2.1. Carbon Nanotube Structure
In order to digitally synthesize carbon nanotubes, it is essential to understand the fundamental geometry of how the nanotubes naturally form. The structure of carbon nanotubes is intuitively related to graphene. Graphene is a two-dimensional sheet of carbon atoms, bonded in a hexagonal lattice. For each carbon atom in the planar sheet, the atoms’ 2
s and two of the three 2
p orbitals combine into three planar orbitals, 120° apart [
17]. These orbitals represent the very strong
sp2 bonds which give graphene such a stable two-dimensional structure. If the carbon atoms are spread in the
x–
y plane, the remaining carbon valence electron’s orbital, the 2
pz orbital, comes out of the plane in the perpendicular direction to the graphene sheet. This layer of probable electron density provides the relatively weaker bonds between separate sheets of graphene that make up graphite. It is therefore intuitive to imagine that a carbon nanotube is a rolled-up graphene sheet that will tend to focus the interaction potential to act normally to the curved surface of the nanotube. Either van der Waals forces between atoms or transient delocalization of the lingering 2
pz orbital from the carbon atoms contributes to how carbon nanotubes attract and repel matter around them, including other nanotubes (inter-CNT attraction) and even different parts of the same nanotube (intra-CNT attraction) [
17].
These tendencies to attract or repel are why carbon nanotubes are rarely straight in nature, especially single-walled carbon nanotubes (SWCNTs) that are structurally less rigid than multi-walled carbon nanotubes (MWCNTs). When the SWCNTs are long, the curvature happens over length scales that are significantly longer than the nanotube diameter, so the localized compression and tension that could resist bending is relatively low. Nanotubes, therefore, intertwine into tangled masses. As the entangled nanotube structure grows, the likelihood of attraction-inducing curvature increases, and this results in the large, looping, and anisotropic structures typically observed in carbon nanotube clusters [
18,
19,
20].
