In Vitro Probiotic Characterization of Lactiplantibacillus plantarum Strains Isolated from Traditional Fermented Dockounou Paste
Abstract
1. Introduction
2. Materials and Methods
2.1. LAB Strains and Growth Conditions
2.2. Identification of LAB Strains via 16S rDNA Sequencing
2.3. In Vitro Tolerance of LAB Strains to Simulated Gastrointestinal Conditions
2.3.1. Resistance to Low pH
2.3.2. Resistance to Pepsin and Low pH
2.3.3. Tolerance to Bile Salts
2.3.4. Phenol Tolerance
2.4. Safety Assessment
2.4.1. Hemolysis Test
2.4.2. Antibiotic Sensitivity
2.5. Cell Surface Characteristics
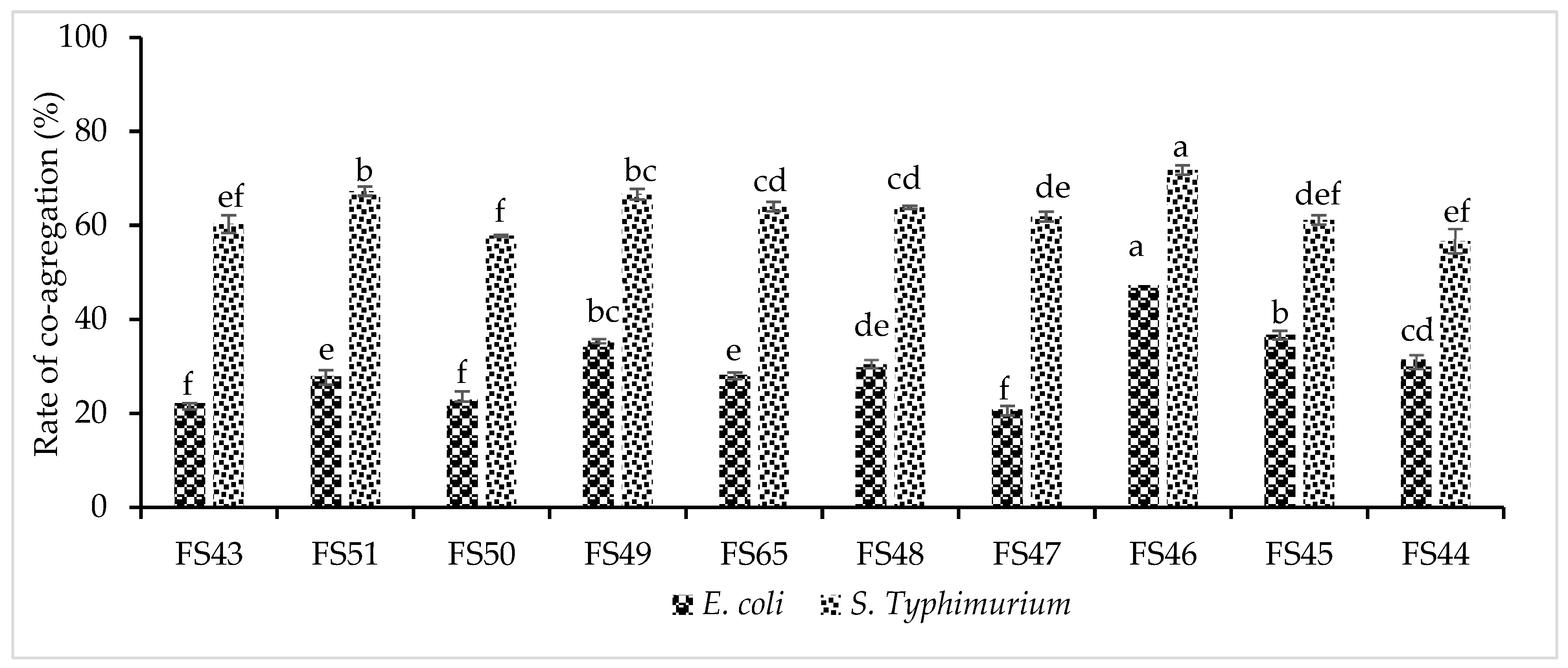
2.5.1. Co-Aggregation Test
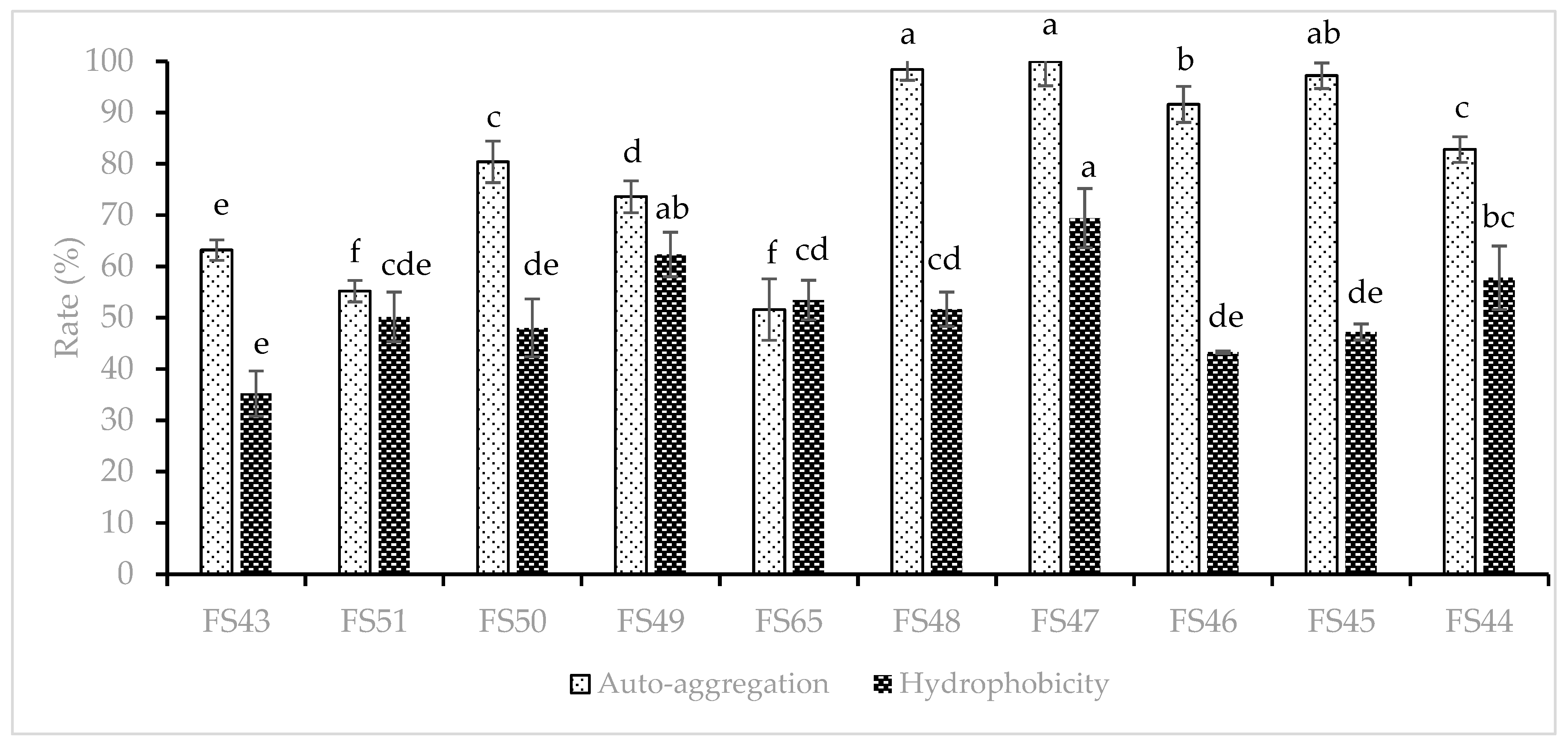
2.5.2. Cell Surface Hydrophobicity
2.5.3. Auto Aggregation Test
2.6. Study of the Probiotic Properties of Lactic Acid Bacteria
2.6.1. Antibacterial Activity
2.6.2. DPPH Free Radical Scavenging Activity
2.6.3. Plate Screening of LAB Isolates for Hydrolytic Enzymes
2.7. Statistical Analysis
3. Results
3.1. Identification of LAB Isolates
3.2. Safety Criteria of LAB Strains
3.3. Exploration of Probiotic Properties
3.3.1. Resistance to Simulated Gastrointestinal Tract Conditions
3.3.2. Cell Surface Characteristics
3.4. Exploration of Probiotic Properties
3.4.1. Antibacterial Spectrum
3.4.2. Antioxidant activity
3.4.3. Enzymatic Activity of LAB Strains
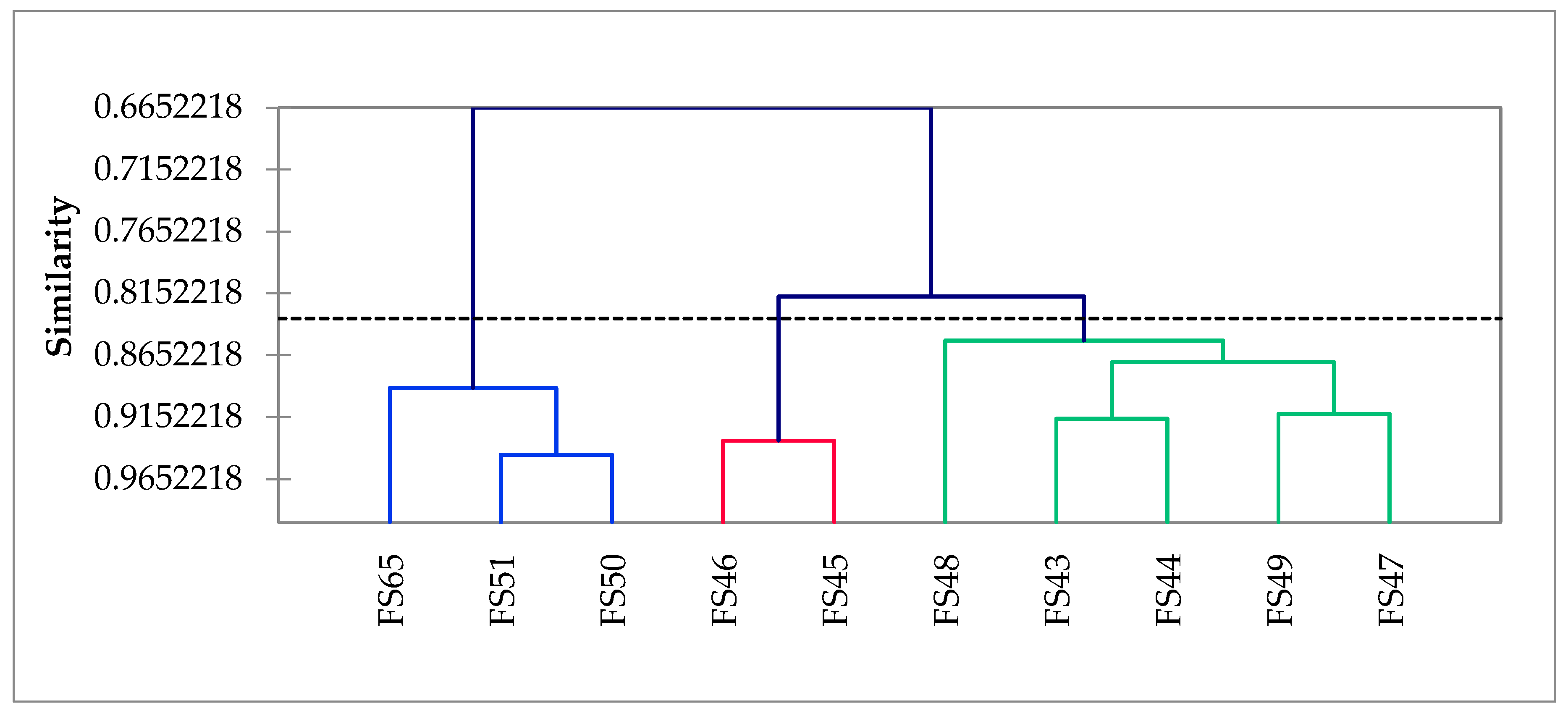
3.5. Principal Component Analysis (PCA)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kra, K.A.S.; Akoa, E.E.F.; Mégnanou, R.M.; Yéboué, K.; Akpa, E.E.; Niamké, S.L. Physicochemical and nutritional characteristics assessments of two different traditional foods prepared with senescent plantain. Afr. J. Food Sci. 2013, 7, 51–55. [Google Scholar]
- Akoa, E.E.F.; Kra, K.A.S.; Mégnanou, R.M.; Kouadio, N.J.; Niamké, L.S. Optimization of dockounou manufacturing process parameters. Sustain. Agric. Res. 2014, 3, 67–75. [Google Scholar]
- Carboni, A.D.; Martins, G.N.; Gómez-Zavaglia, A.; Castilho, P.C. Lactic acid bacteria in the production of traditional fermented foods and beverages of Latin America. Fermentation 2023, 9, 315. [Google Scholar] [CrossRef]
- Petrova, P.; Petrov, K. Lactic acid fermentation of cereals and pseudocereals: Ancient nutritional biotechnologies with modern applications. Nutrients 2020, 12, 1118. [Google Scholar] [CrossRef]
- Sionek, B.; Szydłowska, A.; Küçükgöz, K.; Kołożyn-Krajewska, D. Traditional and new microorganisms in lactic acid fermentation of food. Fermentation 2023, 9, 1019. [Google Scholar] [CrossRef]
- Kayitesi, E.; Onojakpor, O.; Moyo, S.M. Highlighting the impact of lactic-acid-bacteria-derived flavours or aromas on sensory perception of african fermented cereals. Fermentation 2023, 9, 111. [Google Scholar] [CrossRef]
- Achi, O.K.; Asamudo, N.U. Cereal-based fermented foods of Africa as functional foods. Bioact. Mol. Food 2018, 1–32. [Google Scholar]
