Heterologous Expression and Adaptive Evolution of ε-Poly-lysine Synthase Gene in Corynebacterium glutamicum
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganism, Plasmids, and Primers
2.2. Culture and Fermentation Media Composition
2.3. Construction of the Recombinant Strains
2.4. Single Factor Optimization of Shake Flask Culture of Recombinant Bacterial Strains
- -
- Inoculum volume: 2%, 4%, 6%, 8%, and 10%.
- -
- Fermentation temperature: 26°C, 28°C, 30°C, 32°C, and 34°C.
- -
- Initial pH: 6.6, 6.8, 7.0, 7,2, 7.4.
- -
- Shaker speed: 140, 160, 180, 200, and 220 rpm.
2.5. Adaptive Evolution of Recombinant Strain
2.6. Analytical Methods
2.7. Statistical Analysis
3. Results
3.1. Construction of Recombinant C. glutamicum
3.2. Fermentation Yield Determination and Mass Spectrometry Identification of Recombinant Strains
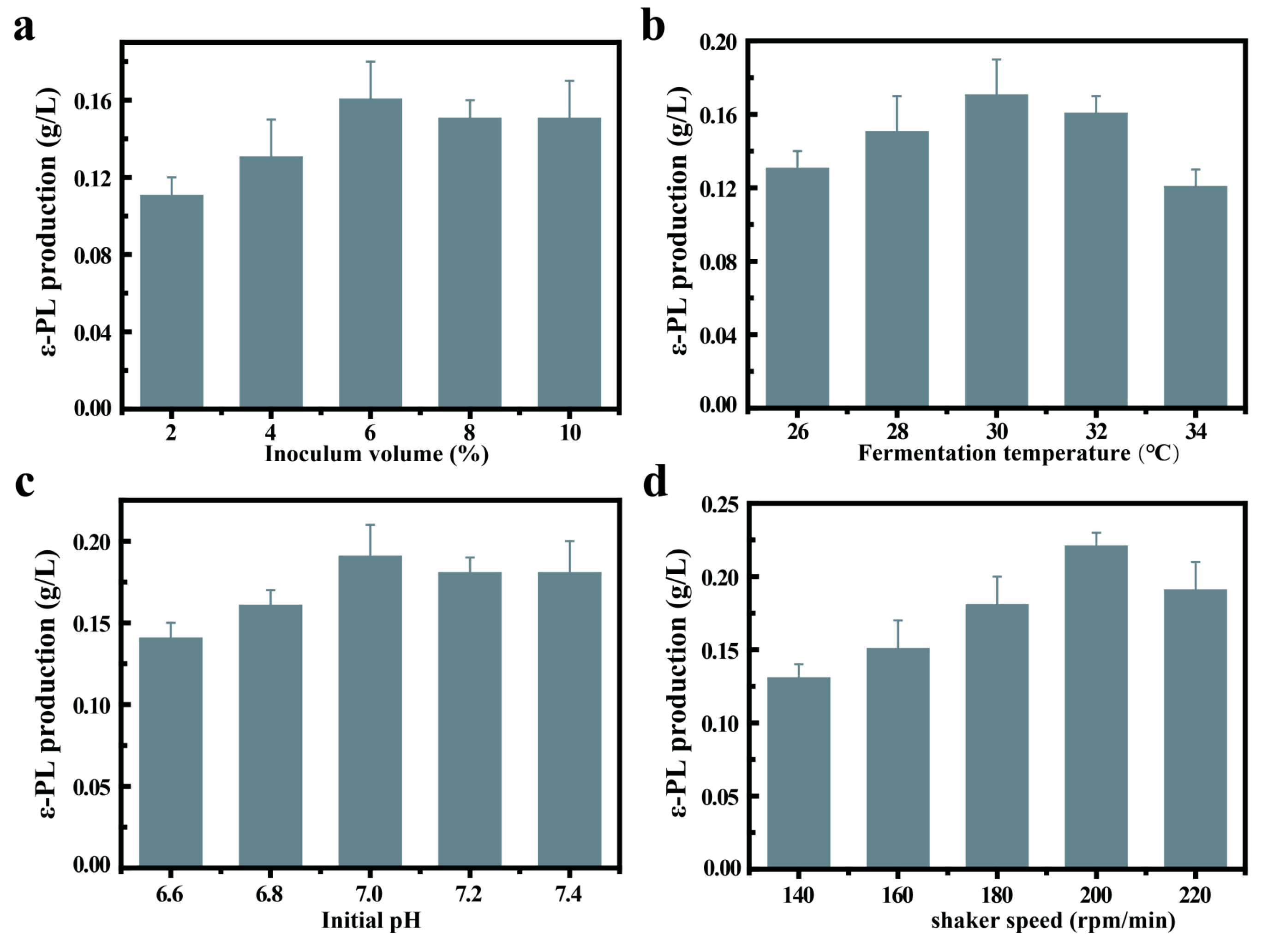
3.3. Single Factor Experiment of the Fermentation by Recombinant C. glutamicum-pXMJ19-pls
3.3.1. Effect of Inoculum Volume on ε-PL Production
3.3.2. Effect of Fermentation Temperature on ε-PL Production
3.3.3. Effect of Initial pH on ε-PL Production
3.3.4. Effect of Shaker Speed on ε-PL Production
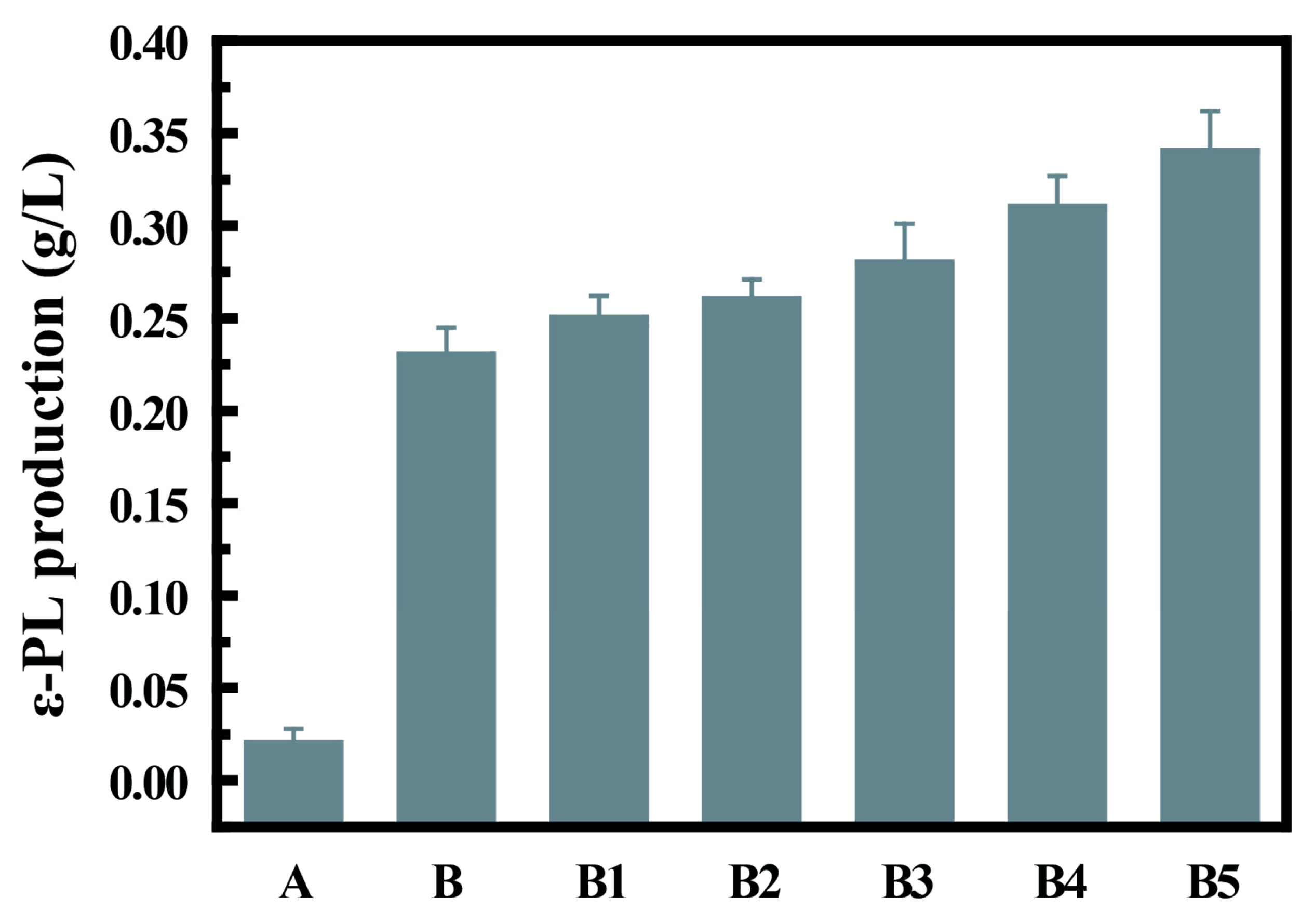
3.4. Adaptive Evolution and Fermentation Level Analysis of C. glutamicum-pXMJ19-pls
3.5. The Effect of Adaptive Evolution on the Growth of Recombinant Bacterial Strains and the Saturation of Cell Membrane Fatty Acids
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saimura, M.; Takehara, M.; Mizukami, S.; Kataoka, K.; Hirohara, H. Biosynthesis of nearly monodispersed poly(ε-l-lysine) in Streptomyces species. Biotechnol. Lett. 2008, 30, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-S.; Ren, X.-D.; Zeng, X.; Zhao, F.-L.; Tang, L.; Zhang, H.-J.; Zhang, J.