Abstract
Recombinant protein expression in Escherichia coli is a fundamental technique in molecular biology and biotechnology. This review provides a comprehensive overview of various additivities to enhance the expression levels of soluble recombinant proteins in E. coli. The discussion encompasses five key aspects. Inducer Optimization: strategies for optimizing the inducer concentration to enhance protein expression. Autoinduction system optimization: the examination of glucose, lactose, and glycerol optimization within autoinduction systems to improve protein production. Osmolytes and osmoprotectants: an analysis of the use of osmolytes and osmoprotectants, such as sorbitol and glycine-betaine, to overcome with ease osmotic stress and enhance protein solubility. Ethanol additives: the impact of ethanol on E. coli physiology and its potential to improve recombinant protein expression. Cofactors and metabolic precursors: insights into the addition of cofactors, such as pyridoxal phosphate, riboflavin, thiamine, and pyridoxine, and the utilization of metabolic precursors to enhance the corresponding protein expression. This review highlights both the successful strategies and challenges in recombinant protein expression and provides insights into potential future research directions. Understanding and optimizing these factors is crucial for the efficient production of recombinant proteins for various applications in biotechnology. Furthermore, based on the analyzed data, we propose a straightforward scheme to optimize the additives in the cultivation medium.
1. Introduction
Recombinant protein expression has revolutionized the production of valuable proteins for various applications, ranging from therapeutic drugs to industrial enzymes. Among the numerous expression systems available, Escherichia coli remains a popular choice [1,2] due to its fast growth, well-characterized genetics, wide experimental experience and robustness in operation. However, one significant challenge in E. coli expression is the formation of insoluble aggregates called inclusion bodies, which hinder the recovery of functional, soluble proteins [3,4]. While the formation of inclusion bodies can simplify the purification of proteins, it does not guarantee that the process of in vitro refolding will result in high quantities of a biologically active product. Inclusion bodies are commonly formed when the overexpressed recombinant protein exceeds the host’s folding capacity or encounters unfavorable conditions in the cell [4,5]. Traditionally, refolding these proteins from inclusion bodies has been a labor-intensive and low-yield process. Despite advances in in vitro refolding strategies, achieving high yields of correctly folded, soluble proteins from inclusion bodies remains a complex task [6]. It is worth noting that reports of unsuccessful attempts at refolding are rare in the literature.
To address this challenge, researchers have explored various strategies to enhance the solubility and yield of recombinant proteins in E. coli. One promising approach involves the use of the co-expression of chaperones, such as heat shock proteins, which assist in the folding and stabilization of proteins [7,8]. Chaperones, such as DnaK, DnaJ and GroEL/ES, interact with the newly synthesized polypeptide chains, preventing misfolding and promoting correct folding, thus increasing the likelihood of obtaining soluble proteins [9]. In addition to chaperones, fusion tags have been employed to enhance protein solubility and facilitate purification [9]. These tags, such as maltose-binding protein (MBP), glutathione S-transferase (GST), or polyhistidine (His-tag), can improve protein stability, prevent aggregation, and provide affinity handles for purification techniques. The choice of fusion tag depends on the specific requirements of the protein and downstream applications. Another aspect of optimizing recombinant protein expression in E. coli involves codon optimization. E. coli has biased codon usage, and the use of codons rarely found in the host organism can lead to inefficient translation and protein misfolding. Codon optimization involves redesigning the DNA sequence of the target gene to incorporate the codons preferred by E. coli, thereby enhancing the translation efficiency and protein production. This approach can be used to improve protein solubility and yield in E. coli expression systems [10]. On the other hand, an increased translation rate can, on the contrary, lead to protein misfolding and insolubility [11]. A simple and rapid approach to reduce the number of inclusion bodies is to lower the temperature during induction. Typically, the temperature is lowered to 15–25 °C. There are also examples of successful cultivation at temperatures below 10 °C [12]. Special strains are developed for cultivation at reduced temperatures, such as E. coli ArcticExpress, which co-expresses the cold-adapted chaperonins Cpn10 and Cpn60 from the psychrophilic bacterium Oleispira antarctica [13].
Another straightforward method for influencing the expression levels of soluble protein forms is to introduce additives to the culture medium. These additives can modify the cellular environment and facilitate the correct folding of recombinant proteins in the cytoplasm, ultimately leading to an increased soluble protein expression. This article aims to provide an overview of the recent studies on the additives for the E. coli cultivation enhancing soluble recombinant protein expression in the cytoplasm. We will explore the role of different additives in promoting proper protein folding, preventing aggregation, and improving the overall efficiency of protein production in E. coli.
2. Inducer
Recombinant protein expression can be achieved through self-induction or by adding an inducer. Increasing the inducer concentration is expected to lead to higher expression yields, but increased expression levels can result in the formation of inclusion bodies (Figure 1A). Conversely, reducing the inducer concentration may lead to a decrease in the protein synthesis rate and a reduction in the number of inclusion bodies. Therefore, when expressing proteins prone to inclusion body formation, strategies aimed at both increasing and decreasing the inducer concentration should be considered.
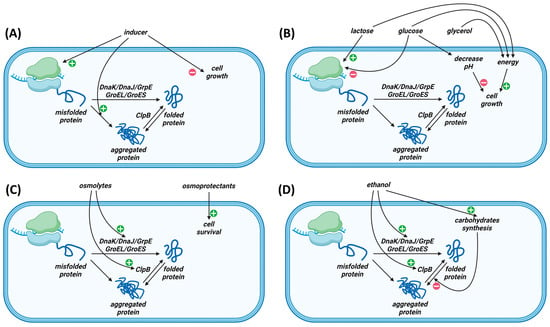
Figure 1.
Possible mechanism of influence of the inducer (A), glucose/lactose/glycerol (B), osmolytes (C) and ethanol (D) on recombinant protein expression in E. coli.