- Malongane, F.; Berejena, T. Exploring the microbiome present in fermented indigenous African foods and their potential impact on human health. J. Agric. Food Res. 2024, 16, 101101. [Google Scholar] [CrossRef]
- Assohoun, M.C.N.; Djéni, T.N.; N’Guessan, F.K.; Koussemon, M. Preliminary study on antimicrobial properties of lactic acid bacteria involved in the fermentation of corn dough during doklu processing in Côte D’Ivoire. Food 2012, 6, 65–70. [Google Scholar]
- Soro-Yao, A.A.; Kouakou, B.; Koffi-Nevry, R.; Djè, K.M. Microbiology of Ivorian fermented products: A review. Asian J. Agric. Food Sci. 2013, 1, 37–47. [Google Scholar]
- Kouadio, N.J.; Goualie, G.B.; Ouattara, G.H.; Kra, K.A.S.; Niamke, L.S. Microorganisms associated with traditional plantain-based food “dockounou” during spontaneous fermentation. Food Environ. Saf. 2014, 13, 276–282. [Google Scholar]
- Assohoun-Djeni, N.M.C.; Djeni, N.T.; Messaoudi, S.; Lhomme, E.; Koussemon-Camara, M.; Ouassa, T.; Chobert, J.M.; Onno, B.; Dousset, X. Biodiversity, dynamics and antimicrobial activity of lactic acid bacteria involved in the fermentation of maize flour for doklu production in Côte d’Ivoire. Food Control 2016, 62, 397–404. [Google Scholar] [CrossRef]
- Mokoena, M.P.; Mutanda, T.; Olaniran, A.O. Perspectives on the probiotic potential of lactic acid bacteria from African traditional fermented foods and beverages. Food Nutr. Res. 2016, 60, 29630. [Google Scholar] [CrossRef] [PubMed]
- Adesulu-Dahunsi, A.T.; Dahunsi, S.O.; Ajayeoba, T.A. Co-occurrence of Lactobacillus species during fermentation of african indigenous foods: Impact on food safety and shelf-life extension. Front. Microbiol. 2022, 13, 684730. [Google Scholar] [CrossRef] [PubMed]
- Mogmenga, I.; Somda, M.K.; Ouattara, C.A.T.; Keita, I.; Dabiré, Y.; Diguță, C.F.; Toma, R.C.; Ezeogu, L.I.; Ugwuanyi, J.O.; Ouattara, A.S.; et al. Promising Probiotic Properties of the Yeasts Isolated from Rabilé, a Traditionally Fermented Beer Produced in Burkina Faso. Microorganisms 2023, 11, 802. [Google Scholar] [CrossRef] [PubMed]
- Grosu-Tudor, S.; Stefan, I.; Stancu, M.; Cornea, C.; De vuyst, L.; Zamfir, M. Microbial and nutritional characteristics of fermented wheat bran in traditional Romanian borş production. Rom. Biotechnol. Lett. 2018, 24, 440–447. [Google Scholar] [CrossRef]
- Constantin, E.A.; Constantinescu-Aruxandei, D.; Matei, F.; Shaposhnikov, S.; Oancea, F. Biochemical and microbiological characterization of traditional Romanian fermented drinks—Socata and borș—A review. AgroLife Sci. J. 2023, 12, 53–61. [Google Scholar] [CrossRef]
- Ștefan, I.R.; Grosu-Tudor, S.S.; Zamfir, M.; Cornea, C.P. Screening for S-layer production by some lactobacilli from home-made fermented foods. Sci. Bull. Ser. F Biotechnol. 2016, XX, 167–171. [Google Scholar]