The difficulty in characterizing such complicated structures for simulations is that they exhibit anisotropy. It is ordered but not chaotic. Randomly creating strands of nanotubes is therefore unlikely to replicate how the structures come together in the first place. Simulating the natural processes that lead to these structures would incorporate the inherent order to provide more accurate representations, but the required durations of such simulations are unachievable with even massive-scale high performance computing resources. Unfortunately doing so would require simulations of time scales that current atomistic simulations still cannot achieve.
2.2. Multi-Scale Coupling
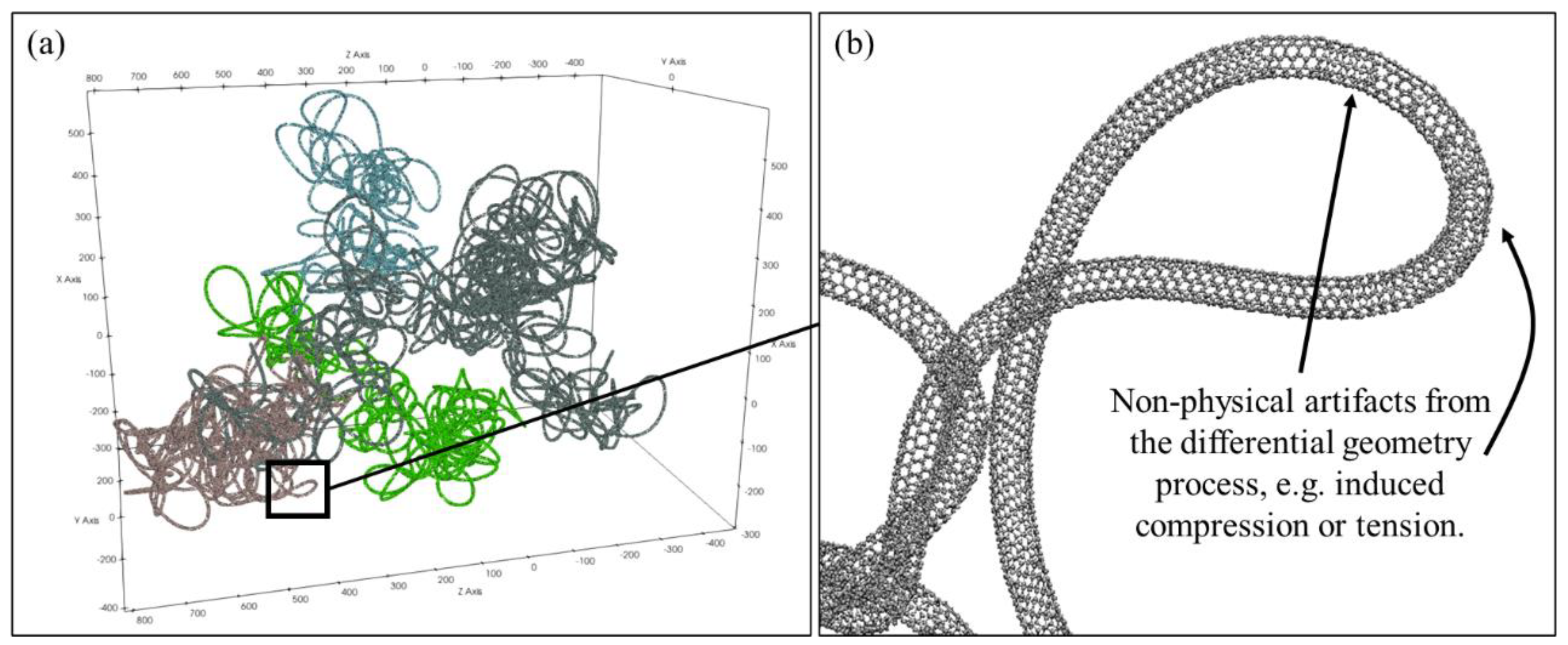
A methodology is presented in this paper to digitally synthesize a three-dimensional nanotube cluster comprised of atomically resolved nanotubes for the purposes of simulating how the cluster would interact with other materials, in particular molten metals. To the authors’ best knowledge, currently there are no carbon nanotube cluster modeling approaches to create atomically resolved clusters of carbon nanotubes that exhibit realistic anisotropy and localized defect details. The digital synthesis of nanotube clusters consists of several phases, including:
Phase I: Use a random walk and coarse-grained molecular model to create a template that serves as the scaffold upon which atomistic nanotubes will be built.
Phase II: Use differential geometry to refine the coarse-grained representations into fully atomistic carbon nanotubes, forming a cluster.
Phase III: Relax the atomistic nanotube cluster using classical molecular dynamics with an accurate potential model that allows for dynamic bonding and bond-breaking, and inter-and intra-nanotube attraction.
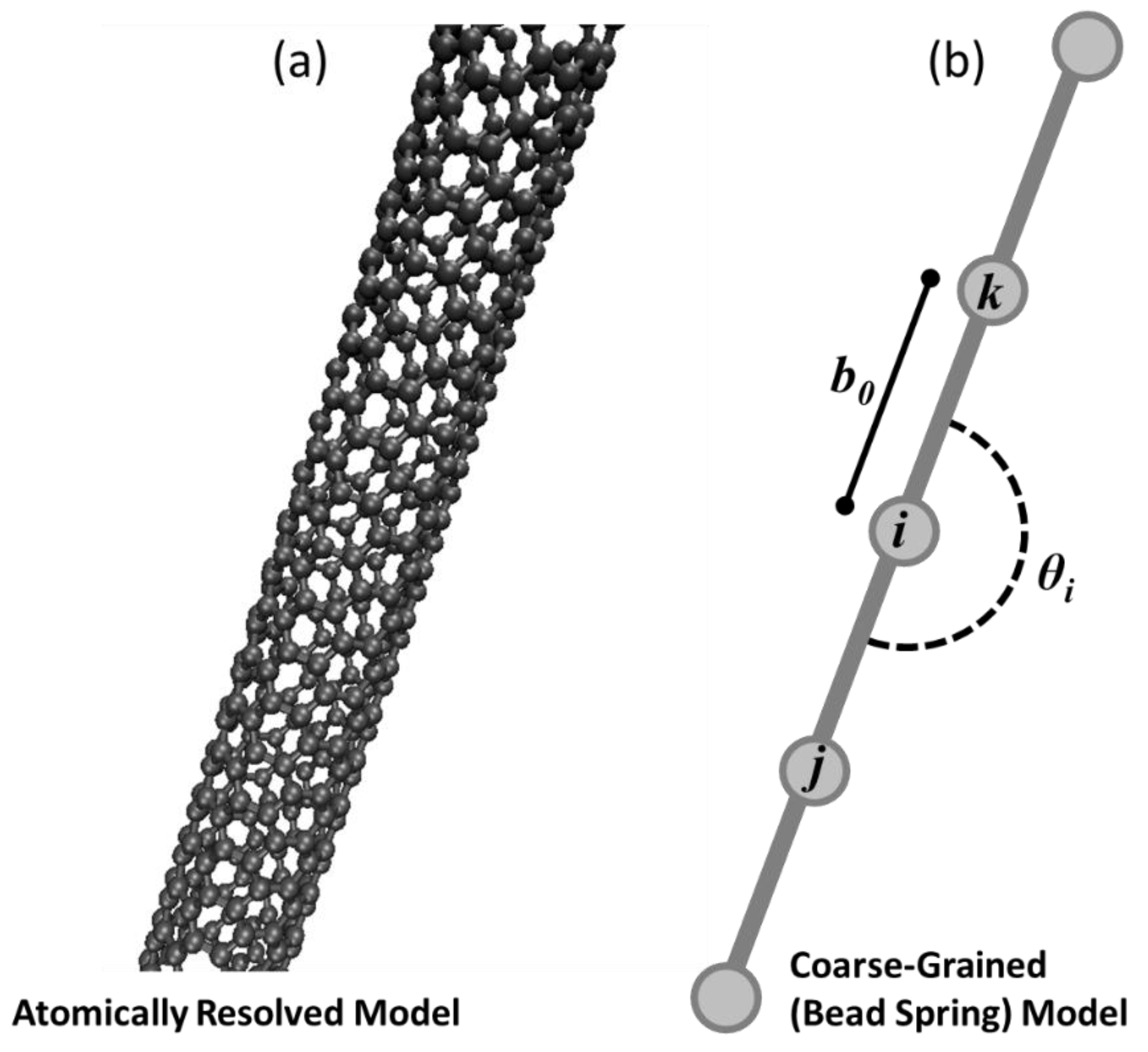
There are two different models for single-walled carbon nanotubes in this work. In Phase I, a Coarse-Grained (CG) bead-spring model [
21], referred to as the CG-CNT model, is used as shown in
Figure 1a. In Phase I, an atomistic model is used that represents carbon nanotubes in full fidelity as bonded carbon atoms shown in
