Investigation of Crypthecodinium cohnii High-Cell-Density Fed-Batch Cultivations
Abstract
1. Introduction
2. Materials and Methods
2.1. Inoculum Preparation
2.2. Bioreactor Cultivations
2.3. Optical Density, Viable Cell Density, Glucose Concentration, and Conductivity Measurements
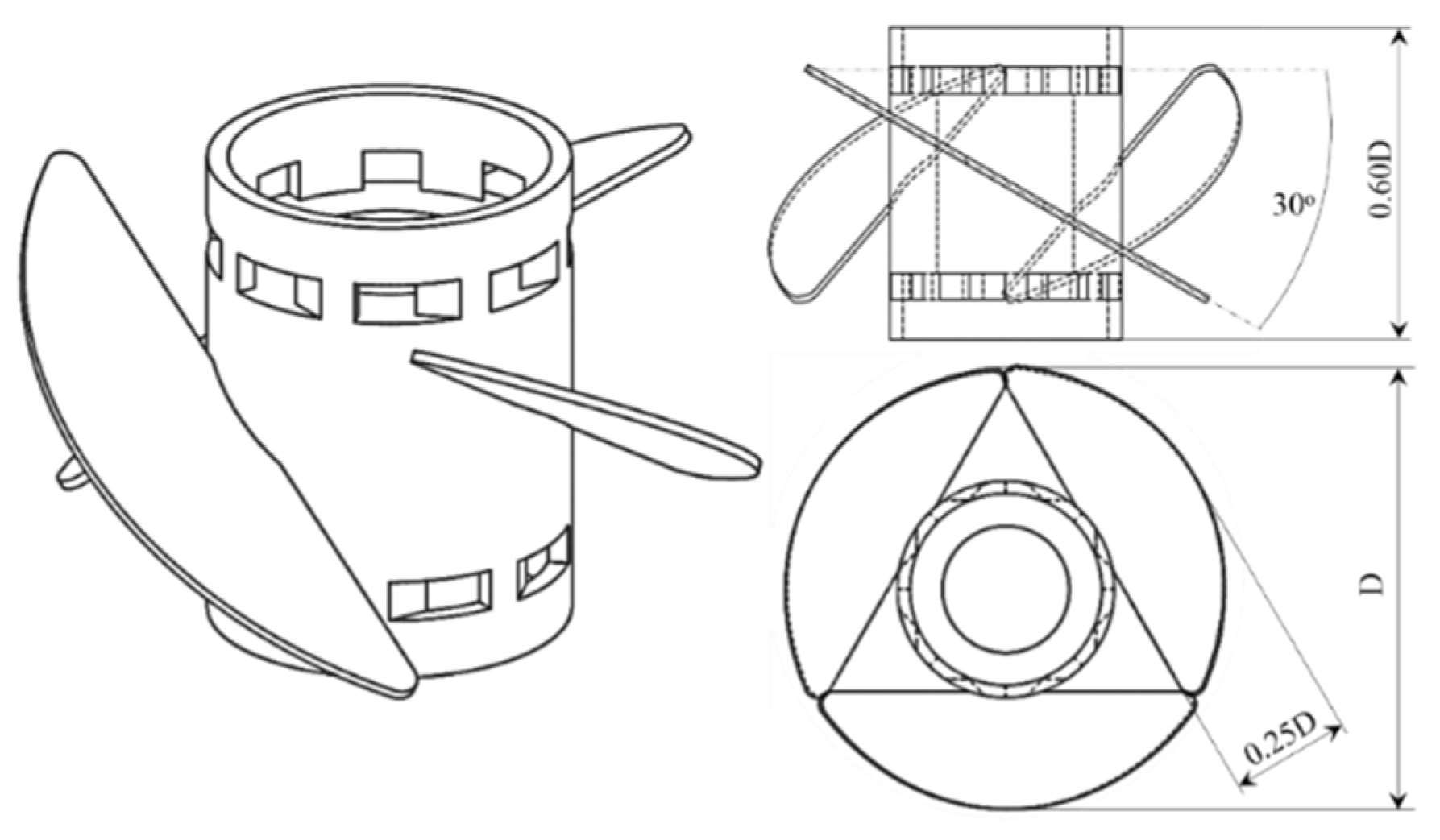
2.4. Mechanical Cell Susceptibility Assessment
2.5. Determination of Biomass Dry Cell Weight
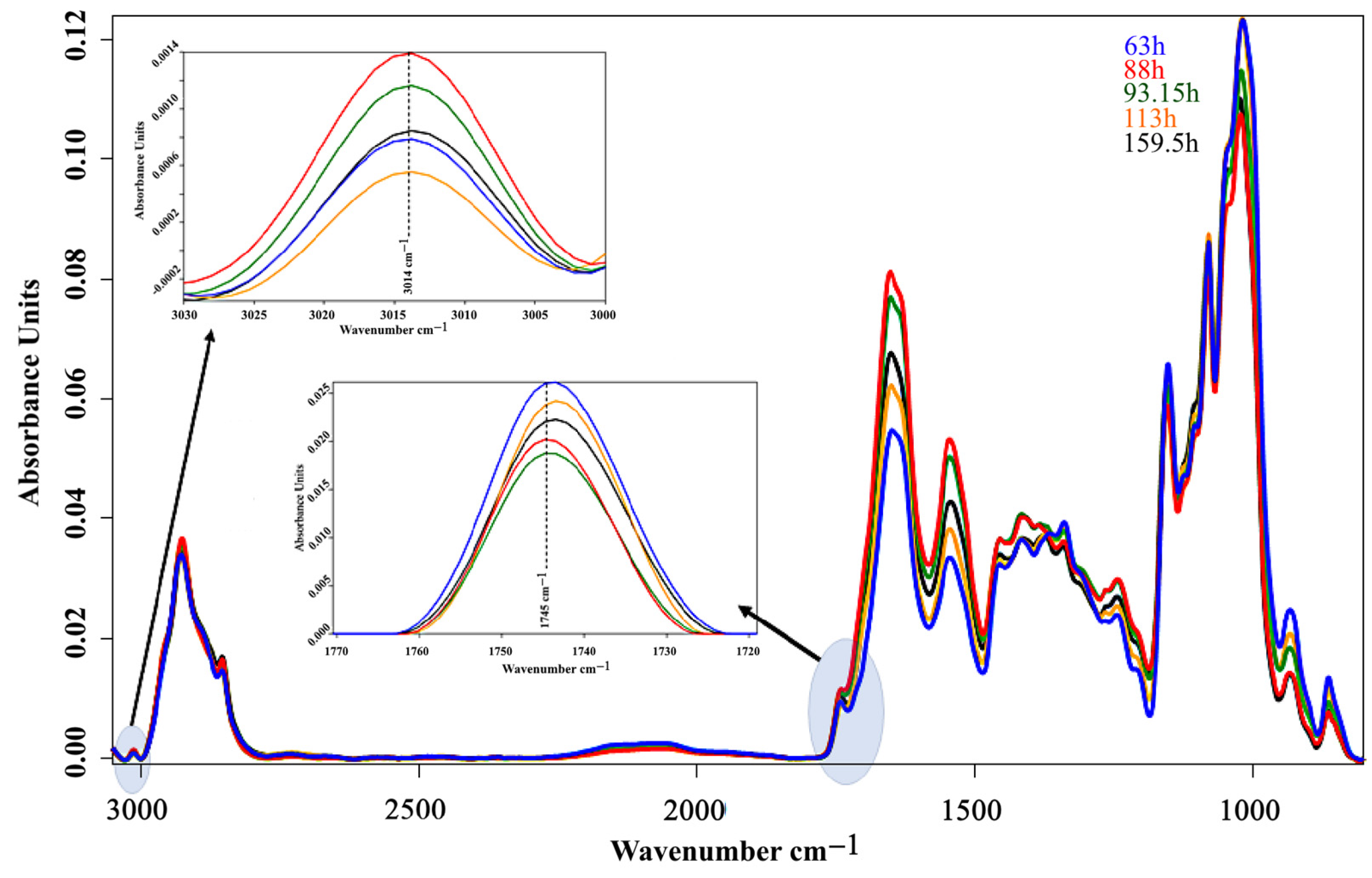
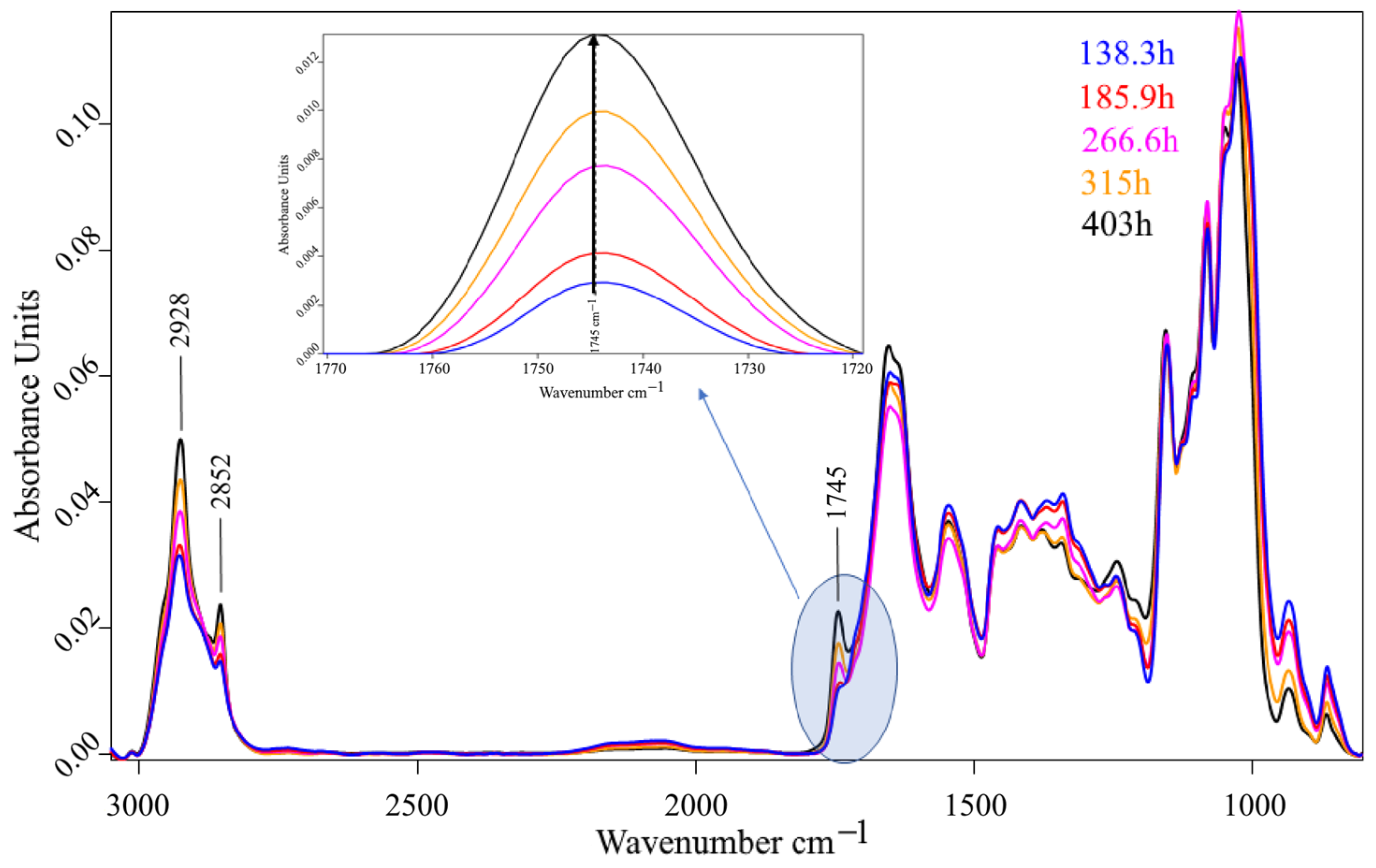
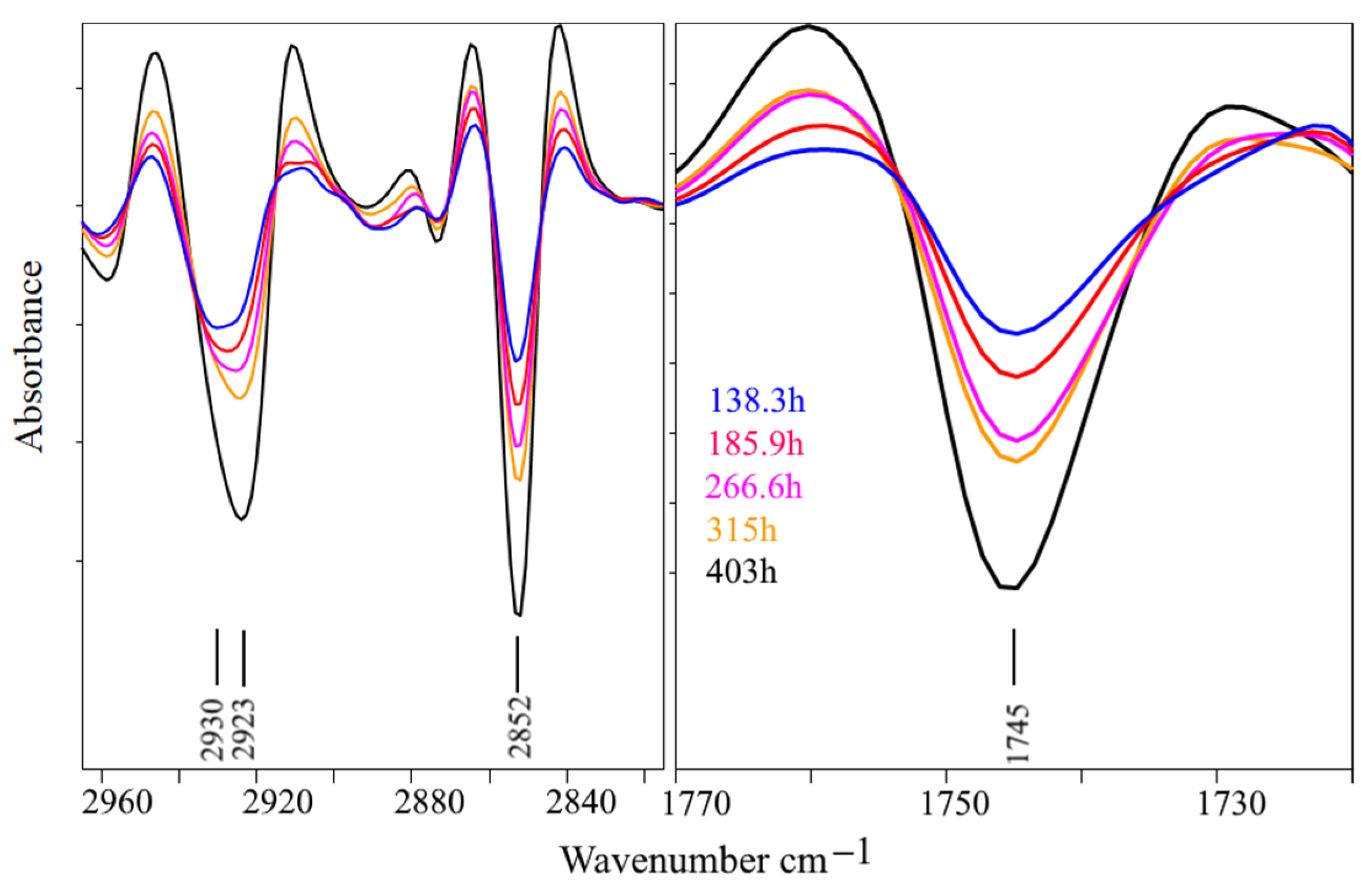
2.6. Fourier Transform Infrared Spectrometry
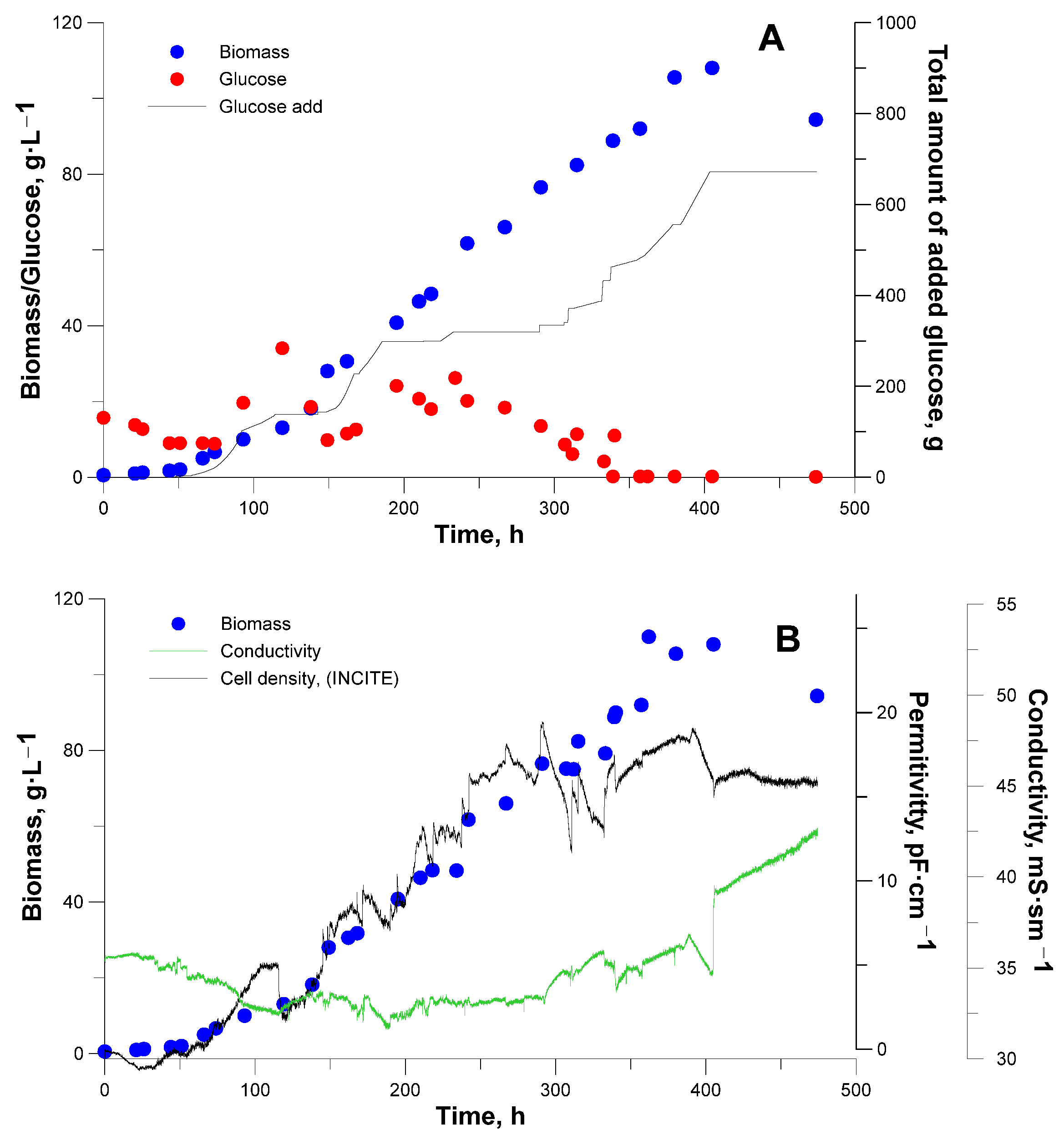
3. Results and Discussion
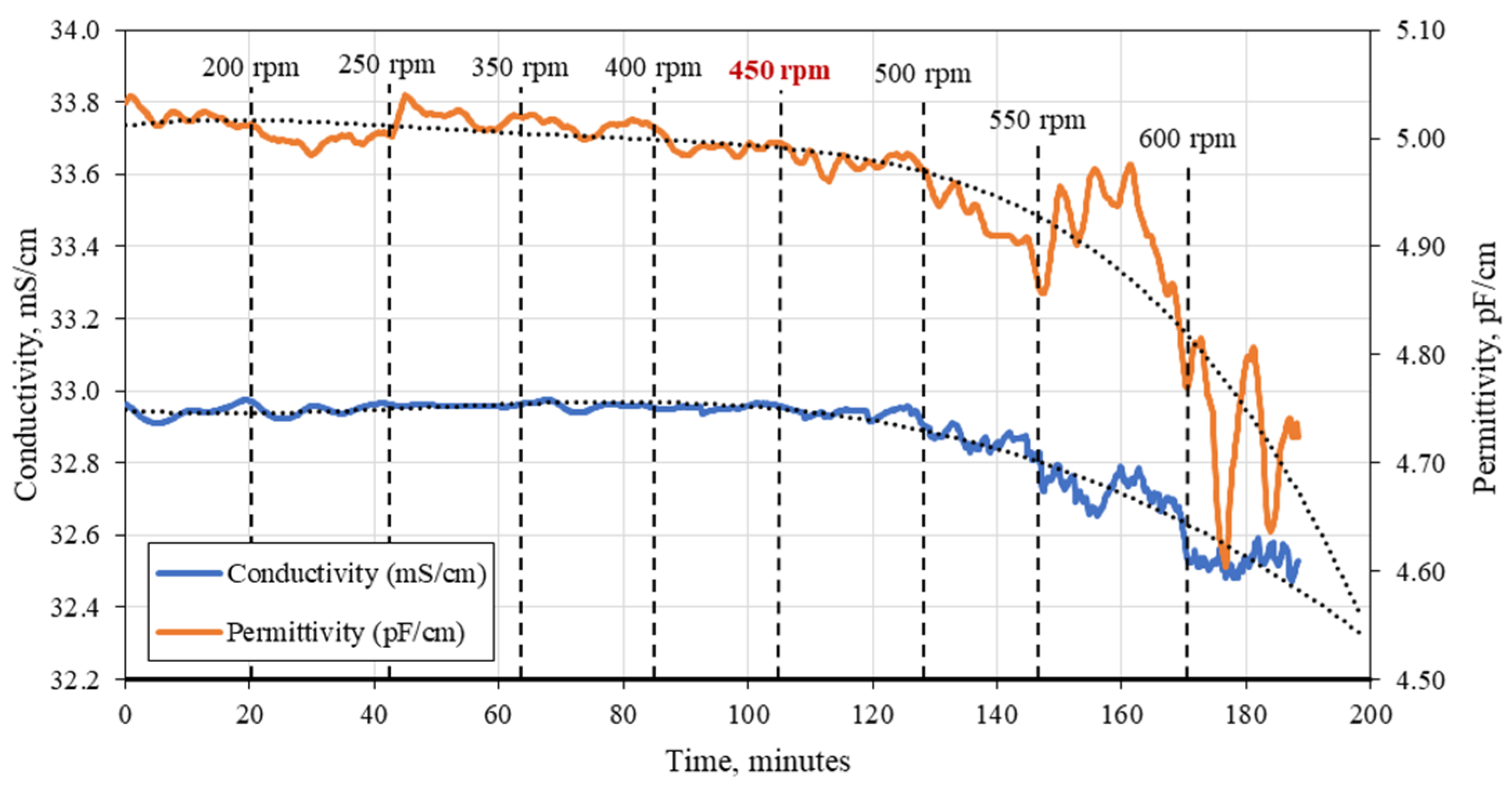
3.1. C. cohnii Susceptibility to Mechanical Damage
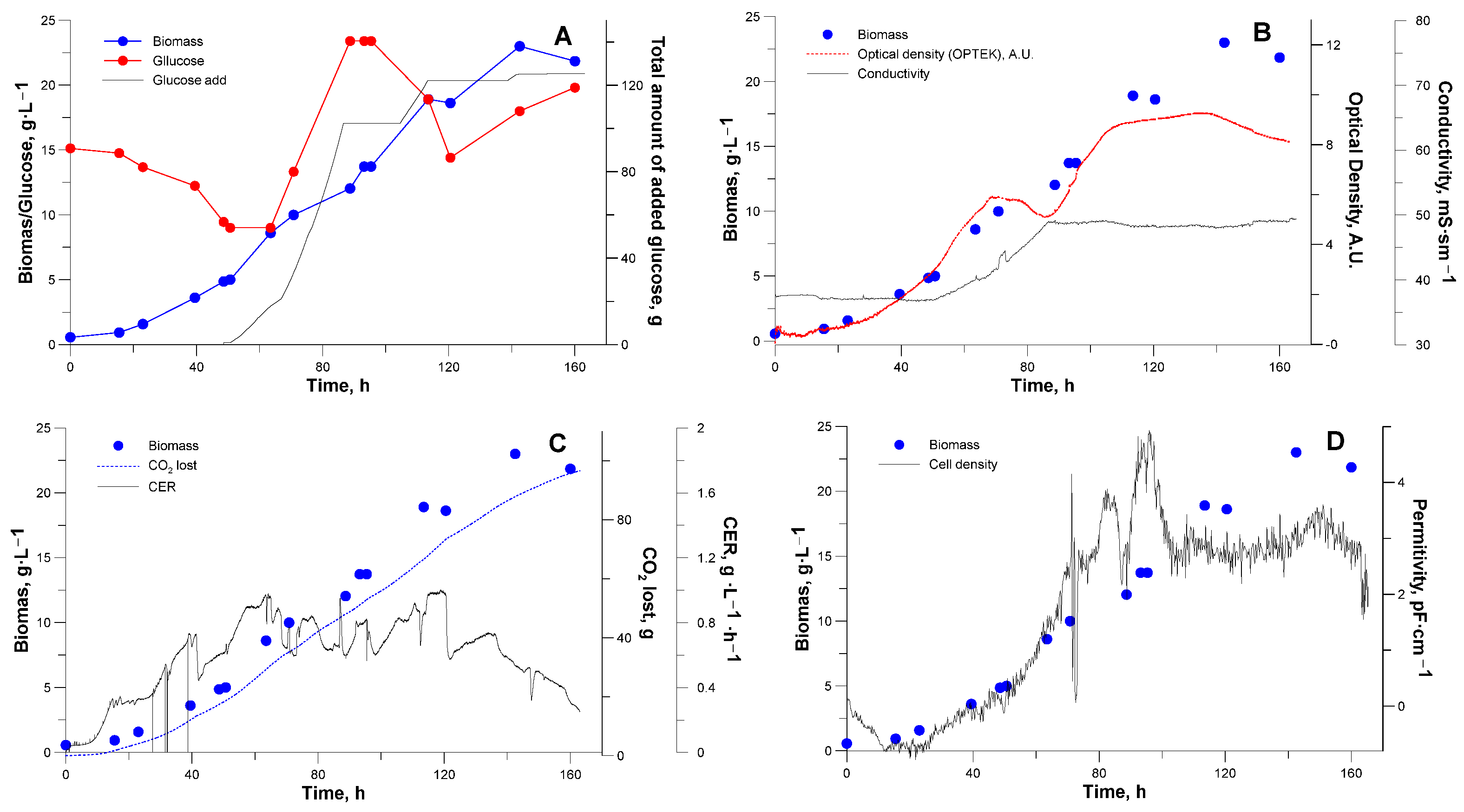
3.2. Bioreactor Cultivation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kumsiri, B.; Pekkoh, J.; Pathom-aree, W.; Lumyong, S.; Pumas, C. Synergistic Effect of Co-Culture of Microalga and Actinomycete in Diluted Chicken Manure Digestate for Lipid Production. Algal Res. 2018, 33, 239–247. [Google Scholar] [CrossRef]