Systems using lac-based promoters are the most potent and well-studied expression systems. Lac-based promoters operate using an “on” or “off” mechanism, which leads to challenges in adjusting the inducer concentration to reduce the enzyme synthesis rate [14]. IPTG is one of the most widely used and effective inducers [15]. Increasing the concentration of IPTG often leads to higher yields of active enzyme forms [16]. However, high concentrations of IPTG can inhibit the growth of E. coli [17,18,19]. Reducing the concentration of IPTG can also improve the yields of soluble protein expression. Reducing the IPTG concentration from 1.2 to 0.3 mM resulted in an increased yield of the soluble form of the recombinant bovine sex-determining region Y protein [20]. During the expression of the leptospiral protein, the highest yield of soluble proteins and the best cell growth were observed at the lowest investigated concentration of IPTG, which was 0.1 mM [21]. When studying the influence of IPTG ranging from 0.25 to 1.25 mM, the maximum expression of the receptor activator of nuclear factor-κB was determined to be at 0.3 mM IPTG [22]. The study on the influence of IPTG concentration (0.25, 0.5, 1, or 2 mM) on the expression levels of the thioredoxin fusion with the epithelial cell adhesion molecule’s extracellular domain showed that the highest yield of the protein was achieved with 0.5 mM of IPTG [23]. The synthesis rate of the yellow fluorescence protein was not dependent on the IPTG concentration, but reducing the IPTG concentration led to an increased delay before protein synthesis [24]. Reducing the IPTG concentration below 0.1 mM can be employed to increase the delay during induction. In some cases, optimizing the IPTG concentration during induction has little effect on protein expression levels [18,19,25,26,27].
A derivative of the BL21 (DE3) expression strain has been engineered by introducing a lacY1 deletion mutation. This modification enables the T7 promoter system to be finely controlled and responsive to varying concentrations of IPTG [28]. The E. coli Tuner (DE3) is a strain containing a lacZY deletion, which allows for the precise modulation of the induction level and, in certain instances, facilitates the production of soluble proteins. The number of studies using the Tuner (DE3) strain and optimizing the inducer concentration is limited. In the optimization of cyclomaltodextrinase expression, the IPTG concentration was varied from 5 μM to 1 mM, and the highest expression yield was observed at a concentration of 50 μM [28].
Another commonly used promoter is the pBAD promoter from the araBAD operon, which enables E. coli cells to transport and metabolize arabinose [29]. Systems utilizing the pBAD promoter are positively controlled, and low basal expression levels are expected, which is particularly important for toxic proteins [30]. When using the pBAD promoter, protein expression levels depend on the concentration of arabinose within two orders of magnitude [31]. This allows for the titration of protein expression levels over a wide range of arabinose concentrations. However, it is worth noting that at low arabinose concentrations, the system forms subpopulations of producing and non-producing cells [30,32,33]. This can lead to a situation where expression levels decrease not due to a reduction in the synthesis rate of the enzyme but due to a decrease in the number of producing cells. When using glucose as the carbon source and arabinose as the inducer, a catabolite repression effect is observed, with glucose having a stronger repression effect compared to glycerol [24]. The catabolite repression effect can be utilized to optimize the type and concentration of the carbon source in inducible systems and autoinduction systems (as discussed below).
3. Glucose, Lactose and Glycerol
Glucose is a readily available and widely used additive in E. coli cultivation. The addition of glucose to the culture medium often reduces the expression rate of recombinant proteins due to the effect known as catabolite repression [34]. The addition of 1% glucose can significantly reduce the inducibility of E. coli BL21 (DE3) cells [35,36]. Reducing the expression rate can lead to increased yields of protein expression in a soluble form. This is especially important when expressing highly toxic proteins, where even low levels of expression can lead to cell death before reaching the required cell density. By optimizing the initial glucose concentration, it was possible to increase the yield of recombinant interferon α-2b by more than 2-fold [37]. The optimal glucose concentration was found to be 2% (20 g/L). Further increases in concentration resulted in a significant reduction in the protein yield.
During the cultivation process, the utilization of glucose by cells leads to a production of unwanted byproducts such as lactate and acetate, which can inhibit cell growth and recombinant protein production. The addition of glucose can significantly decrease pH and halt cell growth, even when using buffering systems (Figure 1B). Adding 2% glucose in combination with 66 mM of phosphate buffer in an M9 medium leads to a decrease in pH and a reduction in cell growth [36]. Therefore, the use of high glucose concentrations is undesirable. The negative impact of pH reduction when using high glucose concentrations can be mitigated by adding succinate, fumarate, aspartate, or glutamate or by controlling the pH during cultivation in bioreactors.
Optimizing glucose concentration is particularly important when using autoinduction systems. The autoinduction system is based on the ability of certain compounds to prevent the induction of the target protein by lactose. The lactose metabolism triggers induction after the depletion of glucose reserves. Glycerol can be used as an additional carbon source, which can be metabolized concurrently with lactose after glucose depletion [38] and has a significantly lower ability to suppress the use of other carbon sources by cells [39]. Glycerol also leads to less acidification of the pH compared to glucose. Therefore, a combination of glucose, glycerol and lactose is often used in autoinduction. The use of the autoinduction system allows cells to grow to a high density before induction is initiated. The use of the autoinduction system can lead to higher yields compared to IPTG induction. When using the ZYM-20052 autoinduction medium (2.5% glycerol, 0.05% glucose and 0.2% lactose), the yield of the soluble form of one of the three studied proteins was significantly higher than when using the nutrient-rich Dynamite medium with IPTG induction [40]. Media such as 5052, containing 0.5% glycerol, 0.05% glucose, and 0.2% lactose, facilitate the autoinduction of a wide range of proteins under various growth conditions [36]. Optimizing the components of the autoinduction medium can significantly increase the yield of the target protein. This optimization becomes particularly important when changing aeration conditions because high levels of aeration can lead to a reduced expression when using the autoinduction system [41]. It appears that increasing the lactose concentration to ~0.5% and the glycerol concentration to ~0.8% may be necessary under high aeration conditions [42]. High concentrations of glycerol and lactose can also enhance expression yields through osmotic shock (see below). The autoinduction system can also be employed with the pBAD promoter in BL21-AI cells [36]. Instead of 0.2% lactose, 0.05% L-arabinose can be used as the starting concentration for optimization in this case.