- Angelescu, I.R.; Zamfir, M.; Stancu, M.M.; Grosu-Tudor, S.S. Identification and probiotic properties of lactobacilli isolated from two different fermented beverages. Ann. Microbiol. 2019, 69, 1557–1565. [Google Scholar] [CrossRef]
- Grosu-Tudor, S.S.; Zamfir, M.; Van der Meulen, R.; De Vuyst, L. Isolation of novel homopolysaccharide-producing lactic acid bacteria from Romanian raw milk and fermented dairy products. Eur. Food Res. Technol. 2013, 237, 609–615. [Google Scholar] [CrossRef]
- Petruţ, Ș.M.; Sârbu, I.; Corbu, V.M.; Pelinescu, D.; Iftime, I.; Dimov, T.V. Screening of lactic acid bacteria from spontaneously fermented products of Romania. Rom. Biotechnol. Lett. 2019, 24, 254–260. [Google Scholar] [CrossRef]
- Grosu-Tudor, S.S.; Zamfir, M. Functional properties of lactic acid bacteria isolated from Romanian fermented vegetables. Food Biotechnol. 2013, 27, 235–248. [Google Scholar] [CrossRef]
- Cornea, C.P.; Israel Roming, F.; Sicuia, O.A.; Voaideș, C.; Zamfir, M.; Grosu-Tudor, S.S. Biosurfactant production by Lactobacillus spp. strains isolated from Romanian traditional fermented food products. Rom. Biotechnol. Lett. 2016, 21, 11312–11320. [Google Scholar]
- FAO/WHO. Guidelines for the Evaluation of Probiotics in Food; Food and Agriculture Organization of the United Nations and World Health Organization Working Group Report; FAO/WHO: London, ON, Canada, 2002. [Google Scholar]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- De Melo Pereira, G.V.; de Oliveira Coelho, B.; Magalhães Júnior, A.I.; Thomaz-Soccol, V.; Soccol, C.R. How to select a probiotic? A review and update of methods and criteria. Biotechnol. Adv. 2018, 36, 2060–2076. [Google Scholar] [CrossRef] [PubMed]
- Chou, L.S.; Weimer, B. Isolation and characterization of acid- and bile-tolerant isolates from strains of Lactobacillus acidophilus. J. Dairy Sci. 1999, 82, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Coulibaly, W.H.; Kouadio, N.R.; Camara, F.; Diguță, C.; Matei, F. Functional properties of lactic acid bacteria isolated from Tilapia (Oreochromis niloticus) in Ivory Coast. BMC Microbiol. 2023, 23, 152. [Google Scholar] [CrossRef] [PubMed]
- Meena, K.K.; Taneja, N.K.; Jain, D.; Ojha, A.; Kumawat, D.; Mishra, V. In vitro assessment of probiotic and technological properties of lactic acid bacteria isolated from indigenously fermented cereal-based food products. Fermentation 2022, 8, 529. [Google Scholar] [CrossRef]
- Ayivi, R.D.; Gyawali, R.; Krastanov, A.; Aljaloud, S.O.; Worku, M.; Tahergorabi, R.; Silva, R.C.D.; Ibrahim, S.A. Lactic Acid Bacteria: Food Safety and Human Health Applications. Dairy 2020, 1, 202–232. [Google Scholar] [CrossRef]
- Picard, C.; Fioramonti, J.; Francois, A.; Robinson, T.; Neant, F.; Matuchansky, C. Review article: Bifidobacteria as probiotic agents–physiological effects and clinical benefits. Aliment. Pharmacol. Ther. 2005, 22, 495–512. [Google Scholar] [CrossRef]