- Santos-Sánchez, N.F.; Valadez-Blanco, R.; Hernández-Carlos, B.; Torres-Ariño, A.; Guadarrama-Mendoza, P.C.; Salas-Coronado, R. Lipids Rich in ω-3 Polyunsaturated Fatty Acids from Microalgae. Appl. Microbiol. Biotechnol. 2016, 100, 8667–8684. [Google Scholar] [CrossRef]
- Cui, J.; Diao, J.; Sun, T.; Shi, M.; Liu, L.; Wang, F.; Chen, L.; Zhang, W. 13C Metabolic Flux Analysis of Enhanced Lipid Accumulation Modulated by Ethanolamine in Crypthecodinium cohnii. Front. Microbiol. 2018, 9, 956. [Google Scholar] [CrossRef]
- Kadalag, N.L.; Pawar, P.R.; Prakash, G. Co-Cultivation of Phaeodactylum Tricornutum and Aurantiochytrium Limacinum for Polyunsaturated Omega-3 Fatty Acids Production. Bioresour. Technol. 2022, 346, 126544. [Google Scholar] [CrossRef]
- Patel, A.; Liefeldt, S.; Rova, U.; Christakopoulos, P.; Matsakas, L. Co-Production of DHA and Squalene by Thraustochytrid from Forest Biomass. Sci. Rep. 2020, 10, 1992. [Google Scholar] [CrossRef]
