals of the Girolando, Gyr, Guzera, and Holstein breeds. Int. Microbiol. 2022, 25, 803–815. [Google Scholar] [CrossRef]
- Biçer, Y.; Telli, A.E.; Sönmez, G.; Telli, N.; Uçar, G. Comparison of microbiota and volatile organic compounds in milk from different sheep breeds. J. Dairy Sci. 2021, 104, 12303–12311. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Lundh, Å.; Höjer, A.; Bernes, G.; Nilsson, D.; Johansson, M.; Hetta, M.; Gustafsson, A.H.; Saedén, K.H.; Dicksved, J. Milking system and premilking routines have a strong effect on the microbial community in bulk tank milk. J. Dairy Sci. 2022, 105, 123–139. [Google Scholar] [CrossRef] [PubMed]
- Desmousseaux, C.; Guilbaud, M.; Jard, G.; Tormo, H.; Oulahal, N.; Hanin, A.; Bourdonnais, E.; Jha, P.K.; Laithier, C. Biofilms in the milking machine, from laboratory scale to on-farm results. J. Dairy Sci. 2025, 108, 8120–8140. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Liedtke, J.; Plattes, S.; Lipski, A. Bacterial community composition of biofilms in milking machines of two dairy farms assessed by a combination of culture-dependent and -independent methods. PLoS ONE 2019, 14, e0222238. [Google Scholar] [CrossRef]
- Doyle, C.J.; Gleeson, D.; O’Toole, P.W.; Cotter, P.D. Impacts of seasonal housing and teat preparation on raw milk microbiota: A high-throughput sequencing study. Appl. Environ. Microbiol. 2017, 83, e02694-16. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, Y.; Siqin, B.; Xilin, T.; Zhang, N.; Li, M. Changes in microbial diversity and nutritional components of mare milk before and after traditional fermentation. Front. Sustain. Food Syst. 2022, 6, 913763. [Google Scholar] [CrossRef]
- Tang, H.; Ma, H.; Hou, Q.; Li, W.; Xu, H.; Liu, W.; Sun, Z.; Haobisi, H.; Menghe, B. Profiling of koumiss microbiota and organic acids and their effects on koumiss taste. BMC Microbiol. 2020, 20, 85. [Google Scholar] [CrossRef]
- Marsh, A.J.; O’Sullivan, O.; Hill, C.; Ross, R.P.; Cotter, P.D. Sequencing-based analysis of the bacterial and fungal composition of kefir grains and milks from multiple sources. PLoS ONE 2013, 8, e69371. [Google Scholar] [CrossRef]
- Wolfe, B.E.; Button, J.E.; Santarelli, M.; Dutton, R.J. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell 2014, 158, 422–433. [Google Scholar] [CrossRef]
- Sessou, P.; Keisam, S.; Gagara, M.; Komagbe, G.; Farougou, S.; Mahillon, J.; Jeyaram, K. Comparative analyses of the bacterial communities present in the spontaneously fermented milk products of Northeast India and West Africa. Front. Microbiol. 2023, 14, 1166518. [Google Scholar] [CrossRef] [PubMed]
- Papadimitriou, K.; Anastasiou, R.; Georgalaki, M.; Bounenni, R.; Paximadaki, A.; Charmpi, C.; Alexandraki, V.; Kazou, M.; Tsakalidou, E. Comparison of the microbiome of artisanal homemade and industrial feta cheese through amplicon sequencing and shotgun metagenomics. Microorganisms 2022, 10, 1073. [Google Scholar] [CrossRef] [PubMed]
- Nelli, A.; Venardou, B.; Skoufos, I.; Voidarou, C.C.; Lagkouvardos, I.; Tzora, A. An insight into goat cheese: The tales of artisanal and industrial Gidotyri microbiota. Microorganisms 2023, 11, 123. [Google Scholar] [CrossRef] [PubMed]
- Bintsis, T.; Mantzouridou, F.T.; Lalou, S.; Alvanoudi, P.; Ordoudi, S.A.; Angelidis, A.S.; Fletouris, D. Comparative analysis of chemical, microbiological, sensory and volatile compound profiles in Manouri PDO and artisanal Manouri cheeses: A preliminary study. Food Bioprocess Technol. 2024, 17, 3561–3575. [Google Scholar] [CrossRef]
- Bintsis, T.; Papademas, P. The evolution of fermented milks, from artisanal to industrial products: A critical review. Fermentation 2022, 8, 679. [Google Scholar] [CrossRef]
- Sindi, A.; Badsha, M.B.; Ünlü, G. Bacterial populations in international artisanal kefirs. Microorganisms 2020, 8, 1318. [Google Scholar] [CrossRef]
- Nejati, F.; Capitain, C.C.; Krause, J.L.; Kang, G.; Riedel, R.; Chang, H.; Kurreck, J.; Junne, S.; Weller, P.; Neubauer, P. Traditional grain-based vs. commercial milk kefirs, how different are they? Appl. Sci. 2022, 12, 3838. [Google Scholar] [CrossRef]
- Serrano, S.; Morais, S.; Semedo-Lemsaddek, T. Tradition unveiled: A comprehensive review of microbiological studies on Portuguese traditional cheeses, merging conventional and OMICs analyses. Front. Ind. Microbiol. 2024, 2, 1420042. [Google Scholar] [CrossRef]
- Moonga, H.B.; Schoustra, S.E.; Van den Heuvel, J.; Linnemann, A.R.; Samad, M.S.; Shindano, J.; Smid, E.J. Composition and diversity of natural bacterial communities in Mabisi, a traditionally fermented milk. Front. Microbiol. 2020, 11, 1816. [Google Scholar] [CrossRef]
- Zhao, X.; Song, L.; Han, D.; Han, P.; Bai, F. Microbiological and physicochemical dynamics in traditional and industrial fermentation processes of koumiss. Fermentation 2024, 10, 66. [Google Scholar] [CrossRef]