- Chen, J.; Chen, X.; Ho, C.L. Recent development of probiotic bifidobacteria for treating human diseases. Front. Bioeng. Biotechnol. 2021, 9, 770248. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Vauterin, L.; Vauterin, P. Computer-aided objective comparison of electrophoresis patterns for grouping and identification of microorganisms. Eur. Microbiol. 1992, 1, 37–41. [Google Scholar]
- Carr, F.J.; Chill, D.; Maida, N. The lactic acid bacteria: A literature survey. Crit. Rev. Microbiol. 2002, 28, 281–370. [Google Scholar] [CrossRef] [PubMed]
- Diguță, C.F.; Nițoi, G.D.; Matei, F.; Luță, G.; Cornea, C.P. The Biotechnological potential of Pediococcus spp. isolated from Kombucha microbial consortium. Foods 2020, 9, 1780. [Google Scholar] [CrossRef] [PubMed]
- Xanthopoulos, V.; Litopoulou-Tzanetaki, E.; Tzanetakis, N. Characterization of Lactobacillus isolates from infant faeces as dietary adjuncts. Food Microbiol. 2000, 17, 205–215. [Google Scholar] [CrossRef]
- Yadav, R.; Puniya, A.K.; Shukla, P. Probiotic Properties of Lactobacillus plantarum RYPR1 from an Indigenous Fermented Beverage Raabadi. Front. Microbiol. 2016, 7, 1683. [Google Scholar] [CrossRef] [PubMed]
- CLSI. CLSI Supplement M100S. Performance Standards for Antimicrobial Susceptibility Testing, 26th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Collado, M.C.; Meriluoto, J.; Salminen, S. Measurement of aggregation properties between probiotics and pathogens: In vitro evaluation of different methods. J. Microbiol. Methods 2007, 71, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, M.; Gutnick, D. and Rosenberg, E. Adherence of bacteria to hydrocarbons: A simple method for measuring cell-surface hydrophobicity. FEMS Microbiol. Lett. 1980, 9, 29–33. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.; Berset, C. Use of a free radical method to evaluate antioxidant activity. LWT 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Proca, I.G.; Diguta, C.F.; Jurcoane, S.; Matei, F. Screening of halotolerant bacteria producing hydrolytic enzymes with biotechnological applications. Sci. Bull. Ser. F Biotechnol. 2020, XXIV, 197–204. [Google Scholar]
- Sanni, A.; Morlon-Guyot, J.; Guyot, J.P. New efficient amylase-producing strains of Lactobacillus plantarum and L. fermentum isolated from different Nigerian traditional fermented foods. Int. J. Food Microbiol. 2002, 72, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Adesulu-Dahunsi, A.T.; Sanni, A.I.; Jeyaram, K.; Banwo, K. Genetic diversity of Lactobacillus plantarum strains from some indigenous fermented foods in Nigeria. LWT 2017, 82, 199–206. [Google Scholar] [CrossRef]
- Oguntoyinbo, F.A.; Narbad, A. Multifunctional properties of Lactobacillus plantarum strains isolated from fermented cereal foods. J. Funct. Foods 2015, 17, 621–631. [Google Scholar] [CrossRef]