- Mendes, A.; Reis, A.; Vasconcelos, R.; Guerra, P.; Lopes Da Silva, T. Crypthecodinium cohnii with Emphasis on DHA Production: A Review. J. Appl. Phycol. 2009, 21, 199–214. [Google Scholar] [CrossRef]
- Taborda, T.; Moniz, P.; Reis, A.; da Silva, T.L. Evaluating Low-Cost Substrates for Crypthecodinium cohnii Lipids and DHA Production, by Flow Cytometry. J. Appl. Phycol. 2021, 33, 263–274. [Google Scholar] [CrossRef]
- Liu, H.; Li, H.; Zhong, Y.; Lu, Q.; Zhang, X.; Wang, Q.; Liu, H.; Diao, Z.; Yao, C. Co-Cultivation of: Rhodotorula glutinis and Chlorella pyrenoidosa to Improve Nutrient Removal and Protein Content by Their Synergistic Relationship. RSC Adv. 2019, 9, 14331–14342. [Google Scholar] [CrossRef]
- Kalia, S.; Lei, X.G. Dietary Microalgae on Poultry Meat and Eggs: Explained versus Unexplained Effects. Curr. Opin. Biotechnol. 2022, 75, 102689. [Google Scholar] [CrossRef]
- de Swaaf, M.E.; de Rijk, T.C.; Eggink, G.; Sijtsma, L. Optimisation of Docosahexaenoic Acid Production in Batch Cultivations by Crypthecodinium cohnii. Prog. Ind. Microbiol. 1999, 35, 185–192. [Google Scholar] [CrossRef]