-H.; Mao, Z.-G. Enhancement of ε-poly-l-lysine production coupled with precursor l-lysine feeding in glucose–glycerol co-fermentation by Streptomyces sp. M-Z18. Bioprocess Biosyst. Eng. 2013, 36, 1843–1849. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Nagasawa, T. ε-Poly-l-lysine: Microbial production, biodegradation and application potential. Appl. Microbiol. Biotechnol. 2003, 62, 21–26. [Google Scholar] [CrossRef]
- Chen, X.S.; Ren, X.D.; Dong, N.; Li, S.; Li, F.; Zhao, F.L.; Tang, L.; Zhang, J.H.; Mao, Z.G. Culture medium containing glucose and glycerol as a mixed carbon source improves ε-poly-l-lysine production by Streptomyces sp. M-Z18. Bioprocess Biosyst. Eng. 2012, 35, 469–475. [Google Scholar] [CrossRef]
- Xu, Z.; Feng, X.; Sun, Z.; Cao, C.; Li, S.; Xu, Z.; Xu, Z.; Bo, F.; Xu, H. Economic process to co-produce poly (ε-l-lysine) and poly (l-diaminopropionic acid) by a pH and dissolved oxygen control strategy. Bioresour. Technol. 2015, 187, 70–76. [Google Scholar] [CrossRef]
- Pan, L.; Chen, X.S.; Wang, K.F.; Mao, Z.G. Mechanisms of response to pH shock in microbial fermentation. Bioprocess Biosyst. Eng. 2020, 43, 361–372. [Google Scholar] [CrossRef]
- Liu, S.; Wu, Q.; Zhang, J.; Mo, S. Production of ε-poly-L-lysine by Streptomyces sp. using resin-based, in situ product removal. Biotechnol. Lett. 2011, 33, 1581–1585. [Google Scholar] [CrossRef]
- Kahar, P.; Kobayashi, K.; Iwata, T.; Hiraki, J.; Kojima, M.; Okabe, M. Production of ε-polylysine in an airlift bioreactor (ABR). J. Biosci. Bioeng. 2002, 93, 274–280. [Google Scholar] [CrossRef]
- Zhang, Y.; Feng, X.; Xu, H.; Yao, Z.; Ouyang, P. ε-Poly-L-lysine production by immobilized cells of Kitasatospora sp. MY 5-36 in repeated fed-batch cultures. Bioresour. Technol. 2010, 101, 5523–5527. [Google Scholar] [CrossRef]