- Mathara, J.M.; Schillinger, U.; Kutima, P.M.; Mbugua, S.K.; Guigas, C.; Franz, C.; Holzapfel, W.H. Functional properties of Lactobacillus plantarum strains isolated from Maasai traditional fermented milk products in Kenya. Curr. Microbiol. 2008, 56, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Omar, N.B.; Abriouel, H.; Lucas, R.; Martínez-Cañamero, M.; Guyot, J.-P.; Gálvez, A. Isolation of bacteriocinogenic Lactobacillus plantarum strains from Ben Saalga, a traditional fermented Gruel from Burkina Faso. Int. J. Food Microbiol. 2006, 112, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Muyanja, C.; Narvhus, J.A.; Treimo, J.; Langsrud, T. Isolation, characterisation and identification of lactic acid bacteria from bushera: A Ugandan traditional fermented beverage. Int. J. Food Microbiol. 2003, 80, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Aka, S.; N’Guessan, K.F.; Nanga, Y.Z.; Loukou, Y.G.; Mazabraud, A.I.; Djè, K.M. Characterization of Lactobacillus species isolated from mash, sour wort and tchapalo produced in Côte d’Ivoire. Food 2010, 4, 49–54. [Google Scholar]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP); Rychen, G.; Aquilina, G.; Azimonti, G.; Bampidis, V.; De Lourdes Bastos, M.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; Flachowsky, G.; et al. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018, 16, e05206. [Google Scholar]
- Sharma, P.; Tomar, S.K.; Goswami, P.; Sangwan, V.; Singh, R. Antibiotic resistance among commercially available probiotics. Food Res. Int. 2014, 57, 176–195. [Google Scholar] [CrossRef]
- Wong, A.; Ngu, D.Y.S.; Dan, L.A.; Ooi, A.; Lim, R.L.H. Detection of antibiotic resistance in probiotics of dietary supplements. Nutr. J. 2015, 14, 95. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Teng, D.; Mao, R.; Hao, Y.; Wang, X.; Wang, J. A critical review of antibiotic resistance in probiotic bacteria. Food Res. Int. 2020, 136, 109571. [Google Scholar] [CrossRef] [PubMed]
- Dou, W.; Abdalla, H.B.; Chen, X.; Sun, C.; Chen, X.; Tian, Q.; Wang, J.; Zhou, W.; Chi, W.; Zhou, X.; et al. ProbResist: A database for drug-resistant probiotic bacteria. Database 2022, 2022, baac064. [Google Scholar] [CrossRef] [PubMed]
- Lei, V.; Jakobsen, M. Microbiological characterization and probiotic potential of koko and koko sour water, African spontaneously fermented millet porridge and drink. J. Appl. Microbiol. 2004, 96, 384–397. [Google Scholar] [CrossRef] [PubMed]
- Negasi, A.; Fassil, A.; Asnake, D. In vitro evaluation of lactic acid bacteria isolated from traditional fermented Shamita and Kocho for their desirable characteristics as probiotics. Afr. J. Biotechnol. 2017, 16, 594–606. [Google Scholar] [CrossRef]
- Matei, B.; Salzat, J.; Diguță, C.F.; Cornea, C.P.; Luta, G.; Utoiu, E.R.; Matei, F. Lactic acid bacteria strains isolated from Kombucha with potential probiotic effect. Rom. Biotechnol. Lett. 2018, 23, 13592–13598. [Google Scholar]