4. Osmolytes and Osmoprotectants
The most extensively studied prokaryotic protein disaggregation system in cytoplasm consists of heat shock proteins DnaK/DnaJ/GrpE, GroEL/ES, and ClpB [43,44,45]. Under osmotic shock conditions, the expression of heat shock proteins increases [46]. Due to the enhanced synthesis of chaperone proteins, one can anticipate an increase in the expression levels of soluble recombinant proteins under osmotic shock conditions (Figure 1C). One of the adaptation mechanisms of non-halophilic bacteria to high salt concentrations is the accumulation of osmoprotectants or compatible solutes, which prevents water loss due to osmotic pressure [47]. Compatible solutes are organic compounds that can accumulate within the cell and do not harm the biochemical processes inside [48]. During the cultivation of E. coli in high osmolarity media, high levels of glycine-betaine uptake have been observed [49]. Additionally, it was found that the addition of glycine-betaine and proline-betaine stimulated the growth of E. coli cells under osmotic stress conditions [50]. The high energetic cost of producing osmoprotectants necessitates their addition to the culture medium [51]. For recombinant protein expression in E. coli, NaCl and sorbitol are commonly used for inducing osmotic stress, with glycine-betaine being the most frequently used osmoprotectant.
Numerous studies provide compelling evidence for the efficacy of osmotic shock in increasing the expression levels of poorly soluble enzymes in E. coli. In Oganesyan’s work, six out of nine proteins showed improved expression in an LB medium with the addition of 0.5 M of NaCl together with 1 mM of betaine [52]. The addition of 660 mM of sorbitol and 2.5 mM of betaine resulted in a 2.4-fold increase in the yield of dimethylallyl pyrophosphate:5’-AMP dimethylallyltransferase at 37 °C, whereas the addition of 1000 mM of sorbitol with betaine at 25 °C resulted in a 6.5-fold increase [53]. An amount of 500 mM of sorbitol without betaine increased the soluble forms of three out of eight enzymes by approximately 1.5 to 2 times [54]. Co-expression with chaperones and supplementation with 0.5 M of sorbitol increased the yields of the transforming growth factor beta 3 [55]. Further enhancement was achieved with 1 M of trehalose, while other osmolytes like ethylene glycol, arginine hydrochloride, and sucrose did not increase the soluble protein yield. In addition to NaCl and sorbitol, high concentrations of glycerol and arginine are often used for osmotic shock. The addition of 0.4% glycerol during induction increased the yields of human phenylalanine hydroxylase wild-type and mutant enzymes [56]. The optimal concentration of sorbitol during the expression of the diphtheria toxin variant with its N-terminus fused to a SUMO tag was 200 mM [57]. Adding 0.3 M of sorbitol or 0.2 M of arginine increased the yield of soluble proteins [20]. The addition of 2% glycerol or 0.2 M of sorbitol increased the yield of active cholesterol oxidase [58]. Supplements of 0.5 M of sorbitol and 0.2 M of arginine in culture media resulted in an increased number of active inclusion bodies of GFP [59]. In some cases, the addition of osmolytes and osmoprotectants has been ineffective in promoting soluble protein expression. For instance, it was not possible to express aminotransferase from Sphingopyxis sp. MTA144 in a soluble form with the addition of 2.5 mM of betaine or 600 mM of sorbitol [27]. Furthermore, the addition of betaine did not lead to increased expression levels of porphyrinogen IX oxidase [60]. In certain instances, such as in the case of the human serotonin transporter, the addition of 1 M of sorbitol and 250 mM of betaine even resulted in a decrease in the yield of the membrane protein [61]. The impact of sorbitol, arginine, trehalose, and NaCl additives on the yield of the soluble flagellin of Salmonella enterica serovar Enteritidis was investigated. The maximum yield of soluble proteins was observed when 200 mM of sorbitol was added. A comparable result was observed with the addition of 100 mM and 250 mM of arginine [62].
The enhancement of soluble enzyme expression levels through the addition of osmolytes may not solely be attributed to the activation of chaperone expression. Osmolytes such as sorbitol, glycerol, and trehalose are often used as protein stabilizers [59,63]. They have the ability to inhibit the unfolding of native conformations into unfolded/incorrectly folded forms through a mechanism similar to that of other polyatomic alcohols [64].
5. Ethanol
The addition of ethanol to the cultivation medium is one of the approaches used to increase the expression levels of soluble protein forms. The introduction of ethanol to bacteria results in significant physiological changes, such as a protein and ion leakage from membranes or an increased membrane permeability [65,66]. Proteomic analysis shows that the addition of ethanol leads to an increase in the quantity of heat shock proteins [67] (Figure 1D). Numerous proteins related to carbohydrate synthesis and transport show a significant increase in expression when exposed to ethanol [68,69]. Carbohydrates are known as stabilizers for most proteins, and altering their levels can lead to increased expression yields in a soluble form. Transcriptomic analysis indicates the presence of ethanol-induced oxidative stress, leading to hypoxia and a reduced aerobic metabolism [69,70]. This results in a slowdown of macromolecule biosynthesis, which, in turn, reduces the quantity of misfolded proteins.
The number of studies with successful applications of ethanol additives to obtain soluble protein forms is significantly lower compared to the number of studies using osmolytes and osmoprotectors. Typically, 3% ethanol is added to the cultivation medium. The addition of ethanol led to a 2-fold increase in the yield of Ranibizumab [71]. Adding 3% ethanol resulted in increased expression levels of four out of six examined proteins [72,73]. The addition of 3% ethanol during the cultivation of the fusion protein preS2-S’-b-galactosidase increased cultivation yields during induction at 30 and 42 °C. In contrast, induction at 37 °C with 3% ethanol did not change the cultivation yields [74]. The addition of ethanol at 37 °C did not significantly affect the yield of CT26-poly-neoepitopes, whereas at 22 °C, the addition of 2% ethanol resulted in increased expression yields. [75]. The addition of 3% ethanol at 20 °C resulted in an increase in the total quantity of the infectious hematopoietic necrosis virus nucleoprotein in both the soluble and insoluble fractions [76]. The addition of 3% ethanol did not alter the expression level of Protoporphyrinogen IX Oxidase [60].
6. Cofactors
Various enzymes with prosthetic groups often require sufficient quantities of corresponding cofactors or their precursors. Their addition can increase the yield of such enzymes [77]. One of the problems limiting the use of many cofactors as additives in expression is their high cost. In such cases, a metabolic precursor of the cofactor can be used.