- Xu, D.; Yao, H.; Xu, Z.; Wang, R.; Xu, Z.; Li, S.; Feng, X.; Liu, Y.; Xu, H. Production of ε-poly-lysine by Streptomyces albulus PD-1 via solid-state fermentation. Bioresour. Technol. 2017, 223, 149–156. [Google Scholar] [CrossRef]
- Kahar, P.; Iwata, T.; Hiraki, J.; Park, E.Y.; Okabe, M. Enhancement of ε-polylysine production by Streptomyces albulus strain 410 using pH control. J. Biosci. Bioeng. 2001, 91, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.-D.; Chen, X.-S.; Zeng, X.; Wang, L.; Tang, L.; Mao, Z.-G. Acidic pH shock induced overproduction of ε-poly-l-lysine in fed-batch fermentation by Streptomyces sp. M-Z18 from agro-industrial by-products. Bioprocess Biosyst. Eng. 2015, 38, 1113–1125. [Google Scholar] [CrossRef] [PubMed]
- Geng, W.; Yang, C.; Gu, Y.; Liu, R.; Wang, S. Cloning of ε-poly-L-lysine (ε-PL) synthetase gene from a newly isolated ε-PL-producing Streptomyces albulus NK660 and its heterologous expression in Streptomyces lividans. Microb. Biotechnol. 2014, 7, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Claudine, I.K.M.; Qiao, Z.; Xu, M.; Chen, X.S.; Yang, T.W.; Shao, M.L.; Rao, Z.M. Heterologous expression of Streptomyces albulus ε-poly-L-lysine synthase and optimization of whole-cell biocatalyst for ε-poly-L-lysine synthesis. Ind. Food Ferment. 2020, 46, 6. [Google Scholar]
- Wieschalka, S.; Blombach, B.; Bott, M.; Eikmanns, B.J. Bio-based production of organic acids with Corynebacterium glutamicum. Microb. Biotechnol. 2013, 6, 631–640. [Google Scholar] [CrossRef]
- Henke, N.A.; Wiebe, D.; Pérez-García, F.; Peters-Wendisch, P.; Wendisch, V.F. Coproduction of cell-bound and secreted value-added compounds: Simultaneous production of carotenoids and amino acids by Corynebacterium glutamicum. Bioresour. Technol. Biomass Bioenergy Biowastes Convers. Technol. Biotransformations Prod. Technol. 2018, 9, 167. [Google Scholar] [CrossRef]