- Xanthopoulos, V.; Hatzikamari, M.; Adamidis, T.; Tsakalidou, E.; Tzanetakis, N.; Litopoulou-Tzanetaki, E. Heterogeneity of Lactobacillus plantarum isolates from Feta cheese throughout ripening. J. Appl. Microbiol. 2000, 88, 1056–1064. [Google Scholar] [CrossRef]
- Divyashree, S.; Ramu, R.; Sreenivasa, M.Y. Evaluation of new candidate probiotic Lactobacillus strains isolated from a traditional fermented food- multigrain-millet dosa batter. Food Biosci. 2024, 57, 103450. [Google Scholar] [CrossRef]
- Krausova, G.; Hyrslova, I.; Hynstova, I. In Vitro Evaluation of adhesion capacity, hydrophobicity, and auto-aggregation of newly isolated potential probiotic strains. Fermentation 2019, 5, 100. [Google Scholar] [CrossRef]
- Kos, B.; Susković, J.; Vuković, S.; Simpraga, M.; Frece, J.; Matosić, S. Adhesion and Aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 2003, 94, 981–987. [Google Scholar] [CrossRef] [PubMed]
- García-Cayuela, T.; Korany, A.M.; Bustos, I.; de Cadinanos, L.P.G.; Requena, T.; Peláez, C.; Martínez-Cuesta, M.C. Adhesion abilities of dairy Lactobacillus plantarum strains showing an aggregation phenotype. Food Res. Int. 2014, 57, 44–50. [Google Scholar] [CrossRef]
- Yasmin, I.; Saeed, M.; Khan, W.A.; Khaliq, A.; Chughtai, M.F.J.; Iqbal, R.; Tehseen, S.; Naz, S.; Liaqat, A.; Mehmood, T.; et al. In vitro probiotic potential and safety evaluation (hemolytic, cytotoxic activity) of Bifidobacterium strains isolated from raw camel milk. Microorganisms 2020, 8, 354. [Google Scholar] [CrossRef] [PubMed]
- Honey, C.C.; Keerthi, T.R. Probiotic potency of Lactobacillus plantarum kx519413 and kx519414 isolated from honey bee gut. FEMS Microbiol. Lett. 2018, 365, fnx285. [Google Scholar]
- Mani-López, E.; Arrioja-Bretón, D.; López-Malo, A. The impacts of antimicrobial and antifungal activity of cell-free supernatants from lactic acid bacteria in vitro and foods. Compr. Rev. Food Sci. Food Saf. 2022, 21, 604–641. [Google Scholar] [CrossRef]
- Rocchetti, M.T.; Russo, P.; Capozzi, V.; Drider, D.; Spano, G.; Fiocco, D. Bioprospecting antimicrobials from Lactiplantibacillus plantarum: Key factors underlying its probiotic action. Int. J. Mol. Sci. 2021, 22, 12076. [Google Scholar] [CrossRef]
- Gómez, N.C.; Ramiro, J.M.P.; Quecan, B.X.V.; de Melo Franco, B.D.G. Use of potential probiotic lactic acid bacteria biofilms for the control of Listeria monocytogenes, Salmonella Typhimurium, and Escherichia coli O157:H7 biofilms formation. Front. Microbiol. 2016, 7, 863. [Google Scholar] [CrossRef]
- Arena, M.P.; Silvain, A.; Normanno, G.; Grieco, F.; Drider, D.; Spano, G.; Fiocco, D. Use of Lactobacillus plantarum strains as a bio-control strategy against food-borne pathogenic microorganisms. Front. Microbiol. 2016, 7, 464. [Google Scholar] [CrossRef]
- Feng, T.; Wang, J. Oxidative stress tolerance and antioxidant capacity of lactic acid bacteria as probiotic: A systematic review. Gut Microbes 2020, 12, 1801944. [Google Scholar] [CrossRef]