- Walsh, L.H.; Coakley, M.; Walsh, A.M.; Crispie, F.; O’Toole, P.W.; Cotter, P.D. Analysis of the milk kefir pan-metagenome reveals four community types, core species, associated metabolic pathways. iScience 2023, 26, 108004. [Google Scholar] [CrossRef]
- Alraddadi, F.A.; Ross, T.; Powell, S.M. Evaluation of the microbial communities in kefir grains and kefir over time. Int. Dairy J. 2023, 136, 105490. [Google Scholar] [CrossRef]
- Motoshima, H.; Fujioka, I.; Uchida, K. Identification of dominant species common to kefir grains from seven origins for kefir grain reconstruction. J. Dairy Sci. 2025, 108, 8222–8236. [Google Scholar] [CrossRef]
- Kazou, M.; Grafakou, A.; Tsakalidou, E.; Georgalaki, M. Zooming into the microbiota of home-made and industrial kefir produced in Greece using classical microbiological and amplicon-based metagenomics analyses. Front. Microbiol. 2021, 12, 621069. [Google Scholar] [CrossRef]
- Garofalo, C.; Osimani, A.; Milanović, V.; Aquilanti, L.; De Filippis, F.; Stellato, G.; Clementi, F. Bacteria and yeast microbiota in milk kefir grains from different Italian regions. Food Microbiol. 2015, 49, 123–133. [Google Scholar] [CrossRef]
- Walsh, A.M.; Crispie, F.; Kilcawley, K.; O’Sullivan, O.; O’Sullivan, M.G.; Claesson, M.J.; Cotter, P.D. Microbial succession and flavor production in the fermented dairy beverage kefir. mSystems 2016, 1, e00052-16. [Google Scholar] [CrossRef] [PubMed]
- Maughan, L.; Koolman, L.; Macori, G.; Killian, C.; Fanning, S.; Whyte, P.; Bolton, D. Characterization of bacterial and fungal populations in retail kefirs in Ireland. J. Dairy Sci. 2025, 108, 8187–8204. [Google Scholar] [CrossRef]
- Güney, H.D.; Altundağ, Ö.Ö.; Çolak, M. Microbiological changes of kefir traditionally produced from different milks according to storage time. Front. Life Sci. Relat. Technol. 2024, 6, 9–14. [Google Scholar] [CrossRef]
- La Torre, C.; Caputo, P.; Cione, E.; Fazio, A. Comparing nutritional values and bioactivity of kefir from different types of animal milk. Molecules 2024, 29, 2710. [Google Scholar] [CrossRef]
- Wang, H.; Sun, X.; Song, X.; Guo, M. Effects of kefir grains from different origins on proteolysis and volatile profile of goat milk kefir. Food Chem. 2021, 339, 128099. [Google Scholar] [CrossRef]
- Baars, T.; van Esch, B.; van Ooijen, L.; Zhang, Z.; Dekker, P.; Boeren, S.; Diks, M.; Garssen, J.; Hettinga, K.; Kort, R. Raw milk kefir: Microbiota, bioactive peptides, immune modulation. Food Funct. 2023, 14, 1648–1661. [Google Scholar] [CrossRef] [PubMed]
- Baars, T.; van Esch, B.; Diks, M.; van Ooijen, L.; Zhang, Z.; Dekker, P.; Kort, R. Bacterial diversity, bioactive peptides, enhanced immunomodulatory effects in raw milk kefir made with defined starter cultures versus backslopping. Int. Dairy J. 2025, 164, 106202. [Google Scholar] [CrossRef]
- Kesmen, Z.; Kacmaz, N. Determination of lactic microflora of kefir grains and kefir beverage by using culture-dependent and culture-independent methods. J. Food Sci. 2011, 76, M276–M283. [Google Scholar] [CrossRef]
- Ozturkoğlu Budak, S.; Figge, M.J.; Houbraken, J.; de Vries, R.P. The diversity and evolution of microbiota in traditional Turkish Divle Cave cheese during ripening. Int. Dairy J. 2016, 58, 50–53. [Google Scholar] [CrossRef]
- Halıcı Demir, F.; Kaptan, B. Identification of lactic acid bacteria isolated from the protected geographical indication Edirne white cheese using MALDI-TOF MS: Impact of ripening time and type of milk on microbial diversity. Int. Dairy J. 2025, 162, 106156. [Google Scholar] [CrossRef]
- Dolci, P.; De Filippis, F.; La Storia, A.; Ercolini, D.; Cocolin, L. rRNA-based monitoring of the microbiota involved in Fontina PDO cheese production in relation to different stages of cow lactation. Int. J. Food Microbiol. 2014, 185, 127–135. [Google Scholar] [CrossRef]
- Calasso, M.; Ercolini, D.; Mancini, L.; Stellato, G.; Minervini, F.; Di Cagno, R.; Gobbetti, M. Relationships among house, rind and core microbiotas during manufacture of traditional Italian cheeses at the same dairy plant. Food Microbiol. 2016, 54, 115–126. [Google Scholar] [CrossRef]
- Merchán, A.V.; Ruiz-Moyano, S.; Benito, M.J.; Hernández, M.V.; Cabañas, C.M.; Román, Á.C. Metabarcoding analysis reveals a differential bacterial community profile associated with ‘Torta del Casar’ and ‘Queso de la Serena’ PDO cheeses. Food Biosci. 2024, 57, 103491. [Google Scholar] [CrossRef]
- Lutin, J.; Dufrene, F.; Guyot, P.; Palme, R.; Achilleos, C.; Bouton, Y.; Buchin, S. Microbial composition and viability of natural whey starters used in PDO Comté cheese-making. Food Microbiol. 2024, 121, 104521. [Google Scholar] [CrossRef]