- Egbune, E.O.; Ezedom, T.; Odeghe, O.B.; Osuvwe, C.O.; Egbune, O.U.; Ehwarieme, A.D.; Aganbi, E.; Ebuloku, C.S.; Chukwuegbo, A.O.; Bogard, E.; et al. Solid-state fermentation production of L-lysine by Corynebacterium glutamicum (ATCC 13032) using agricultural by-products as substrate. World J. Microbiol. Biotechnol. 2024, 40, 20. [Google Scholar] [CrossRef]
- Pan, L.; Chen, X.; Wang, K.; Mao, Z. A temporal transcriptomic dynamics study reveals the reason of enhanced ε-poly-L-lysine production in Streptomyces albulus M-Z18 by pH shock. Process Biochem. 2019, 85, 1–11. [Google Scholar] [CrossRef]
- Ren, X.D.; Chen, X.S.; Tang, L.; Zeng, X.; Wang, L.; Mao, Z.G. Physiological mechanism of the overproduction of ε-poly-l-lysine by acidic pH shock in fed-batch fermentation. Bioprocess Biosyst. Eng. 2015, 38, 2085–2094. [Google Scholar] [CrossRef]
- Itzhaki, R.F. Calorimetric method for estimating polylysine and poly-arginine. Anal. Biochem. 1972, 50, 569–574. [Google Scholar] [CrossRef]
- Nishikawa, M.; Kobayashi, K. Streptomyces roseoverticillatus produces two different poly(amino acid)s: Lariat-shaped gamma-poly(L-glutamic acid) and epsilon-poly(L-lysine). Microbiology 2009, 155, 2988–2993. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Xu, P.; Sun, J.; Yuan, J.; Zhao, J. Effects of ethanol stress on epsilon-poly-l-lysine (ε-PL) biosynthesis in Streptomyces albulus X-18. Enzym. Microb. Technol. 2022, 153, 109907. [Google Scholar] [CrossRef] [PubMed]
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids; MIDI Inc.: Newark, DE, USA, 1990. [Google Scholar]