During the expression of recombinant human hemoglobin, the addition of hemin resulted in an increase in the amount of soluble proteins [78]. Positive effects on the expression of heme-containing proteins may be achieved by adding thiamine and δ-aminolevulinic acid. During the expression of recombinant Cytochrome P450 1B1, the addition of thiamine did not lead to an increase in expression levels, whereas the addition of δ-aminolevulinic acid at concentrations of up to 1 mM resulted in a significant increase in the protein yield [79]. Thiamine supplements can be particularly important when cultivating strains of E. coli derived from E. coli K12, as they may be deficient in enzymes involved in thiamine anabolism [80,81].
Adding 1 µM of riboflavin 2 h before induction during the expression of FAD-containing protoporphyrinogen oxidase increased the amount of the recovered enzyme by approximately 4-fold [82]. The positive effect on enzyme expression upon the addition of riboflavin in E. coli may be related to the presence of a riboflavin transmembrane import system (YpaA protein in E. coli) and an endogenous riboflavin biosynthesis pathway [83,84,85]. Notably, E. coli BL21 (derived from E. coli B) is more prone to accumulate riboflavin than E. coli MG1655 (similar to E. coli K-12) [86].
The addition of cofactors can increase the yield of activity not only by increasing solubility, but also by increasing the specific enzymatic activity. The addition of 0.02 mM of pyridoxal phosphate (pyridoxine-5-phosphate) increased the yield of active glutamate decarboxylase by 2–2.5 times and simultaneously double increased in glutamate decarboxylase specific activity [87]. Pyridoxine can be taken up by E. coli cells and used for the synthesis of pyridoxal phosphate [88]. The addition of pyridoxine at concentrations above 0.05 mM allows for an almost 1.8-fold increase in the yield of active glutamate decarboxylase and a 1.5-fold increase in the specific activity of the enzyme [89]. The addition of pyridoxine also resulted in a 2.8-fold increase in the stability of glutamate decarboxylase.
Vitamin additives or their precursors cannot always increase the yield of the active soluble form of a protein. For example, pyridoxal phosphate, which is a co-enzyme of aminotransferase FumI, has no effect when added to the medium at a concentration of 1 mM [27]. Additionally, adding 0.1 M of FAD, FMN, and riboflavin during cultivation did not lead to increased expression yields of human D-amino acid oxidase [90].
7. Optimization
Based on this analysis of existing approaches to enhance expression levels, we propose a simple and versatile scheme for optimizing additives in the culture medium to increase the yield of soluble protein forms (Figure 2). The first step involves optimizing the inducer concentration for induction systems or the ratios of lactose/glucose/glycerol components for autoinduction systems. The literature’s data indicate that the optimal inducer concentration significantly varies depending on the target protein. Therefore, we suggest optimizing within a wide range of inducer concentrations and narrowing it down in case of success. For IPTG induction, concentrations can be optimized in the range of 0.1 to 1.0 mM. When using autoinduction systems, a starting point could be a medium like 5052 supplemented with 0.5% glycerol, 0.05% glucose, and 0.2% lactose. To reduce the number of experiments, it is advisable to simultaneously vary each component within a wide range as part of parallel optimization. For instance, in a series of nine parallel experiments, test additions of 0.2%, 0.5%, and 1.0% glycerol, 0.02%, 0.05%, and 0.1% glucose, and 0.1%, 0.2%, and 0.5% lactose while keeping the other components constant.
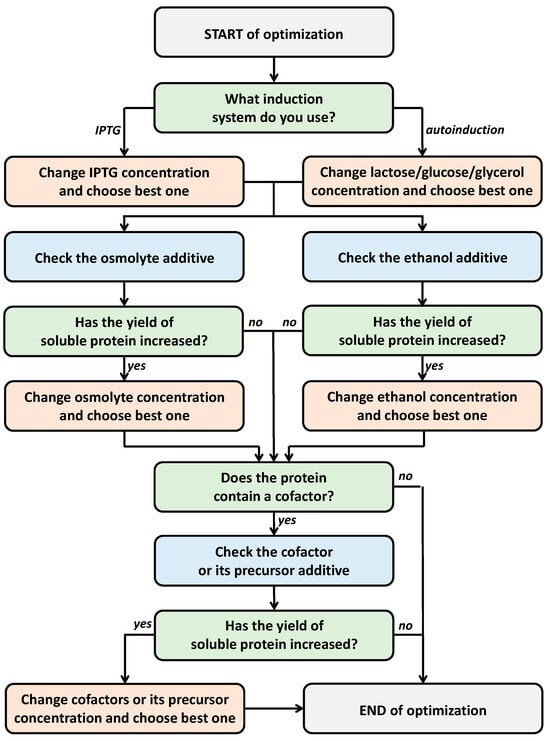
Figure 2.
Scheme for optimizing additives.
The second step involves the parallel assessment of the effects of osmolytes/osmoprotectants and ethanol additives. For an osmolyte, we recommend using 0.5 M sorbitol, 0.5 M sodium chloride, or 2% glycerol. According to the literature, sorbitol is the most popular additive for conducting osmotic shock, but sodium chloride and glycerol are more cost-effective and readily available in laboratory practice. For an osmoprotectant, you can use a 2.5 mM betaine additive. In the absence of betaine, you can evaluate the impact of an osmolyte addition without an osmoprotectant. To assess the influence of ethanol, we propose using a 3% ethanol additive. If osmolyte/osmoprotectant or ethanol additives lead to increased expression levels, further optimization of the respective additive concentrations should be conducted.
In cases where the studied protein contains a cofactor, the optimization of the cofactor or its metabolic precursor additives can be considered. Such additives are often costly, so it is advisable to conduct this step in the final stages of optimization. If the addition of a cofactor or its precursor leads to increased expression yields, further optimization of the added quantities can be pursued. The proposed optimization scheme is simple and does not require a large number of experiments.
8. Conclusions and Future Perspectives
Enhancing the expression level of soluble recombinant proteins is an important task for molecular biology and biotechnology. Introducing additives in a culture medium is a technically straightforward method for optimizing the expression levels of recombinant proteins. Expression systems based on E. coli are among the most popular for obtaining recombinant proteins. This review provides a comprehensive overview of the use of additives to enhance the expression levels of recombinant proteins in E. coli in soluble forms.
The most popular and researched approaches include changing the inducer concentration and adding osmolytes. Based on studies [52,54], approximately half of the cases show increased expression levels of recombinant proteins in soluble forms under osmotic shock conditions. This effectiveness is comparable to the efficiency of a co-expression with chaperones [91].