- Wu, M.; Gao, G.; Jian, Y.; Xu, J. High CO2 Increases Lipid and Polyunsaturated Fatty Acid Productivity of the Marine Diatom Skeletonema costatum in a Two-Stage Model. J. Appl. Phycol. 2022, 34, 43–50. [Google Scholar] [CrossRef]
- Jiang, X.; Han, Q.; Gao, X.; Gao, G. Conditions Optimising on the Yield of Biomass, Total Lipid, and Valuable Fatty Acids in Two Strains of Skeletonema menzelii. Food Chem. 2016, 194, 723–732. [Google Scholar] [CrossRef]
- Gao, G.; Wu, M.; Fu, Q.; Li, X.; Xu, J. A Two-Stage Model with Nitrogen and Silicon Limitation Enhances Lipid Productivity and Biodiesel Features of the Marine Bloom-Forming Diatom Skeletonema costatum. Bioresour. Technol. 2019, 289, 121717. [Google Scholar] [CrossRef]
- Didrihsone, E.; Dubencovs, K.; Grube, M.; Shvirksts, K.; Suleiko, A.; Suleiko, A.; Vanags, J. Crypthecodinium cohnii Growth and Omega Fatty Acid Production in Mediums Supplemented with Extract from Recycled Biomass. Mar. Drugs 2022, 20, 68. [Google Scholar] [CrossRef]