- Du, H.; Qiao, J.; Qi, Y.; Li, L.; Xu, N.; Shao, L.; Wei, L.; Liu, J. Reprogramming the sulfur recycling network to improve L-cysteine production in Corynebacterium glutamicum. Green Chem. 2023, 8, 3152–3165. [Google Scholar] [CrossRef]
- Mao, Z.G.; Sun, Q.X.; Ren, X.D.; Chen, X.S. Improvement of epsilon-Poly-L-Lysine Production Through Seed Stage Development Based on In Situ pH Monitoring. Appl. Biochem. Biotechnol. Part A. Enzym. Eng. Biotechnol. 2015, 175, 802–812. [Google Scholar]
- Hamushan, M.; Yu, J.; Jiang, F.; Wang, B.; Li, M.; Hu, Y.; Wang, J.; Wu, Q.; Tang, J.; Han, P. Adaptive evolution of the Clf-Sdr subfamily contributes to Staphylococcus aureus musculoskeletal infection: Evidence from comparative genomics. Microbiol. Res. 2024, 278, 127502. [Google Scholar] [CrossRef]
- Liu, Q.; Lu, M.; Jin, C.; Hou, W.; Zhao, L.; Bao, J. Ultra-centrifugation force in adaptive evolution changes the cell structure of oleaginous yeast Trichosporon cutaneum into a favorable space for lipid accumulation. Biotechnol. Bioeng. 2022, 119, 1509–1521. [Google Scholar] [CrossRef]
- Bromig, L.; Weuster-Botz, D. Accelerated Adaptive Laboratory Evolution by Automated Repeated Batch Processes in Parallelized Bioreactors. Microorganisms 2023, 11, 275. [Google Scholar] [CrossRef]
- Bedau, M.A.; Packard, N.H. Evolution of evolvability via adaptation of mutation rates. Biosystems 2003, 69, 143–162. [Google Scholar] [CrossRef]
- Wang, Y.; Manow, R.; Finan, C.; Wang, J.; Garza, E.; Zhou, S. Adaptive evolution of nontransgenic Escherichia coli KC01 for improved ethanol tolerance and homoethanol fermentation from xylose. J. Ind. Microbiol. Biotechnol. 2011, 38, 1371–1377. [Google Scholar] [CrossRef]
- Cowen, L.E.; Sanglard, D.; Calabrese, D.; Sirjusingh, C.; Anderson, J.B.; Kohn, L.M. Evolution of Drug Resistance in Experimental Populations of Candida albicans. J. Bacteriol. 2000, 182, 1515–1522. [Google Scholar] [CrossRef]