The number of studies focusing on the impact of ethanol on the expression levels of recombinant proteins is significantly lower compared to osmolytes. It would be interesting to see research comparing the effectiveness of adding various osmolytes, ethanol, and other additives on the expression of a wide range of different proteins. Most studies focus on the impact of a single additive. It is unclear how a combination of additives will affect the expression levels of recombinant proteins.
Temperature is an important factor in optimizing the expression of recombinant proteins, and lowering it appears to increase the effectiveness of osmolyte and ethanol additives [53,74,75]. Predicting the impact of any additive in advance is not feasible; therefore, it is most practical to test additives that most commonly lead to increased expression levels of soluble enzyme forms.
It can be assumed that as the experimental knowledge base expands and our understanding of E. coli metabolism and regulation grows, researchers will continue to discover new additives that lead to increased yields of soluble enzymes. The rapidly advancing field of metabolic engineering may also aid in creating E. coli strains with metabolic pathways adapted for the highly efficient expression of recombinant proteins, including those requiring specific additives.
Author Contributions
Conceptualization, D.L.A., E.P.S. and A.A.P.; writing—original draft preparation, D.L.A., E.P.S. and D.I.G.; writing—review and editing, D.L.A.; funding acquisition, D.L.A. and A.A.P. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported in part by the State Budget Project.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Tripathi, N.K.; Shrivastava, A. Recent Developments in Bioprocessing of Recombinant Proteins: Expression Hosts and Process Development. Front. Bioeng. Biotechnol. 2019, 7, 420. [Google Scholar] [CrossRef] [PubMed]
- Mital, S.; Christie, G.; Dikicioglu, D. Recombinant expression of insoluble enzymes in Escherichia coli: A systematic review of experimental design and its manufacturing implications. Microb. Cell Factoties 2021, 20, 208. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Upadhyay, V.; Upadhyay, A.K.; Singh, S.M.; Panda, A.K. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb. Cell Factoties 2015, 14, 41. [Google Scholar] [CrossRef]
- Bhatwa, A.; Wang, W.; Hassan, Y.I.; Abraham, N.; Li, X.Z.; Zhou, T. Challenges Associated With the Formation of Recombinant Protein Inclusion Bodies in Escherichia coli and Strategies to Address Them for Industrial Applications. Front. Bioeng. Biotechnol. 2021, 9, 630551. [Google Scholar] [CrossRef]
- Fink, A.L. Protein aggregation: Folding aggregates, inclusion bodies and amyloid. Fold. Des. 1998, 3, R9–R23. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Yamamoto, E.; Mannen, T.; Nagamune, T. Protein refolding using chemical refolding additives. Biotechnol. J. 2013, 8, 17–31. [Google Scholar] [CrossRef]
- Nishihara, K.; Kanemori, M.; Kitagawa, M.; Yanagi, H.; Yura, T. Chaperone coexpression plasmids: Differential and synergistic roles of DnaK-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli. Appl. Environ. Microbiol. 1998, 64, 1694–1699. [Google Scholar] [CrossRef]
- Tong, Y.; Feng, S.; Xin, Y.; Yang, H.; Zhang, L.; Wang, W.; Chen, W. Enhancement of soluble expression of codon-optimized Thermomicrobium roseum sarcosine oxidase in Escherichia coli via chaperone co-expression. J. Biotechnol. 2016, 218, 75–84. [Google Scholar] [CrossRef]
- Imamoglu, R.; Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. Bacterial Hsp70 resolves misfolded states and accelerates productive folding of a multi-domain protein. Nat. Commun. 2020, 11, 365. [Google Scholar] [CrossRef] [PubMed]
- Burgess-Brown, N.A.; Sharma, S.; Sobott, F.; Loenarz, C.; Oppermann, U.; Gileadi, O. Codon optimization can improve expression of human genes in Escherichia coli: A multi-gene study. Protein Expr. Purif. 2008, 59, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Rosano, G.L.; Ceccarelli, E.A. Rare codon content affects the solubility of recombinant proteins in a codon bias-adjusted Escherichia coli strain. Microb. Cell Factoties 2009, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Song, J.M.; An, Y.J.; Kang, M.H.; Lee, Y.-H.; Cha, S.-S. Cultivation at 6–10 °C is an effective strategy to overcome the insolubility of recombinant proteins in Escherichia coli. Protein Expr. Purif. 2012, 82, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Chernikova, T.N.; Yakimov, M.M.; Golyshin, P.N.; Timmis, K.N. Chaperonins govern growth of Escherichia coli at low temperatures. Nat. Biotechnol. 2003, 21, 1266–1267. [Google Scholar] [CrossRef] [PubMed]
- Chu, I.-T.; Speer, S.L.; Pielak, G.J. Rheostatic Control of Protein Expression Using Tuner Cells. Biochemistry 2020, 59, 733–735. [Google Scholar] [CrossRef] [PubMed]
- Marbach, A.; Bettenbrock, K. Lac operon induction in Escherichia coli: Systematic comparison of IPTG and TMG induction and influence of the transacetylase LacA. J. Biotechnol. 2012, 157, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Behravan, A.; Hashemi, A. RSM-based Model to Predict Optimum Fermentation Conditions for Soluble Expression of the Antibody Fragment Derived from 4D5MOC-B Humanized Mab in SHuffle T7 E. coli. Iran. J. Pharm. Res. 2021, 20, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Jeong, K.J.; Lee, S.Y. High-level production of human leptin by fed-batch cultivation of recombinant Escherichia coli and its purification. Appl. Environ. Microbiol. 1999, 65, 3027–3032. [Google Scholar] [CrossRef]