- Freiberger, F.; Budde, J.; Ateş, E.A.; Schlüter, M.; Pörtner, R.; Möller, J. New Insights from Locally Resolved Hydrodynamics in Stirred Cell Culture Reactors. Processes 2022, 10, 107. [Google Scholar] [CrossRef]
- Suleiko, A.; Vanags, J.; Konuhova, M.; Dubencovs, K.; Grigs, O. The Application of Novel Magnetically Coupled Mixer Drives in Bioreactors of up to 15 m3. Biochem. Eng. J. 2020, 154, 107464. [Google Scholar] [CrossRef]
- Dubencovs, K.; Liepins, J.; Suleiko, A.; Suleiko, A.; Vangravs, R.; Kassaliete, J.; Scerbaka, R.; Grigs, O. Optimization of Synthetic Media Composition for Kluyveromyces Marxianus Fed-Batch Cultivation. Fermentation 2021, 7, 62. [Google Scholar] [CrossRef]
- Berzins, K.; Muiznieks, R.; Baumanis, M.R.; Strazdina, I.; Shvirksts, K.; Prikule, S.; Galvanauskas, V.; Pleissner, D.; Pentjuss, A.; Grube, M.; et al. Kinetic and Stoichiometric Modeling-Based Analysis of Docosahexaenoic Acid (DHA) Production Potential by C. cohnii from Glycerol, Glucose and Ethanol. Mar. Drugs 2022, 20, 115. [Google Scholar] [CrossRef]