Figure 1a. Despite the difference in scale and fidelity, both nanotube variations model interactions of atoms or coarse-grained beads based on the potential energy for a discrete system as a function of relative atom or bead positions with respect to nearby neighbors. This potential energy approach enables the use of classical molecular dynamics to integrate the trajectories of both representations (beads/nodes or atoms) in time, based on force computations. While the models that govern interactions differ for the two representations of CNTs, the overall approach of simulating them both as systems of atoms in a molecular dynamics simulation is the same. Therefore, both Phase I and Phase III require atomic scale simulations to predict cluster relaxation, but with different computational requirements.
The equations of motion for classical molecular dynamics simulations can be rigorously derived from either Lagrangian mechanics [
22] or Hamiltonian dynamics [
23]. The system is fully described by atomic coordinates and atomic momenta, collectively referred to as the system’s phase space. This dynamic system is given thermodynamic meaning through statistical mechanics where ensemble averages of phase space are related to macroscopic thermodynamic variables. In this paper both the micro-canonical ensemble (constant number of atoms, constant volume, constant energy, NVE) and the canonical ensemble (constant number of atoms, constant volume, and constant temperature, NVT) are used. Among the differences of the two are that the NVE ensemble holds energy constant, treating the system as thermodynamically conserved. The NVT ensemble allows the system to interact with a thermal reservoir where energy can be exchanged. We defer the formal derivations to reference texts [
22,
23] and define here the governing equations of motion for the molecular dynamics system as in [
22] and shown in Equation (1) for a single atom
i, where the vector
denotes spatial coordinates of the atom,
is mass of the atom,
represents potential energy of the atom relative to its neighbors that will be defined below,
is a damping coefficient used only for the canonical ensemble simulations and otherwise set to zero, and
is the instantaneous velocity of the atom.
Potential energy for a given atom is defined following [
22] as shown in Equation (2) where
denotes potential energy functions involving 1, 2, 3, …,
q particles at positions
in three-dimensional space relative to each
ith atom of the system.