- Einsfeldt, K.; Severo Junior, J.B.; Correa Argondizzo, A.P.; Medeiros, M.A.; Alves, T.L.; Almeida, R.V.; Larentis, A.L. Cloning and expression of protease ClpP from Streptococcus pneumoniae in Escherichia coli: Study of the influence of kanamycin and IPTG concentration on cell growth, recombinant protein production and plasmid stability. Vaccine 2011, 29, 7136–7143. [Google Scholar] [CrossRef]
- Malik, A.; Alsenaidy, A.M.; Elrobh, M.; Khan, W.; Alanazi, M.S.; Bazzi, M.D. Optimization of expression and purification of HSPA6 protein from Camelus dromedarius in E. coli. Saudi J. Biol. Sci. 2016, 23, 410–419. [Google Scholar] [CrossRef]
- Soleymani, B.; Mostafaie, A. Analysis of Methods to Improve the Solubility of Recombinant Bovine Sex Determining Region Y Protein. Rep. Biochem. Mol. Biol. 2019, 8, 227–235. [Google Scholar]
- Larentis, A.L.; Nicolau, J.F.M.Q.; Esteves, G.d.S.; Vareschini, D.T.; de Almeida, F.V.R.; dos Reis, M.G.; Galler, R.; Medeiros, M.A. Evaluation of pre-induction temperature, cell growth at induction and IPTG concentration on the expression of a leptospiral protein in E. coli using shaking flasks and microbioreactor. BMC Res. Notes 2014, 7, 671. [Google Scholar] [CrossRef]
- Papaneophytou, C.P.; Rinotas, V.; Douni, E.; Kontopidis, G. A statistical approach for optimization of RANKL overexpression in Escherichia coli: Purification and characterization of the protein. Protein Expr. Purif. 2013, 90, 9–19. [Google Scholar] [CrossRef]
- Rasooli, F.; Hashemi, A. Efficient expression of EpEX in the cytoplasm of Escherichia coli using thioredoxin fusion protein. Res. Pharm. Sci. 2019, 14, 554–565. [Google Scholar] [CrossRef]
- Lozano Terol, G.; Gallego-Jara, J.; Sola Martinez, R.A.; Martinez Vivancos, A.; Canovas Diaz, M.; de Diego Puente, T. Impact of the Expression System on Recombinant Protein Production in Escherichia coli BL21. Front. Microbiol. 2021, 12, 682001. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Hall, D.; Eiteman, M.A.; Altman, E. Optimization of recombinant aminolevulinate synthase production in Escherichia coli using factorial design. Appl. Microbiol. Biotechnol. 2003, 63, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Farshdari, F.; Ahmadzadeh, M.; Nematollahi, L.; Mohit, E. The improvement of anti-HER2 scFv soluble expression in Escherichia coli. Braz. J. Pharm. Sci. 2020, 56, e17861. [Google Scholar] [CrossRef]
- Hartinger, D.; Heinl, S.; Schwartz, H.E.; Grabherr, R.; Schatzmayr, G.; Haltrich, D.; Moll, W.-D. Enhancement of solubility in Escherichia coli and purification of an aminotransferase from Sphingopyxis sp. MTA144 for deamination of hydrolyzed fumonisin B1. Microb. Cell Factoties 2010, 9, 62. [Google Scholar] [CrossRef] [PubMed]
- Turner, P.; Holst, O.; Karlsson, E.N. Optimized expression of soluble cyclomaltodextrinase of thermophilic origin in Escherichia coli by using a soluble fusion-tag and by tuning of inducer concentration. Protein Expr. Purif. 2005, 39, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Schleif, R. Regulation of the L-arabinose operon of Escherichia coli. Trends Genet. 2000, 16, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Siegele, D.A.; Hu, J.C. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. USA 1997, 94, 8168–8172. [Google Scholar] [CrossRef] [PubMed]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef]
- Lee, N.; Francklyn, C.; Hamilton, E.P. Arabinose-induced binding of AraC protein to araI2 activates the araBAD operon promoter. Proc. Natl. Acad. Sci. USA 1987, 84, 8814–8818. [Google Scholar] [CrossRef]
- Afroz, T.; Biliouris, K.; Kaznessis, Y.; Beisel, C.L. Bacterial sugar utilization gives rise to distinct single-cell behaviours. Mol. Microbiol. 2014, 93, 1093–1103. [Google Scholar] [CrossRef] [PubMed]
- Deutscher, J. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2008, 11, 87–93. [Google Scholar] [CrossRef]
- Grossman, T.H.; Kawasaki, E.S.; Punreddy, S.R.; Osburne, M.S. Spontaneous cAMP-dependent derepression of gene expression in stationary phase plays a role in recombinant expression instability. Gene 1998, 209, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Studier, F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef] [PubMed]
- Mansey, M.S.; Ghareeb, K.A.; Moghazy, A.N.; Tawfick, M.M.; Fouda, M.M.; Marzugi, N.E.; Othman, N.Z.; El Enshasy, H.A. Glucose concentration affects recombinant interferon α-2b production in Escherichia coli using thermo-induction system. J. Appl. Pharm. Sci. 2014, 4, 1–5. [Google Scholar] [CrossRef]
- Studier, F.W. Stable expression clones and auto-induction for protein production in E. coli. In Structural Genomics. Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2014; Volume 1091, pp. 17–32. [Google Scholar] [CrossRef]
- Eppler, T.; Postma, P.; Schutz, A.; Volker, U.; Boos, W. Glycerol-3-phosphate-induced catabolite repression in Escherichia coli. J. Bacteriol. 2002, 184, 3044–3052. [Google Scholar] [CrossRef]
- Taylor, T.; Denson, J.P.; Esposito, D. Optimizing Expression and Solubility of Proteins in E. coli Using Modified Media and Induction Parameters. In Heterologous Gene Expression in E.coli. Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; Volume 1586, pp. 65–82. [Google Scholar] [CrossRef]
- Ukkonen, K.; Mayer, S.; Vasala, A.; Neubauer, P. Use of slow glucose feeding as supporting carbon source in lactose autoinduction medium improves the robustness of protein expression at different aeration conditions. Protein Expr. Purif. 2013, 91, 147–154. [Google Scholar] [CrossRef]