- Rüstemoğlu, M.; Erkan, M.E.; Cengiz, G.; Hajyzadeh, M. Bacterial metagenome profiling of hand-made herby cheese samples utilizing high-throughput sequencing to detect geographical indication and marketing potential. Heliyon 2023, 9, e13334. [Google Scholar] [CrossRef] [PubMed]
- Ihsan, M.A.; Valdramidis, V.P.; Griffin, S. Bacterial and fungal profiling of Maltese sheep cheese with amplicon metabarcoding. Int. Dairy J. 2025, 170, 106362. [Google Scholar] [CrossRef]
- Aragao, M.D.O.P.; Lima, F.R.; Passamani, F.R.F.; de Aguilar Santos, M.A.; de Paula Rezende, J.; Batista, L.R. Fungal and bacterial diversity present on the rind and core of Natural Bloomy Rind Artisanal Minas Cheese from the Canastra region, Brazil. Food Res. Int. 2025, 202, 115724. [Google Scholar] [CrossRef] [PubMed]
- Santamarina-García, G.; Hernández, I.; Amores, G.; Virto, M. Characterization of microbial shifts during the production and ripening of raw ewe milk-derived Idiazabal cheese by high-throughput sequencing. Biology 2022, 11, 769. [Google Scholar] [CrossRef]
- Moreira, R.V.; Costa, M.P.; Frasao, B.S.; Sobral, V.S.; Cabral, C.C.; Rodrigues, B.L.; Mano, S.B.; Conte-Junior, C.A. Effect of ripening time on bacteriological and physicochemical goat milk cheese characteristics. Food Sci. Biotechnol. 2020, 29, 459–467. [Google Scholar] [CrossRef]
- Jingkai, J.; Jianming, Z.; Zhenmin, L.; Huaxi, Y. Dynamic changes of microbiota and texture properties during the ripening of traditionally prepared cheese of China. Arch. Microbiol. 2020, 202, 2059–2069. [Google Scholar] [CrossRef]
- Busetta, G.; Garofalo, G.; Claps, S.; Sardina, M.T.; Franciosi, E.; Alfonzo, A.; Gaglio, R. The wooden shelf surface and cheese rind mutually exchange microbiota during the traditional ripening process. Int. J. Food Microbiol. 2024, 409, 110478. [Google Scholar] [CrossRef]
- Delcenserie, V.; Taminiau, B.; Delhalle, L.; Nezer, C.; Doyen, P.; Crevecoeur, S.; Daube, G. Microbiota characterization of a Belgian protected designation of origin cheese, Herve cheese, using metagenomic analysis. J. Dairy Sci. 2014, 97, 6046–6056. [Google Scholar] [CrossRef] [PubMed]
- Dopieralska, P.; Barłowska, J.; Teter, A.; Król, J.; Brodziak, A.; Domaradzki, P. Changes in fatty acid and volatile compound profiles during storage of smoked cheese made from the milk of native Polish cow breeds raised in the Low Beskids. Animals 2020, 10, 2103. [Google Scholar] [CrossRef]
- Irlinger, F.; Mounier, J. Microbial interactions in cheese: Implications for cheese quality and safety. Curr. Opin. Biotechnol. 2009, 20, 142–148. [Google Scholar] [CrossRef]
- Bertuzzi, A.S.; Walsh, A.M.; Sheehan, J.J.; Cotter, P.D.; Crispie, F.; McSweeney, P.L.; Rea, M.C. Omics-based insights into flavor development and microbial succession within surface-ripened cheese. mSystems 2018, 3, e00211-17. [Google Scholar] [CrossRef]
- da Silva Duarte, V.; Bjørgan, B.; Franklin, F.V.; Olsen, K.; Abdelghani, A.; Skeie, S.; Porcellato, D. Metataxonomic analysis of bulk tank milk and seasonal impact on starter culture development in aged goat milk cheese. Int. Dairy J. 2025, 163, 106160. [Google Scholar] [CrossRef]
- Fontana, F.; Longhi, G.; Alessandri, G.; Lugli, G.A.; Mancabelli, L.; Tarracchini, C.; Milani, C. Multifactorial microvariability of the Italian raw milk cheese microbiota and implication for current regulatory scheme. mSystems 2023, 8, e01068-22. [Google Scholar] [CrossRef]
- Jin, H.; Mo, L.; Pan, L.; Hou, Q.; Li, C.; Darima, I.; Yu, J. Using PacBio sequencing to investigate the bacterial microbiota of traditional Buryatian cottage cheese and comparison with Italian and Kazakhstan artisanal cheeses. J. Dairy Sci. 2018, 101, 6885–6896. [Google Scholar] [CrossRef]
- Buzzanca, D.; Giordano, M.; Chiarini, E.; Ferrocino, I.; Cocolin, L.; Zeppa, G.; Alessandria, V. Delving into Roccaverano PDO cheese: A comprehensive examination of microbial diversity and flavour profiles compared to non-PDO cheeses. Int. J. Food Microbiol. 2025, 429, 111014. [Google Scholar] [CrossRef]
- Dertli, E.; Çon, A.H. Microbial diversity of traditional kefir grains and their role on kefir aroma. LWT-Food Sci. Technol. 2017, 85, 151–157. [Google Scholar] [CrossRef]
- Purutoğlu, K.; İspirli, H.; Yüzer, M.O.; Serencam, H.; Dertli, E. Diversity and functional characteristics of lactic acid bacteria from traditional kefir grains. Int. J. Dairy Technol. 2020, 73, 57–66. [Google Scholar] [CrossRef]
- Biçer, Y.; Telli, A.E.; Sönmez, G.; Turkal, G.; Telli, N.; Uçar, G. Comparison of commercial and traditional kefir microbiota using metagenomic analysis. Int. J. Dairy Technol. 2021, 74, 528–534. [Google Scholar] [CrossRef]
- Magalhães, K.T.; Pereira, G.V.D.M.; Campos, C.R.; Dragone, G.; Schwan, R.F. Brazilian kefir: Structure, microbial communities and chemical composition. Braz. J. Microbiol. 2011, 42, 693–702. [Google Scholar] [CrossRef]