- Yoshida, S.; Yoshida, H. Noninvasive Analyses of Polyunsaturated Fatty Acids in Human Oral Mucosa in Vivo by Fourier-Transform Infrared Spectroscopy. Biopolymers 2004, 74, 403–412. [Google Scholar] [CrossRef]
- Simopoulos, A.P. The Importance of the Ratio of Omega-6/Omega-3 Essential Fatty Acids. Biomed. Pharmacother. 2002, 56, 365–379. [Google Scholar] [CrossRef]
- Killeen, D.P.; Marshall, S.N.; Burgess, E.J.; Gordon, K.C.; Perry, N.B. Raman Spectroscopy of Fish Oil Capsules: Polyunsaturated Fatty Acid Quantitation Plus Detection of Ethyl Esters and Oxidation. J. Agric. Food Chem. 2017, 65, 3551–3558. [Google Scholar] [CrossRef]
- Karunathilaka, S.R.; Choi, S.H.; Mossoba, M.M.; Yakes, B.J.; Brückner, L.; Ellsworth, Z.; Srigley, C.T. Rapid Classification and Quantification of Marine Oil Omega-3 Supplements Using ATR-FTIR, FT-NIR and Chemometrics. J. Food Compos. Anal. 2019, 77, 9–19. [Google Scholar] [CrossRef]