Appropriate selection of the potential energy functions is of paramount importance for a molecular dynamics simulation in either the micro-canonical or canonical ensemble. The forms of these functions used in this paper are detailed in subsequent sections and vary between Phase I and Phase III of the methodology presented in this paper. Despite the different choices for the potential energy functions, the open source Large-scale Atomic/Molecular Massively Parallel Simulator (LAMMPS) code [
24] is used for both the CG-CNT relaxation (Phase I) as well as the fully atomistic simulations (Phase III) in this work.
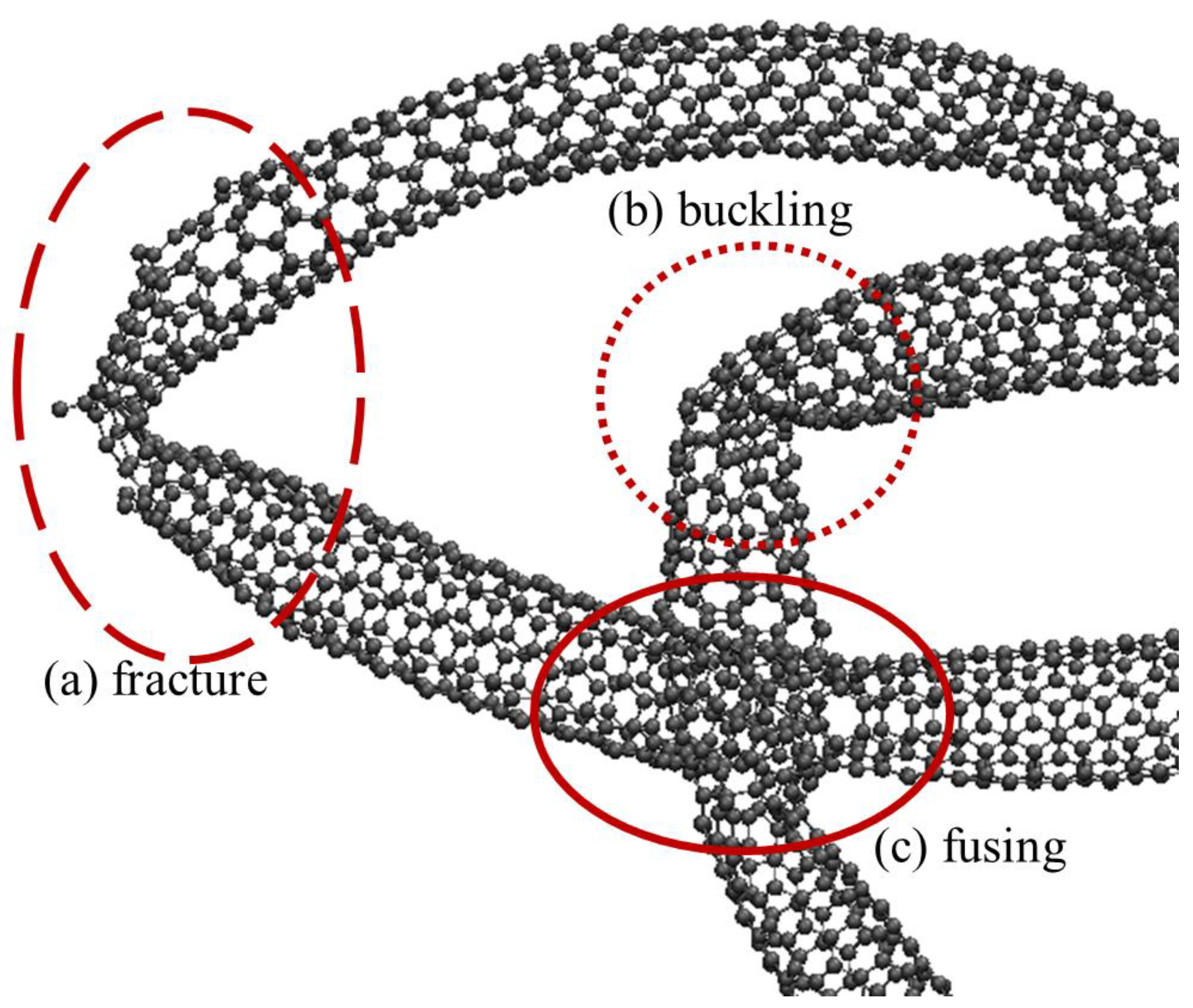
The CG-CNT model is used only in Phase I for creating initial topological scaffolds for the creation of realistic carbon nanotubes and clusters. The CG-CNT model enables large conformational relaxation of fabricated clusters, but does not correctly model CNT defects from buckling, twisting, or bending. The atomistic carbon interactions that are used for all stages of this research beyond initial CNT synthesis are modeled using the Tersoff empirical bond order model for atomistic carbon interactions [
25,
26], detailed in Phase III.