- Fouhy, F.; Clooney, A.G.; Stanton, C.; Claesson, M.J.; Cotter, P.D. 16S rRNA gene sequencing of mock microbial populations—Impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol. 2016, 16, 123. [Google Scholar] [CrossRef]
- Salonen, A.; Nikkilä, J.; Jalanka-Tuovinen, J.; Immonen, O.; Rajilić-Stojanović, M.; Kekkonen, R.A.; de Vos, W.M. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: Effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J. Microbiol. Methods 2010, 81, 127–134. [Google Scholar] [CrossRef]
- Reuben, R.C.; Langer, D.; Eisenhauer, N.; Jurburg, S.D. Universal drivers of cheese microbiomes. iScience 2023, 26, 105744. [Google Scholar] [CrossRef]
- Henri-Dubernet, S.; Desmasures, N.; Guéguen, M. Culture-dependent and culture-independent methods for molecular analysis of the diversity of lactobacilli in “Camembert de Normandie” cheese. Lait 2004, 84, 179–189. [Google Scholar] [CrossRef]
- Ercolini, D. High-throughput sequencing and metagenomics: Moving forward in the culture-independent analysis of food microbial ecology. Appl. Environ. Microbiol. 2013, 79, 3148–3155. [Google Scholar] [CrossRef] [PubMed]
- Stellato, G.; La Storia, A.; De Filippis, F.; Borriello, G.; Villani, F.; Ercolini, D. Overlap of spoilage-associated microbiota between meat and the meat processing environment in small-scale and large-scale retail distributions. Appl. Environ. Microbiol. 2016, 82, 4045–4054. [Google Scholar] [CrossRef]
- McIlwaine, D.B.; Moore, M.; Corrigan, A.; Niemaseck, B.; Nicoletti, D. A comparison of the microbial populations in a culture-dependent and a culture-independent analysis of industrial water samples. Appl. Microbiol. 2024, 4, 1079–1090. [Google Scholar] [CrossRef]
- Virtanen, S.; Saqib, S.; Kanerva, T.; Ventin-Holmberg, R.; Nieminen, P.; Holster, T.; Salonen, A. Metagenome-validated combined amplicon sequencing and text mining-based annotations for simultaneous profiling of bacteria and fungi: Vaginal microbiota and mycobiota in healthy women. Microbiome 2024, 12, 273. [Google Scholar] [CrossRef]
- Curiale, M.S.; Lepper, W.; Robison, B. Enzyme-linked immunoassay for detection of Listeria monocytogenes in dairy products, seafoods, and meats: Collaborative study. J. AOAC Int. 1994, 77, 1472–1489. [Google Scholar] [CrossRef] [PubMed]
- Nicolaou, N.; Goodacre, R. Rapid and quantitative detection of the microbial spoilage in milk using Fourier transform infrared spectroscopy and chemometrics. Analyst 2008, 133, 1424–1431. [Google Scholar] [CrossRef] [PubMed]
- Nieuwoudt, M.K.; Holroyd, S.E.; McGoverin, C.M.; Simpson, M.C.; Williams, D.E. Rapid, sensitive, and reproducible screening of liquid milk for adulterants using a portable Raman spectrometer and a simple, optimized sample well. J. Dairy Sci. 2016, 99, 7821–7831. [Google Scholar] [CrossRef]
- Ampuero, S.; Bosset, J.O. The electronic nose applied to dairy products: A review. Sens. Actuators B Chem. 2003, 94, 1–12. [Google Scholar] [CrossRef]
- Yap, M.; Feehily, C.; Walsh, C.J.; Fenelon, M.; Murphy, E.F.; McAuliffe, F.M.; van Sinderen, D.; O’Toole, P.W.; O’Sullivan, O.; Cotter, P.D. Evaluation of methods for the reduction of contaminating host reads when performing shotgun metagenomic sequencing of the milk microbiome. Sci. Rep. 2020, 10, 21665. [Google Scholar] [CrossRef]
- You, L.; Yang, C.; Jin, H.; Kwok, L.; Lv, R.; Ma, T.; Zhao, Z.; Zhang, H.; Sun, Z. Shotgun metagenomic analysis of microbiota dynamics during long-term backslopping fermentation of traditional fermented milk in a controlled laboratory environment. J. Dairy Sci. 2024, 107, 7619–7630. [Google Scholar] [CrossRef]
- Lv, X.C.; Jia, R.B.; Li, Y.; Chen, F.; Chen, Z.; Liu, B.; Chen, S.; Rao, P.; Ni, L. Characterization of the dominant bacterial communities of traditional fermentation starters for Hong Qu glutinous rice wine by means of MALDI-TOF mass spectrometry fingerprinting, 16S rRNA gene sequencing and species-specific PCRs. Food Control 2016, 67, 292–302. [Google Scholar] [CrossRef]
- Kluz, M.I.; Waszkiewicz-Robak, B.; Kačániová, M. The applications of MALDI-TOF MS in the diagnosis of microbiological food contamination. Appl. Sci. 2025, 15, 7863. [Google Scholar] [CrossRef]
- Elcheninov, A.G.; Zayulina, K.S.; Klyukina, A.A.; Kremneva, M.K.; Kublanov, I.V.; Kochetkova, T.V. Metagenomic insights into the taxonomic and functional features of traditional fermented milk products from Russia. Microorganisms 2023, 12, 16. [Google Scholar] [CrossRef]
- Santamarina-García, G.; Yap, M.; Crispie, F.; Amores, G.; Lordan, C.; Virto, M.; Cotter, P.D. Shotgun metagenomic sequencing reveals the influence of artisanal dairy environments on the microbiomes, quality, safety of Idiazabal, a raw ewe milk PDO cheese. Microbiome 2024, 12, 262. [Google Scholar] [CrossRef]