- Fong, S.S.; Burgard, A.P.; Herring, C.D.; Knight, E.M.; Palsson, B.O. In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnol. Bioeng. 2010, 91, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.M.; Rock, C.O. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 2008, 6, 222. [Google Scholar] [CrossRef] [PubMed]
- Giotis, E.S.; Mcdowell, D.A.; Blair, I.S.; Wilkinson, B.J. Role of Branched-Chain Fatty Acids in pH Stress Tolerance in Listeria monocytogenes. Appl. Environ. Microbiol. 2007, 73, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Kolbeck, S.; Behr, J.; Vogel, R.F.; Ludwig, C.; Ehrmann, M.A. Acid stress response of Staphylococcus xylosus elicits changes in the proteome and cellular membrane. J. Appl. Microbiol. 2019, 126, 1480–1495. [Google Scholar] [CrossRef]
- Wang, L.; Li, S.; Zhao, J.J.; Chen, X.S.; Mao, Z.G. Efficiently activated ε-poly-L-lysine production by multiple antibiotic-resistance mutations and acidic pH shock optimization in Streptomyces albulus. MicrobiologyOpen 2019, 3, e00728. [Google Scholar] [CrossRef]
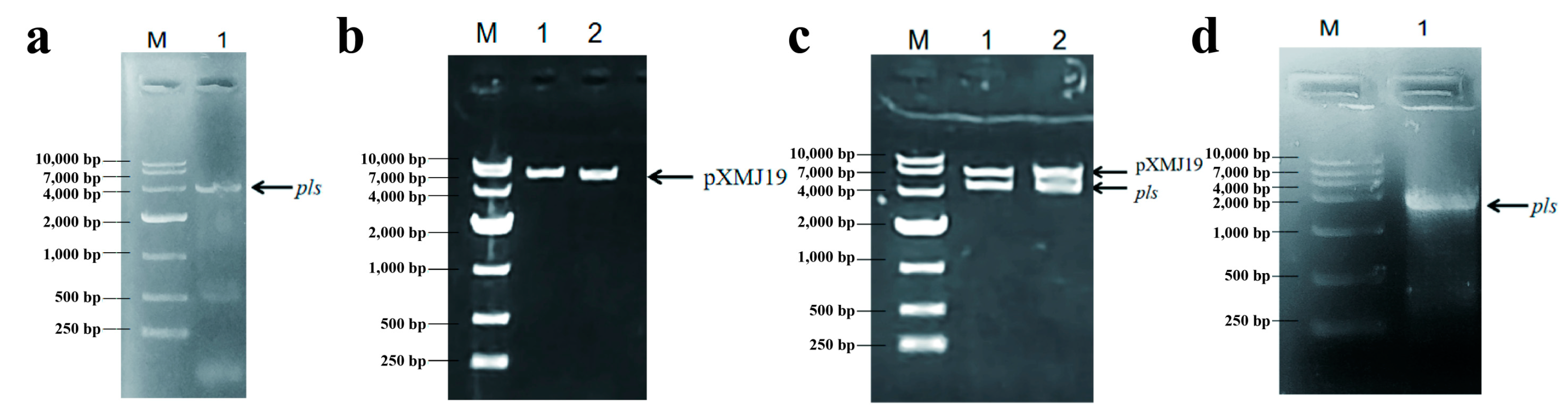
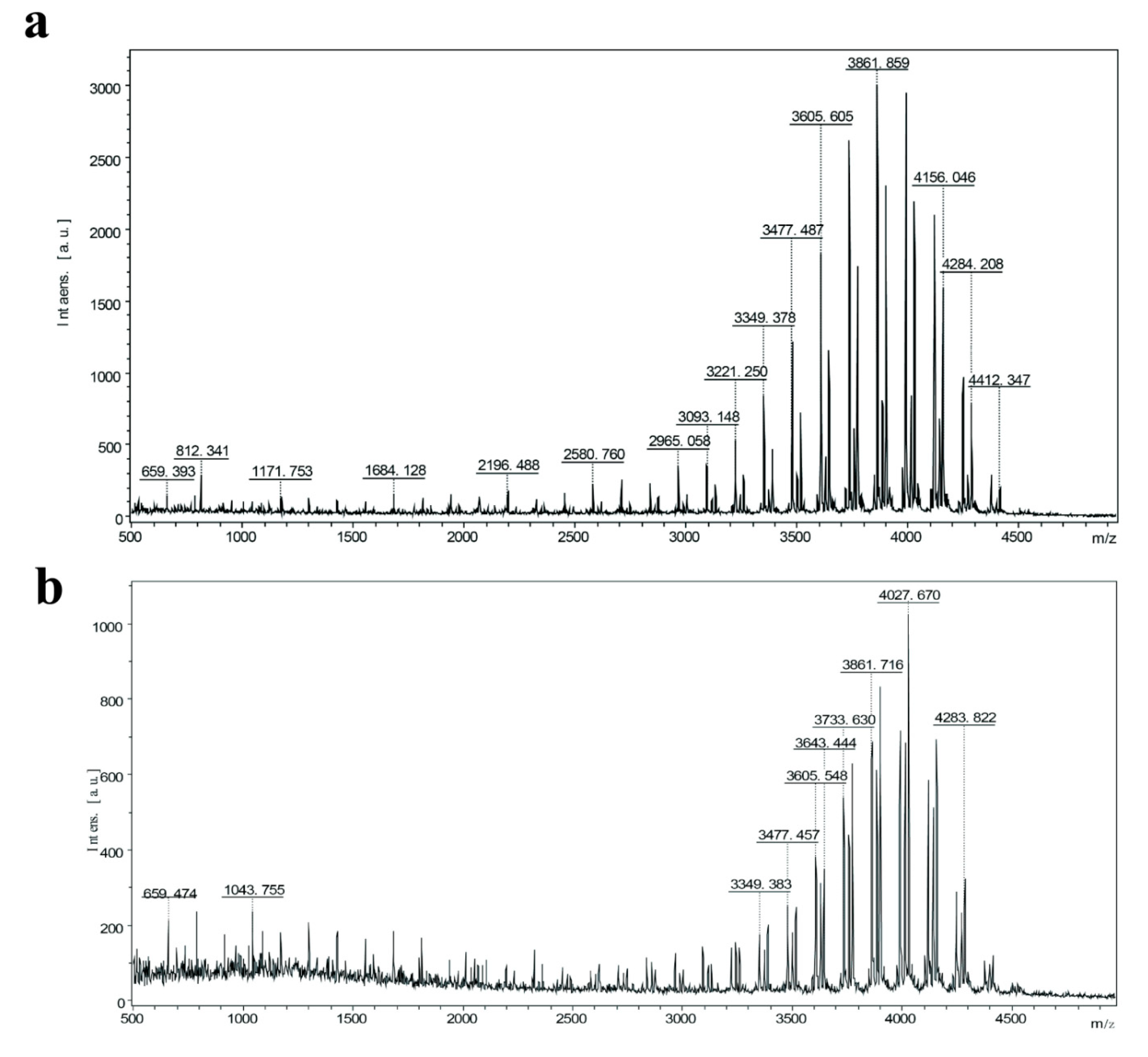
| Bacterial Strains/Plasmids/Primers | Function | Source |
|---|---|---|
| Bacterial strains | ||
| S. albulus IFO 14147 | Provide genomic templates | CICC |
| C. glutamicum CICC 10064 | Heterologous expression of host bacteria | CICC |
| C. glutamicum-pXMJ19 | C. glutamicum with empty plasmid | Constructed in this study |
| C. glutamicum-pXMJ19-pls | C. glutamicum heterologous expression with pls | Constructed in this study |
| E. coli DH5α | Host bacteria for clone | Wuhan Miaoling Biotechnology Co., Ltd. |
| Plasmids | ||