2.4. Phase Ib: Coarse-Grained Carbon Nanotube Modeling
There is no accounting for physically preferential orientations of the discrete curve elements generated during the random walk, but the intended effect of these curves is to serve as something resembling a relatively rigid carbon nanotube. The intuition is therefore that the very tangled and cramped random walk realizations will want to stretch out, and the CG-CNT methodology is employed to govern that relaxation. The relaxation is a process to allow the atoms to settle into equilibrium positions that minimize the system’s potential energy.
There is considerable work in the literature on modeling single- and double-walled carbon nanotubes as coarse-grained filaments, attempting to replicate how clusters of carbon nanotubes exist in nature as individual elements and as interacting structures [
21,
27,
28,
29]. These methods attempt to incorporate more realistic physical modeling of the natural nanotube clustering, but still lack the effects of nanotube fracture, crimping, and torsion in such relatively larger structures. The model used in this paper is based on [
21] where the CG-CNT model coarsens a single-walled carbon nanotube into a filament discretized into evenly spaced beads or nodes, as illustrated in
Figure 1.
The beads are subject to inter- and intra-bead attraction, bending, and bonding forces that are deliberately designed to be analogous with conventional molecular dynamics potentials. Referring to
Figure 1b for graphical depictions of parameters defined in the following equations, three terms contribute to an overall potential energy relationship for any bead
i as a function of its explicitly bonded neighbors (
j, and
k) and any nearby beads within a cutoff radius as shown in Equation (3).
Stretching between bonded neighbors is modeled using a harmonic potential shown in Equation (4) where an equilibrium distance between a bead and its neighbors is defined as
and
is a defined parameter that dictates the magnitude of the potential energy for atomic positions that are not at the equilibrium distance. In this paper,
denotes the non-normalized distance in Cartesian space from coordinate
to coordinate
.
Each bead except the first and final bead of a coarse-grained carbon nanotube model experience this stretching potential for two neighbors, in the illustration in
Figure 1b this would mean that bead
i experiences stretching contributions to the net potential energy from its bond with bead
j and bead
k.
The bending function is a three-body potential, measured about a bead and its two bonded neighbors. An angle defined as
in
Figure 1b is computed using the Law of Cosines and the coordinates of bead
i and its two bonded neighbors. Displacement of that computed angle
from an equilibrium angle
contributes to a harmonic potential energy function weighted by a bending stiffness coefficient,
as shown in Equation (5).
The angle term is only computed for beads with two bonded neighbors.
The final term in the coarse-grained model potential energy function shown in Equation (3) is a pair-wise potential with any non-bonded beads within a cutoff radius. The distance between a bead
i and any non-bonded neighbor bead
m is computed using a Lennard-Jones 12-6 potential as shown in Equation (6) where
and
are the potential energy well depth and equilibrium distance respectively.
For computational efficiency, the pair-wise interactions modeled using Equation (6) are modeled only for as many neighbors as fall within a certain radial distance from a bead, a distance referred to as the Lennard-Jones (LJ) cutoff distance. The contribution of this term rapidly diminishes for atomic distances on the order of two or three times the equilibrium distance σ, so truncating the number of neighbors based on a cutoff radius is a widely accepted procedure for this type of a potential model. The pair-wise potential accounts for inter- and intra-nanotube attraction and is the mechanism through which carbon nanotubes tend to agglomerate into clusters and exhibit looping structures.
The free parameters of the bead-spring model defined in Equations (4)–(6) are determined so that the coarse-grained model will match the tensile strength, bending modulus, and persistence length of a (5,5) single-walled carbon nanotube (
Figure 1a). The parameters used in this paper are shown in
Table 1 and consistent with those reported in [
21,
30,
31].