- Emerson, J.B.; Adams, R.I.; Román, C.M.B.; Brooks, B.; Coil, D.A.; Dahlhausen, K.; Ganz, H.H.; Hartmann, E.M.; Hsu, T.; Justice, N.B.; et al. Schrödinger’s microbes: Tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 2017, 5, 86. [Google Scholar] [CrossRef] [PubMed]
- Mancabelli, L.; Milani, C.; Anzalone, R.; Alessandri, G.; Lugli, G.A.; Tarracchini, C.; Fontana, F.; Turroni, F.; Ventura, M. Free DNA and metagenomics analyses: Evaluation of free DNA inactivation protocols for shotgun metagenomics analysis of human biological matrices. Front. Microbiol. 2021, 12, 749373. [Google Scholar] [CrossRef]
- Dinu, L.D.; Al-Zaidi, Q.J.; Matache, A.G.; Matei, F. Improving the efficiency of viability-qPCR with lactic acid enhancer for the selective detection of live pathogens in foods. Foods 2024, 13, 1021. [Google Scholar] [CrossRef] [PubMed]
- Luo, B.; Dong, F.; Liu, Y.; Du, J.; Sun, H.; Ni, Y.; Zhang, Y. Insights into the microbiota of raw milk from seven breeds animals distributing in Xinjiang, China. Front. Microbiol. 2024, 15, 1382286. [Google Scholar] [CrossRef] [PubMed]
- McHugh, A.J.; Feehily, C.; Fenelon, M.A.; Gleeson, D.; Hill, C.; Cotter, P.D. Tracking the dairy microbiota from farm bulk tank to skimmed milk powder. mSystems 2020, 5, e00226-20. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, Y.; Jay-Russell, M.; Lemay, D.G.; Mills, D.A. Reservoirs of antimicrobial resistance genes in retail raw milk. Microbiome 2020, 8, 99. [Google Scholar] [CrossRef]
- Li, N.; Wang, Y.; You, C.; Ren, J.; Chen, W.; Zheng, H.; Liu, Z. Variation in raw milk microbiota throughout 12 months and the impact of weather conditions. Sci. Rep. 2018, 8, 2371. [Google Scholar] [CrossRef]
- Breitenwieser, F.; Doll, E.V.; Clavel, T.; Scherer, S.; Wenning, M. Complementary use of cultivation and high-throughput amplicon sequencing reveals high biodiversity within raw milk microbiota. Front. Microbiol. 2020, 11, 1557. [Google Scholar] [CrossRef]
- Esteban-Blanco, C.; Gutiérrez-Gil, B.; Puente-Sánchez, F.; Marina, H.; Tamames, J.; Acedo, A.; Arranz, J.J. Microbiota characterization of sheep milk and its association with somatic cell count using 16S rRNA gene sequencing. J. Anim. Breed. Genet. 2020, 137, 73–83. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, Z.; Lei, F.; Wang, B.; Jiang, S.; Peng, Q.; Shao, Y. Bacterial diversity in goat milk from the Guanzhong area of China. J. Dairy Sci. 2017, 100, 7812–7824. [Google Scholar] [CrossRef]
- Korsak, N.; Taminiau, B.; Leclercq, M.; Nezer, C.; Crevecoeur, S.; Ferauche, C.; Daube, G. Evaluation of the microbiota of kefir samples using metagenetic analysis targeting the 16S and 26S ribosomal DNA fragments. J. Dairy Sci. 2015, 98, 3684–3689. [Google Scholar] [CrossRef]
- Silva, M.H.; Batista, L.L.; Malta, S.M.; Santos, A.C.; Mendes-Silva, A.P.; Bonetti, A.M.; Ueira-Vieira, C.; dos Santos, A.R. Unveiling the Brazilian kefir microbiome: Discovery of a novel Lactobacillus kefiranofaciens (LkefirU) genome and in silico prospection of bioactive peptides with potential anti-Alzheimer properties. BMC Genom. 2024, 25, 884. [Google Scholar] [CrossRef] [PubMed]
- Demirci, T.; Akın, N.; Öztürk, H.İ.; Oğul, A. A metagenomic approach to homemade back-slopped yogurts produced in mountainous villages of Turkey with the potential next-generation probiotics. LWT 2022, 154, 112860. [Google Scholar] [CrossRef]
- Frétin, M.; Martin, B.; Rifa, E.; Isabelle, V.M.; Pomiès, D.; Ferlay, A.; Delbès, C. Bacterial community assembly from cow teat skin to ripened cheeses is influenced by grazing systems. Sci. Rep. 2018, 8, 200. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Valentino, V.; Yap, M.; Cabrera-Rubio, R.; Barcenilla, C.; Carlino, N.; Cobo-Díaz, J.F.; Quijada, N.M.; Calvete-Torre, I.; Ruas-Madiedo, P.; et al. Microbiome mapping in dairy industry reveals new species and genes for probiotic and bioprotective activities. npj Biofilms Microbiomes 2024, 10, 67. [Google Scholar] [CrossRef]
- Park, H.; Yeo, S.; Ryu, C.B.; Huh, C.S. A streamlined culturomics case study for the human gut microbiota research. Sci. Rep. 2024 14, 20361. [CrossRef]
- Walsh, A.M.; Leech, J.; Huttenhower, C.; Delhomme-Nguyen, H.; Crispie, F.; Chervaux, C.; Cotter, P.D. Integrated molecular approaches for fermented food microbiome research. FEMS Microbiol. Rev. 2023, 47, fuad001. [Google Scholar] [CrossRef]
- Mayo, B.; Rodríguez, J.; Vázquez, L.; Flórez, A.B. Microbial interactions within the cheese ecosystem and their application to improve quality and safety. Foods 2021, 10, 602. [Google Scholar] [CrossRef]
- Hegazi, F.Z.; Abo-Elnaga, I.G. Production of acetoin and diacetyl by lactic acid bacteria in skimmed milk with added citrate and pyruvate. Z. Lebensm. Unters. Forsch. 1980, 171, 367–370. [Google Scholar] [CrossRef]