- Blommel, P.G.; Becker, K.J.; Duvnjak, P.; Fox, B.G. Enhanced bacterial protein expression during auto-induction obtained by alteration of lac repressor dosage and medium composition. Biotechnol. Prog. 2007, 23, 585–598. [Google Scholar] [CrossRef]
- Mogk, A.; Tomoyasu, T.; Goloubinoff, P.; Rudiger, S.; Roder, D.; Langen, H.; Bukau, B. Identification of thermolabile Escherichia coli proteins: Prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999, 18, 6934–6949. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.G.; Baneyx, F. ClpB and HtpG facilitate de novo protein folding in stressed Escherichia coli cells. Mol. Microbiol. 2000, 36, 1360–1370. [Google Scholar] [CrossRef] [PubMed]
- Konieczny, I.; Liberek, K. Cooperative action of Escherichia coli ClpB protein and DnaK chaperone in the activation of a replication initiation protein. J. Biol. Chem. 2002, 277, 18483–18488. [Google Scholar] [CrossRef] [PubMed]
- Meury, J.; Kohiyama, M. Role of heat shock protein DnaK in osmotic adaptation of Escherichia coli. J. Bacteriol. 1991, 173, 4404–4410. [Google Scholar] [CrossRef] [PubMed]
- Amezaga, M.R.; Booth, I.R. Osmoprotection of Escherichia coli by peptone is mediated by the uptake and accumulation of free proline but not of proline-containing peptides. Appl. Environ. Microbiol. 1999, 65, 5272–5278. [Google Scholar] [CrossRef] [PubMed]
- Bremer, E.; Kramer, R. Responses of Microorganisms to Osmotic Stress. Annu. Rev. Microbiol. 2019, 73, 313–334. [Google Scholar] [CrossRef]
- Perroud, B.; Le Rudulier, D. Glycine betaine transport in Escherichia coli: Osmotic modulation. J. Bacteriol. 1985, 161, 393–401. [Google Scholar] [CrossRef]
- Larsen, P.I.; Sydnes, L.K.; Landfald, B.; Strom, A.R. Osmoregulation in Escherichia coli by accumulation of organic osmolytes: Betaines, glutamic acid, and trehalose. Arch. Microbiol. 1987, 147, 1–7. [Google Scholar] [CrossRef]
- Oren, A. Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev. 1999, 63, 334–348. [Google Scholar] [CrossRef]
- Oganesyan, N.; Ankoudinova, I.; Kim, S.H.; Kim, R. Effect of osmotic stress and heat shock in recombinant protein overexpression and crystallization. Protein Expr. Purif. 2007, 52, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, J.R.; Horgan, R. A novel strategy for production of a highly expressed recombinant protein in an active form. FEBS Lett. 1991, 295, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Zou, R.; Stephanopoulos, G.; Too, H.-P. Enhancing solubility of deoxyxylulose phosphate pathway enzymes for microbial isoprenoid production. Microb. Cell Factoties 2012, 11, 148. [Google Scholar] [CrossRef]
- Dawn, A.; Deep, S. An improved strategy of TGFβ3 expression in Escherichia coli: Exploiting folding modulators for a switch from misfolded to folded form. Int. J. Biol. Macromol. 2021, 167, 787–795. [Google Scholar] [CrossRef]
- Leandro, P.; Lechner, M.C.; de Almeida, I.T.; Konecki, D. Glycerol increases the yield and activity of human phenylalanine hydroxylase mutant enzymes produced in a prokaryotic expression system. Mol. Genet. Metab. 2001, 73, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Tarahomjoo, S.; Bandehpour, M.; Aghaebrahimian, M.; Ahangaran, S. Soluble Diphtheria Toxin Variant, CRM 197 was Obtained in Escherichia coli at High Productivity Using SUMO Fusion and an Adjusted Expression Strategy. Protein Pept. Lett. 2022, 29, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, H.E.; El-Far, S.W.; Embaby, A.M. Cloning, expression, and in silico structural modeling of cholesterol oxidase of Acinetobacter sp. strain RAMD in E. coli. FEBS Open Bio. 2021, 11, 2560–2575. [Google Scholar] [CrossRef] [PubMed]
- Prasad, S.; Khadatare, P.B.; Roy, I. Effect of chemical chaperones in improving the solubility of recombinant proteins in Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 4603–4609. [Google Scholar] [CrossRef]
- de Marco, A.; Volrath, S.; Bruyere, T.; Law, M.; Fonne-Pfister, R. Recombinant maize protoporphyrinogen IX oxidase expressed in Escherichia coli forms complexes with GroEL and DnaK chaperones. Protein Expr. Purif. 2000, 20, 81–86. [Google Scholar] [CrossRef]
- Worms, D.; Maertens, B.; Kubicek, J.; Subhramanyam, U.K.T.; Labahn, J. Expression, purification and stabilization of human serotonin transporter from E. coli. Protein Expr. Purif. 2019, 164, 105479. [Google Scholar] [CrossRef]
- Bakhtiarvand, B.; Sadeghi, Z.; Tarahomjoo, S.; Yaghmaie, S. Chaperones Promote Remarkable Solubilization of Salmonella enterica serovar Enteritidis Flagellin Expressed in Escherichia coli. Protein Pept. Lett. 2020, 27, 210–218. [Google Scholar] [CrossRef]
- Filatova, L.Y.; Becker, S.C.; Donovan, D.M.; Gladilin, A.K.; Klyachko, N.L. LysK, the enzyme lysing Staphylococcus aureus cells: Specific kinetic features and approaches towards stabilization. Biochimie 2010, 92, 507–513. [Google Scholar] [CrossRef]
- Kaushik, J.K.; Bhat, R. Why is trehalose an exceptional protein stabilizer? An analysis of the thermal stability of proteins in the presence of the compatible osmolyte trehalose. J. Biol. Chem. 2003, 278, 26458–26465. [Google Scholar] [CrossRef]
- Ingram, L.O. Ethanol tolerance in bacteria. Crit. Rev. Biotechnol. 1990, 9, 305–319. [Google Scholar] [CrossRef]
- Chiou, R.Y.; Phillips, R.D.; Zhao, P.; Doyle, M.P.; Beuchat, L.R. Ethanol-mediated variations in cellular fatty acid composition and protein profiles of two genotypically different strains of Escherichia coli O157:H7. Appl. Environ. Microbiol. 2004, 70, 2204–2210. [Google Scholar] [CrossRef]
- Soufi, B.; Krug, K.; Harst, A.; Macek, B. Characterization of the E. coli proteome and its modifications during growth and ethanol stress. Front. Microbiol. 2015, 6, 103. [Google Scholar] [CrossRef]