- Kullisaar, T.; Zilmer, M.; Mikelsaar, M.; Vihalemm, T.; Annuk, H.; Kairane, C. and Kilk, A. Two antioxidative lactobacilli strains as promising probiotics. Int. J. Food Microbiol. 2002, 72, 215–224. [Google Scholar] [CrossRef]
- Maske, B.L.; Pereira, G.V.D.; Vale, A.D.; Neto, D.P.D.; Karp, S.G.; Viesser, J.A.; Lindner, J.D.; Pagnoncelli, M.G.; Soccol, V.T.; Soccol, C.R. A review on enzyme-producing lactobacilli associated with the human digestive process: From metabolism to application. Enzym. Microb. Technol. 2021, 149, 109836. [Google Scholar] [CrossRef] [PubMed]
- Govindaraj, K.; Samayanpaulraj, V.; Narayanadoss, V.; Uthandakalaipandian, R. Isolation of lactic acid bacteria from intestine of freshwater fishes and elucidation of probiotic potential for aquaculture application. Probiotics Antimicrob. Proteins 2021, 13, 1598–1610. [Google Scholar] [CrossRef] [PubMed]



| Isolates | Parameters | Origin of Isolate | ||||
|---|---|---|---|---|---|---|
| Shape | Gram Reaction | Cell | Catalase Test | Oxidase Test | ||
| FS43 | Smooth | + | rod-shape | - | - | Rice dockounou paste |
| FS51 | Round | + | rod-shape | - | - | Rice dockounou paste |
| FS50 | Smooth | + | rod-shape | - | - | Maize dockounou paste |
| FS49 | Round | + | rod-shape | - | - | Maize dockounou paste |
| FS65 | Smooth | + | rod-shape | - | - | Maize dockounou paste |
| FS48 | Smooth | + | rod-shape | - | - | Maize dockounou paste |
| FS47 | Round | + | rod-shape | - | - | Millet dockounou paste |
| FS46 | Round | + | rod-shape | - | - | Millet dockounou paste |
| FS45 | Smooth | + | rod-shape | - | - | Millet dockounou paste |
| FS44 | Round | + | rod-shape | - | - | Cassava dockounou paste |
| Strains | FS43 | FS51 | FS50 | FS49 | FS65 | FS48 | FS47 | FS46 | FS45 | FS44 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Class/Antibiotics | |||||||||||
| Chloramphenicol | CHL | 20S | 24S | 24S | 28S | 24S | 24S | 26S | 25S | 24S | 29S |
| Tetracyclin | T | 20S | 16I | 14R | 12R | 12R | 14R | 12R | 16I | 14R | 18I |
| Macrolides | E | 12R | 18I | 20S | 19I | 18I | 18I | 21S | 20S | 18I | 16I |
| L | 26S | 24S | 0R | 26S | 20S | 26S | 0R | 28S | 30S | 0R | |
| Beta-lactam | AM | 20S | 24S | 26S | 22S | 20S | 20S | 24S | 22S | 22S | 20S |
| AMC | 18S | 20S | 26S | 30S | 30S | 28S | 34S | 32S | 28S | 26S | |
| P | 16I | 18I | 18I | 16I | 14R | 20S | 20S | 18I | 20S | 16I | |
| Nitrofuran | F | 22S | 28S | 22S | 26S | 22S | 25S | 28S | 29S | 28S | 24S |
| Sulfamide + diaminopyrimidine | SXT | 18I | 20S | 20S | 22S | 20S | 22S | 20S | 20S | 20S | 20S |
| Hemolysis activity | γ | γ | γ | γ | γ | γ | γ | γ | γ | γ |
| Survival Rate (%) | ||||
|---|---|---|---|---|
| Isolates | pH 1.5 | Pepsin (0.1%)/pH 2.5 | Phenol (0.4%) | Bile Salts (0.3%) |
| FS43 | 84.35 ± 0.85 d | 91.58 ± 3.43 def | 94.58 ± 3.70 c | 82.25 ± 0.75 c |
| FS51 | 83.33 ± 0.91 d | 89.98 ± 1.81 ef | 40.59 ± 2.80 g | 93.09 ± 1.25 b |