- Thierry, A.; Pogačić, T.; Weber, M.; Lortal, S. Production of flavor compounds by lactic acid bacteria in fermented foods. In Biotechnology of Lactic Acid Bacteria: Novel Applications; Mozzi, F., Raya, R.R., Vignolo, G.M., Eds.; Wiley-Blackwell: Oxford, UK, 2015; pp. 314–340. [Google Scholar] [CrossRef]
- Fröhlich-Wyder, M.T.; Guggisberg, D.; Bisig, W.; Jakob, E.; Schmidt, R.S. The total eye volume of cheese is influenced by different fat levels. Int. Dairy J. 2023, 144, 105690. [Google Scholar] [CrossRef]
- Thierry, A.; Maillard, M.B.; Richoux, R.; Kerjean, J.R.; Lortal, S. Propionibacterium freudenreichii strains quantitatively affect production of volatile compounds in Swiss cheese. Lait 2005, 85, 57–74. [Google Scholar] [CrossRef]
- Saygili, D.; Yerlikaya, O.; Akpinar, A. The effect of using different yeast species on the composition of carbohydrates and volatile aroma compounds in kefir drinks. Food Biosci. 2023, 54, 102867. [Google Scholar] [CrossRef]
- Patton, S. The methyl ketones of blue cheese and their relation to its flavor. J. Dairy Sci. 1950, 33, 680–684. [Google Scholar] [CrossRef]
- Walker, V.; Mills, G.A. 2-Pentanone production from hexanoic acid by Penicillium roqueforti from blue cheese: Is this the pathway used in humans? Sci. World J. 2014, 2014, 215783. [Google Scholar] [CrossRef]
- Berger, C.; Khan, J.A.; Molimard, P.; Martin, N.; Spinnler, H.E. Production of sulfur flavors by ten strains of Geotrichum candidum. Appl. Environ. Microbiol. 1999, 65, 5510–5514. [Google Scholar] [CrossRef]
- Pracharova, P.; Lieben, P.; Pollet, B.; Beckerich, J.M.; Bonnarme, P.; Landaud, S.; Swennen, D. Geotrichum candidum gene expression and metabolite accumulation inside the cells reflect the strain oxidative stress sensitivity and ability to produce flavour compounds. FEMS Yeast Res. 2019, 19, foy111. [Google Scholar] [CrossRef]
- Duensing, P.W.; Hinrichs, J.; Schieberle, P. Formation of key aroma compounds during 30 weeks of ripening in Gouda-type cheese produced from pasteurized and raw milk. J. Agric. Food Chem. 2024, 72, 11072–11079. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Shi, X.; Wang, B. A review on the general cheese processing technology, flavor biochemical pathways and the influence of yeasts in cheese. Front. Microbiol. 2021, 12, 703284. [Google Scholar] [CrossRef]
- Alexa, E.A.; Cobo-Díaz, J.F.; Renes, E.; O’Callaghan, T.F.; Kilcawley, K.; Mannion, D.; Skibinska, I.; Ruiz, L.; Margolles, A.; Fernández-Gómez, P.; et al. The detailed analysis of the microbiome and resistome of artisanal blue-veined cheeses provides evidence on sources and patterns of succession linked with quality and safety traits. Microbiome 2024, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhao, S.; Hao, G.; Yu, H.; Tian, H.; Zhao, G. Role of lactic acid bacteria on the yogurt flavour: A review. Int. J. Food Prop. 2017, 20, S316–S330. [Google Scholar] [CrossRef]
- Leite, A.M.O.; Miguel, M.A.L.; Peixoto, R.S.; Rosado, A.S.; Silva, J.T.; Paschoalin, V.M.F. Microbiological, technological and therapeutic properties of kefir: A natural probiotic beverage. Braz. J. Microbiol. 2013, 44, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Prado, M.R.; Blandón, R.; Vandenberghe, S.; Rodrigues, A.; Castro-Gómez, L.; Soccol, C. Milk kefir: Composition, microbial cultures, biological activities, related products. Front. Microbiol. 2015, 6, 1177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhang, T.T.; Guo, R.R.; Ye, Q.; Zhao, H.L.; Huang, X.H. The regulation of key flavor of traditional fermented food by microbial metabolism: A review. Food Chem. X 2023, 19, 100871. [Google Scholar] [CrossRef]
- Cheng, T.; Zhang, T.; Zhang, P.; He, X.; Sadiq, F.A.; Li, J.; Sang, Y.; Gao, J. The complex world of kefir: Structural insights and symbiotic relationships. Compr. Rev. Food Sci. Food Saf. 2024, 23, e13364. [Google Scholar] [CrossRef] [PubMed]
- Nachtigall, C.; Surber, G.; Wefers, D.; Vogel, C.; Rohm, H.; Jaros, D. Capsular exopolysaccharides from two Streptococcus thermophilus strains differ in their moisture sorption behavior. Foods 2023, 12, 596. [Google Scholar] [CrossRef]
- Ramos, I.M.; Seseña, S.; Poveda, J.M.; Palop, M.L. Screening of lactic acid bacteria strains to improve the properties of non-fat set yogurt by in situ EPS production. Food Bioprocess Technol. 2023, 16, 2541–2558. [Google Scholar] [CrossRef]
- Rosa, D.D.; Dias, M.M.; Grześkowiak, Ł.M.; Reis, S.A.; Conceição, L.L.; Peluzio, M.D.C.G. Milk kefir: Nutritional, microbiological and health benefits. Nutr. Res. Rev. 2017, 30, 82–96. [Google Scholar] [CrossRef]