- Vongsvivut, J.; Heraud, P.; Zhang, W.; Kralovec, J.A.; McNaughton, D.; Barrow, C.J. Quantitative Determination of Fatty Acid Compositions in Micro-Encapsulated Fish-Oil Supplements Using Fourier Transform Infrared (FTIR) Spectroscopy. Food Chem. 2012, 135, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Guillén, M.D.; Cabo, N. Characterization of Edible Oils and Lard by Fourier Transform Infrared Spectroscopy. Relationships between Composition and Frequency of Concrete Bands in the Fingerprint Region. JAOCS J. Am. Oil Chem. Soc. 1997, 74, 1281–1286. [Google Scholar] [CrossRef]
- Ripoche, A.; Guillard, A.S. Determination of Fatty Acid Composition of Pork Fat by Fourier Transform Infrared Spectroscopy. Meat Sci. 2001, 58, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Meng, W.; Jiang, Y.; Rothschild, D.; Lipke, M.; Hall, G.; Wang, L. Modeling the Structure and Infrared Spectra of Omega-3 Fatty Acid Esters. J. Chem. Phys. 2020, 153, 035101. [Google Scholar] [CrossRef]
- Susi, H.; Byler, D.M. [13] Resolution-Enhanced Fourier Transform Infrared Spectroscopy of Enzymes. Methods Enzym. 1986, 130, 290–311. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dubencovs, K.; Suleiko, A.; Suleiko, A.; Didrihsone, E.; Grube, M.; Shvirksts, K.; Vanags, J. Investigation of Crypthecodinium cohnii High-Cell-Density Fed-Batch Cultivations. Fermentation 2024, 10, 203. https://doi.org/10.3390/fermentation10040203
Dubencovs K, Suleiko A, Suleiko A, Didrihsone E, Grube M, Shvirksts K, Vanags J. Investigation of Crypthecodinium cohnii High-Cell-Density Fed-Batch Cultivations. Fermentation. 2024; 10(4):203. https://doi.org/10.3390/fermentation10040203
Chicago/Turabian StyleDubencovs, Konstantins, Arturs Suleiko, Anastasija Suleiko, Elina Didrihsone, Mara Grube, Karlis Shvirksts, and Juris Vanags. 2024. "Investigation of Crypthecodinium cohnii High-Cell-Density Fed-Batch Cultivations" Fermentation 10, no. 4: 203. https://doi.org/10.3390/fermentation10040203
APA StyleDubencovs, K., Suleiko, A., Suleiko, A., Didrihsone, E., Grube, M., Shvirksts, K., & Vanags, J. (2024). Investigation of Crypthecodinium cohnii High-Cell-Density Fed-Batch Cultivations. Fermentation, 10(4), 203. https://doi.org/10.3390/fermentation10040203