- Richarme, G.; Caldas, T.D. Chaperone properties of the bacterial periplasmic substrate-binding proteins. J. Biol. Chem. 1997, 272, 15607–15612. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Yu, Z.; Shu, W.; Fu, X.; Zhao, X.; Yang, S.; Tan, M.; Xu, J.; Liu, Y.; Song, H. Ethanol effects on the overexpression of heterologous catalase in Escherichia coli BL21 (DE3). Appl. Microbiol. Biotechnol. 2019, 103, 1441–1453. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Wei, D.; Yang, Y.; Shang, Y.; Li, G.; Zhou, Y.; Ma, Q.; Xu, Y. Systems-level understanding of ethanol-induced stresses and adaptation in E. coli. Sci. Rep. 2017, 7, 44150. [Google Scholar] [CrossRef] [PubMed]
- Priyanka, P.; Patil, R.S.; Meshram, P.; Gupta, J.A.; Banerjee, M.; Rathore, A.S. Ethanol as additive enhances expression of Ranibizumab in Escherichia coli: Impact on cellular physiology and transcriptome. Process. Biochem. 2022, 112, 167–176. [Google Scholar] [CrossRef]
- Chhetri, G.; Kalita, P.; Tripathi, T. An efficient protocol to enhance recombinant protein expression using ethanol in Escherichia coli. MethodsX 2015, 2, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Chhetri, G.; Pandey, T.; Kumar, B.; Akhtar, M.S.; Tripathi, T. Recombinant expression, purification and preliminary characterization of the mRNA export factor MEX67 of Saccharomyces cerevisiae. Protein Expr. Purif. 2015, 107, 56–61. [Google Scholar] [CrossRef]
- Thomas, J.G.; Baneyx, F. Protein misfolding and inclusion body formation in recombinant Escherichia coli cells overexpressing Heat-shock proteins. J. Biol. Chem. 1996, 271, 11141–11147. [Google Scholar] [CrossRef]
- Movahed, Z.; Sharif, E.; Ahmadzadeh, M.; Nezafat, N.; Jahandar, H.; Mohit, E. Different strategies for expression and purification of the CT26-poly-neoepitopes vaccine in Escherichia coli. Mol. Biol. Rep. 2022, 49, 859–873. [Google Scholar] [CrossRef]
- Mohammadinezhad, R.; Farahmand, H.; Jalali, S.A.H.; Mirvaghefi, A. Efficient osmolyte-based procedure to increase expression level and solubility of infectious hematopoietic necrosis virus (IHNV) nucleoprotein in E. coli. Appl. Microbiol. Biotechnol. 2018, 102, 4087–4100. [Google Scholar] [CrossRef]
- Sorensen, H.P.; Mortensen, K.K. Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb. Cell Factoties 2005, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Weickert, M.J.; Pagratis, M.; Glascock, C.B.; Blackmore, R. A mutation that improves soluble recombinant hemoglobin accumulation in Escherichia coli in heme excess. Appl. Environ. Microbiol. 1999, 65, 640–647. [Google Scholar] [CrossRef] [PubMed]
- Faiq, M.A.; Ali, M.; Dada, T.; Dada, R.; Saluja, D. A novel methodology for enhanced and consistent heterologous expression of unmodified human cytochrome P450 1B1 (CYP1B1). PLoS ONE 2014, 9, e110473. [Google Scholar] [CrossRef][Green Version]
- Kawasaki, T.; Nakata, T.; Nose, Y. Genetic mapping with a thiamine-requiring auxotroph of Escherichia coli K-12 defective in thiamine phosphate pyrophosphorylase. J. Bacteriol. 1968, 95, 1483–1485. [Google Scholar] [CrossRef] [PubMed]
- Newman, E.B.; Miller, B.; Colebrook, L.D.; Walker, C. A mutation in Escherichia coli K-12 results in a requirement for thiamine and a decrease in L-serine deaminase activity. J. Bacteriol. 1985, 161, 272–276. [Google Scholar] [CrossRef] [PubMed]
- Dailey, H.A.; Dailey, T.A. Protoporphyrinogen oxidase of Myxococcus xanthus. Expression, purification, and characterization of the cloned enzyme. J. Biol. Chem. 1996, 271, 8714–8718. [Google Scholar] [CrossRef]
- Vitreschak, A.G.; Rodionov, D.A.; Mironov, A.A.; Gelfand, M.S. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 2002, 30, 3141–3151. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Angulo, V.A. Overlapping riboflavin supply pathways in bacteria. Crit. Rev. Microbiol. 2017, 43, 196–209. [Google Scholar] [CrossRef] [PubMed]
- Novakova, Z.; Khuntsaria, D.; Gresova, M.; Mikesova, J.; Havlinova, B.; Shukla, S.; Kolarova, L.; Vesela, K.; Martasek, P.; Barinka, C. Heterologous expression and purification of recombinant human protoporphyrinogen oxidase IX: A comparative study. PLoS ONE 2021, 16, e0259837. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Q.; Qi, Q. Identification of riboflavin: Revealing different metabolic characteristics between Escherichia coli BL21(DE3) and MG1655. FEMS Microbiol. Lett. 2015, 362, fnv071. [Google Scholar] [CrossRef] [PubMed]
- Plokhov, A.Y.; Gusyatiner, M.M.; Yampolskaya, T.A.; Kaluzhsky, V.E.; Sukhareva, B.S.; Schulga, A.A. Preparation of γ-aminobutyric acid using E. coli cells with high activity of glutamate decarboxylase. Appl. Biochem. Biotechnol. 2000, 88, 257–265. [Google Scholar] [CrossRef]
- Ito, T.; Downs, D.M. Pyridoxal Reductase, PdxI, Is Critical for Salvage of Pyridoxal in Escherichia coli. J. Bacteriol. 2020, 202, e00056-00020. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Su, L.; Wu, J. Pyridoxine Supplementation Improves the Activity of Recombinant Glutamate Decarboxylase and the Enzymatic Production of Gama-Aminobutyric Acid. PLoS ONE 2016, 11, e0157466. [Google Scholar] [CrossRef]
- Romano, D.; Molla, G.; Pollegioni, L.; Marinelli, F. Optimization of human D-amino acid oxidase expression in Escherichia coli. Protein Expr. Purif. 2009, 68, 72–78. [Google Scholar] [CrossRef]
- de Marco, A.; Deuerling, E.; Mogk, A.; Tomoyasu, T.; Bukau, B. Chaperone-based procedure to increase yields of soluble recombinant proteins produced in E. coli. BMC Biotechnol. 2007, 7, 32. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).