- Georgalaki, M.; Zoumpopoulou, G.; Anastasiou, R.; Kazou, M.; Tsakalidou, E. Lactobacillus kefiranofaciens: From isolation and taxonomy to probiotic properties and applications. Microorganisms 2021, 9, 2158. [Google Scholar] [CrossRef]
- Tingirikari, J.M.R.; Sharma, A.; Lee, H.J. Kefir: A fermented plethora of symbiotic microbiome and health. J. Ethn. Foods 2024, 11, 35. [Google Scholar] [CrossRef]
- Voidarou, C.; Antoniadou, M.; Rozos, G.; Tzora, A.; Skoufos, I.; Varzakas, T.; Lagiou, A.; Bezirtzoglou, E. Fermentative foods: Microbiology, biochemistry, potential human health benefits and public health issues. Foods 2020, 10, 69. [Google Scholar] [CrossRef]
- Bisig, W.; Guggisberg, D.; Jakob, E.; Turgay, M.; Irmler, S.; Wechsler, D.; Fröhlich-Wyder, M.T. The effect of NaCl and metabolic profile of propionibacteria on eye formation in experimental Swiss-type cheese. Int. Dairy J. 2019, 89, 86–95. [Google Scholar] [CrossRef]
- Arab, M.; Yousefi, M.; Khanniri, E.; Azari, M.; Ghasemzadeh-Mohammadi, V.; Mollakhalili-Meybodi, N. A comprehensive review on yogurt syneresis: Effect of processing conditions and added additives. J. Food Sci. Technol. 2023, 60, 1656–1665. [Google Scholar] [CrossRef]
- Amani, E.; Eskandari, M.H.; Shekarforoush, S. The effect of proteolytic activity of starter cultures on technologically important properties of yogurt. Food Sci. Nutr. 2017, 5, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Barzideh, Z.; Siddiqi, M.; Mohamed, H.M.; LaPointe, G. Dynamics of starter and non-starter lactic acid bacteria populations in long-ripened cheddar cheese using propidium monoazide (PMA) treatment. Microorganisms 2022, 10, 1669. [Google Scholar] [CrossRef] [PubMed]

| Product/Matrix | Species | Method(s) | Key Findings | References |
|---|---|---|---|---|
| Raw milk (multi-species) | Cow, sheep, goat, donkey, horse, camel, yak | 16S rRNA amplicon (V3–V4) | Clear inter-species differences: horse milk enriched in Bacteroidetes; sheep milk enriched in Gammaproteobacteria | [102] |
| Raw milk | Cow (bulk tank) | 16S rRNA amplicon | Pararhizobium and Agrobacterium abundances were found to be related to spring and Pseudomonas abundance was related to winter and spring seasons; core raw milk microbiota identified | [6] |
| Dairy chain (farm bulk tank → milk powder) | Cow | 16S rRNA + shotgun metagenomics | Microbiota tracked along the processing chain; day-to-day variability noted | [103] |
| Retail raw milk | Cow | Shotgun metagenomics | Dominated by Pseudomonadaceae (mainly Pseudomonas spp.); LAB at very low levels. At 4 °C microbiota remained stable; at room temperature rapid fermentation and bacterial growth occurred Very low LAB; retail raw milk is a reservoir of ARGs | [104] |
| Raw milk | Holstein; monthly samples for 12 months | 16S rRNA gene amplicon sequencing (high-throughput, Illumina) | Dominant genera: Pseudomonas, Lactococcus, Acinetobacter; also Firmicutes, Proteobacteria, Actinobacteria at the phylum level | [105] |
| Raw milk | Cow (bulk tank milk) | Culture-dependent isolation (>500 colonies) + 16S rRNA amplicon sequencing (Illumina MiSeq) | Culture revealed ~70–110 bacterial species; Gram(+) dominant genera: Staphylococcus, Corynebacterium, Streptococcus, Janibacter; Gram(−): Chryseobacterium, Acinetobacter. 16S sequencing detected anaerobes and hard-to-culture species, increasing diversity. | [106] |
| Raw ewe milk | Sheep (Assaf) | 16S rRNA amplicon | Core genera: Staphylococcus, Lactobacillus, Corynebacterium, Streptococcus, Escherichia/Shigella | [107] |
| Raw ewe milk | Sheep (Assaf, Lacaune, Merino) | 16S rRNA amplicon (V3–V4) | Lactobacillus, Jeotgalicoccus, Enterococcus, Corynebacterium | [24] |
| Raw goat milk | Saanen vs. Guanzhong breeds | 16S rRNA gene amplicon sequencing (Illumina, V3–V4) | Proteobacteria dominant phylum (~71%); Enterobacter (~25%) most abundant genus. Saanen goat milk had higher levels of lactose-fermenting LAB (Lactococcus, Lactobacillus, Bifidobacterium, Streptococcus) compared to Guanzhong. | [108] |
| Milk kefir and kefir grains | Cow milk kefir | 16S rRNA (V1–V3 region) pyrosequencing (bacteria) + 26S rDNA pyrosequencing (yeasts) | 16S: ~20 major bacterial species detected; dominant: Lb. kefiranofaciens, Lc. lactis ssp. cremoris, Gluconobacter frateurii, Lb. kefiri, Acetobacter orientalis, A. lovaniensis. 3 main yeasts in grains: Naumovozyma spp., Kluyveromyces marxianus, Kazachastania khefir. | [109] |
| Kefir (home-made vs. industrial) | Cow milk kefir | Culture + qPCR + 16S/ITS amplicon | Grains: Lb. kefiranofaciens, Lb. kefiri; Beverage: Streptococcus, Lactobacillus, Lactococcus; yeasts: Kluyveromyces, Debaryomyces | [46] |
| Kefir (home-made vs. industrial) | Cow milk kefir | 16S rRNA amplicon (V3–V4) | Industrial: Lactococcus and Streptococcus. Home-made: Lactobacillus | [98] |