| FS50 | 87.05 ± 1.95 c | 94.88 ± 2.41 bcd | 44.12 ± 3.01 fg | 99.58 ± 2.07 a |
| FS49 | 81.18 ± 1.05 e | 91.45 ± 1.78 def | 85.12 ± 6.60 d | 79.63 ± 3.90 cd |
| FS65 | 98.15 ± 0.95 a | 95.60 ± 0.84 bc | 48.09 ± 2.80 f | 77.791 ± 0.40 de |
| FS48 | 88.30 ± 0.91 c | 99.78 ± 2.14 a | 128.24 ± 1.08 a | 77.49 ± 0.59 de |
| FS47 | 91.38 ± 0.88 b | 92.63 ± 1.78 cde | 112.64 ± 1.60 b | 75.32 ± 2.15 e |
| FS46 | 90.74 ± 1.80 b | 97.96 ± 0.84 ab | 73.72 ± 0.78 e | 68.62 ± 3.64 f |
| FS45 | 85.27 ± 0.57 d | 92.44 ± 1.88 cde | 90.22 ± 0.44 cd | 74.18 ± 1.15 e |
| FS44 | 85.03 ± 0.98 d | 88.54 ± 1.26 f | 109.46 ± 0.51 b | 100.89 ± 3.52 a |
| Isolates | E. coli ATCC 8739 | L. monocytogenes ATCC 7644 | S. enterica Serovar Typhimurium ATCC 14028 | S. aureus ATCC 33592 |
|---|---|---|---|---|
| FS43 | +++ | +++ | +++ | +++ |
| FS51 | ++ | +++ | +++ | +++ |
| FS50 | ++ | +++ | +++ | +++ |
| FS49 | ++ | +++ | +++ | +++ |
| FS65 | +++ | +++ | +++ | +++ |
| FS48 | +++ | +++ | +++ | +++ |
| FS47 | ++ | +++ | +++ | +++ |
| FS46 | +++ | +++ | +++ | +++ |
| FS45 | ++ | +++ | +++ | +++ |
| FS44 | +++ | +++ | +++ | +++ |
| Parameters | Enzymatic Activities | |||||
|---|---|---|---|---|---|---|
| Strains | Proteolytic | Lipolytic | Pectinolytic | Cellulolytic | Amylolytic | |
| FS43 | + | + | + | + | - | |
| FS51 | + | - | + | + | + | |
| FS50 | + | - | + | + | - | |
| FS49 | + | - | - | + | + | |
| FS65 | + | - | + | + | + | |
| FS48 | + | - | - | + | + | |
| FS47 | + | - | + | + | - | |
| FS46 | + | - | + | + | - | |
| FS45 | + | - | + | + | + | |
| FS44 | + | - | + | + | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kouadio, N.J.; Zady, A.L.O.; Kra, K.A.S.; Diguță, F.C.; Niamke, S.; Matei, F. In Vitro Probiotic Characterization of Lactiplantibacillus plantarum Strains Isolated from Traditional Fermented Dockounou Paste. Fermentation 2024, 10, 264. https://doi.org/10.3390/fermentation10050264
Kouadio NJ, Zady ALO, Kra KAS, Diguță FC, Niamke S, Matei F. In Vitro Probiotic Characterization of Lactiplantibacillus plantarum Strains Isolated from Traditional Fermented Dockounou Paste. Fermentation. 2024; 10(5):264. https://doi.org/10.3390/fermentation10050264
Chicago/Turabian StyleKouadio, Natia Joseph, Alalet Luc Olivier Zady, Kouassi Aboutou Séverin Kra, Filofteia Camelia Diguță, Sébastien Niamke, and Florentina Matei. 2024. "In Vitro Probiotic Characterization of Lactiplantibacillus plantarum Strains Isolated from Traditional Fermented Dockounou Paste" Fermentation 10, no. 5: 264. https://doi.org/10.3390/fermentation10050264
APA StyleKouadio, N. J., Zady, A. L. O., Kra, K. A. S., Diguță, F. C., Niamke, S., & Matei, F. (2024). In Vitro Probiotic Characterization of Lactiplantibacillus plantarum Strains Isolated from Traditional Fermented Dockounou Paste. Fermentation, 10(5), 264. https://doi.org/10.3390/fermentation10050264