| pXMJ19 | Used for heterologous expression vector of C. glutamicum | Wuhan Miaoling Biotechnology Co., Ltd. |
| pXMJ19-pls | Heterologous expression of pls genes in Corynebacterium glutamicum | Constructed in this study |
| Primers | ||
| pls-F | tgcctgcaggtcgactctagaATGTCCAGCCCGCTGCTG | Suzhou Junji Biotechnology Co., Ltd. |
| pls-R | caaaacagccaagctgaattcTGCCGCAGCACCACCTTC | Suzhou Junji Biotechnology Co., Ltd. |
| Strains * | Biomass (OD600) | Membrane Unsaturation (%) |
|---|---|---|
| A | 2.68 ± 0.23 d | 12.63 ± 1.15 c |
| B | 2.52 ± 0.21 de | 11.74 ± 1.69 c |
| B1 | 2.76 ± 0.13 c | 12.88 ± 0.52 c |
| B2 | 2.98 ± 0.11 ab | 14.51 ± 1.76 b |
| B3 | 3.14 ± 0.17 a | 16.14 ± 2.03 ab |
| B4 | 2.94 ± 0.09 ab | 17.33 ± 1.42 a |
| B5 | 2.82 ± 0.12 b | 18.56 ± 2.13 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, L.; Chen, Y.; Liao, A.; Hou, Y.; Huang, J. Heterologous Expression and Adaptive Evolution of ε-Poly-lysine Synthase Gene in Corynebacterium glutamicum. Fermentation 2025, 11, 11. https://doi.org/10.3390/fermentation11010011
Pan L, Chen Y, Liao A, Hou Y, Huang J. Heterologous Expression and Adaptive Evolution of ε-Poly-lysine Synthase Gene in Corynebacterium glutamicum. Fermentation. 2025; 11(1):11. https://doi.org/10.3390/fermentation11010011
Chicago/Turabian StylePan, Long, Yihang Chen, Aimei Liao, Yinchen Hou, and Jihong Huang. 2025. "Heterologous Expression and Adaptive Evolution of ε-Poly-lysine Synthase Gene in Corynebacterium glutamicum" Fermentation 11, no. 1: 11. https://doi.org/10.3390/fermentation11010011
APA StylePan, L., Chen, Y., Liao, A., Hou, Y., & Huang, J. (2025). Heterologous Expression and Adaptive Evolution of ε-Poly-lysine Synthase Gene in Corynebacterium glutamicum. Fermentation, 11(1), 11. https://doi.org/10.3390/fermentation11010011