| Kefir (global collection) | Cow milk kefir | Shotgun metagenomics (pan-metagenome) | Global kefir community types; recurrent LAB (Lb. kefiranofaciens, Lb. kefiri) and yeasts (Kazachstania) | [43] |
| Kefir (Brazil) | Cow milk kefir | Shotgun metagenomics + peptidomics | Species-level LAB profiling; bioactive peptide repertoire described | [110] |
| Yogurt (back-slopped, Turkey) | Cow | 16S rRNA amplicon | Firmicutes-dominated; potential next-generation probiotics highlighted | [111] |
| Cheese | Raw milk cheese production chain (cow teat skin → milk → cheese) | 16S rRNA gene amplicon sequencing (Illumina) | Teat skin microbiota is an important source for milk and cheese microbiota. 85% of raw milk and 27% of ripened cheeses contained teat skin-derived OTUs. Shared taxa included Micrococcales, Staphylococcaceae, and LAB important for cheese flavor. Grazing influenced subdominant LAB levels in cheese. | [112] |
| Cheese plants and cheeses | Cow (industrial plants) | Shotgun metagenomics | Dairy plant microbiomes harbor flavor- and probiotic-related genes; microbial transfer networks from environment to product | [113] |
| Method | Description | Advantages | Disadvantages |
|---|---|---|---|
| Culture-dependent methods (classical microbiology) | Isolation of microorganisms on selective media, colony morphology, biochemical tests. | Allows isolation and characterization of live strains; enables testing of probiotic potential; antimicrobial susceptibility testing possible | Detects only cultivable organisms (many species cannot be cultured); underestimates diversity; time-consuming |
| Molecular PCR-based techniques (16S rRNA PCR, qPCR, RT-PCR) | Amplification and analysis of target genes (mainly 16S rRNA). | Rapid and sensitive; can detect low-abundance organisms; culture-independent detection possible | Not always species-level resolution; sensitive to contamination; limited to known taxa with available primers |
| DGGE/TGGE (Denaturing/Temperature Gradient Gel Electrophoresis) | Separation of PCR products on gradient gels to profile communities. | Provides an overview of community structure and diversity; multiple taxa detectable simultaneously | Low resolution; rare species often missed; time consuming |
| T-RFLP (Terminal Restriction Fragment Length Polymorphism) | Fluorescent-labeled PCR fragments digested with restriction enzymes and analyzed. | Rapid overview of microbial community; provides semi-quantitative diversity information | Limited species-level resolution; band interpretation difficult in complex samples |
| Microarray (PhyloChip, LAB-Chip) | Hybridization of target DNA fragments to taxon-specific probes. | Simultaneous analysis of many taxa; high-throughput and relatively fast | Detects only known species with probes; cannot discover novel taxa |
| NGS (Next-Generation Sequencing—16S rRNA amplicon sequencing) | High-throughput sequencing of specific 16S rRNA regions (e.g., V3–V4). | Culture-independent, comprehensive profiling; detects rare taxa; enables calculation of diversity indices | Species/subspecies resolution sometimes limited; requires bioinformatics; relatively costly |
| Shotgun metagenomics | Sequencing of all genomic DNA from a sample. | Species- and even strain-level resolution; functional gene and metabolic pathway analysis; detects bacteria, fungi, and viruses simultaneously | High costs; computationally demanding; requires expertise in bioinformatics |
| Metatranscriptomics | Sequencing of total microbial RNA, focusing on expressed genes. | Reveals active functional genes; provides insight into metabolic activity | RNA degrades easily; technically challenging; high costs |
| Metaproteomics and Metabolomics | Profiling of microbial proteins and metabolites using MS. | Direct functional readout; links to fermentation quality and aroma compounds | Complex sample preparation; expensive; lack of standardization |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biçer, Y.; Telli, A.E.; Turkal, G.; Telli, N.; Uçar, G. From Raw to Fermented: Uncovering the Microbial Wealth of Dairy. Fermentation 2025, 11, 552. https://doi.org/10.3390/fermentation11100552
Biçer Y, Telli AE, Turkal G, Telli N, Uçar G. From Raw to Fermented: Uncovering the Microbial Wealth of Dairy. Fermentation. 2025; 11(10):552. https://doi.org/10.3390/fermentation11100552
Chicago/Turabian StyleBiçer, Yusuf, Arife Ezgi Telli, Gamze Turkal, Nihat Telli, and Gürkan Uçar. 2025. "From Raw to Fermented: Uncovering the Microbial Wealth of Dairy" Fermentation 11, no. 10: 552. https://doi.org/10.3390/fermentation11100552
APA StyleBiçer, Y., Telli, A. E., Turkal, G., Telli, N., & Uçar, G. (2025). From Raw to Fermented: Uncovering the Microbial Wealth of Dairy. Fermentation, 11(10), 552. https://doi.org/10.3390/fermentation11100552