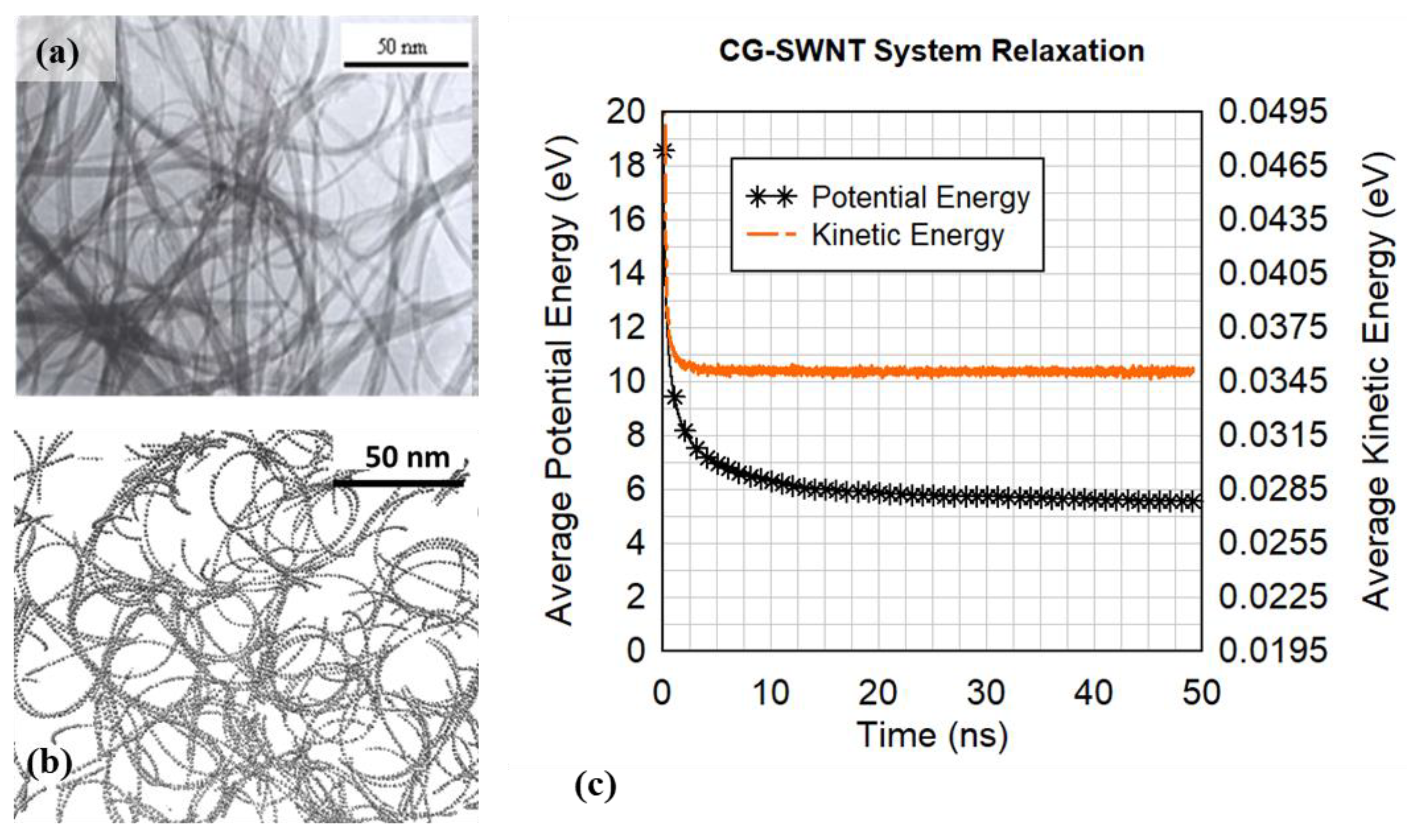
Molecular dynamics simulations using the potential model described in this section relax the previously disordered and energetically unfavorable random walk clusters into filaments that physically resemble carbon nanotubes and nanotube clusters more realistically.
Ensuring the coarse-grained model replicates atomically resolved carbon nanotube properties helps improve the physical accuracy and fidelity of the random walk realizations as they relax into a cluster, improving the behavior of the fully resolved atomic nanotube cluster relaxation in Phase III.
2.5. Phase II: Differential Geometry
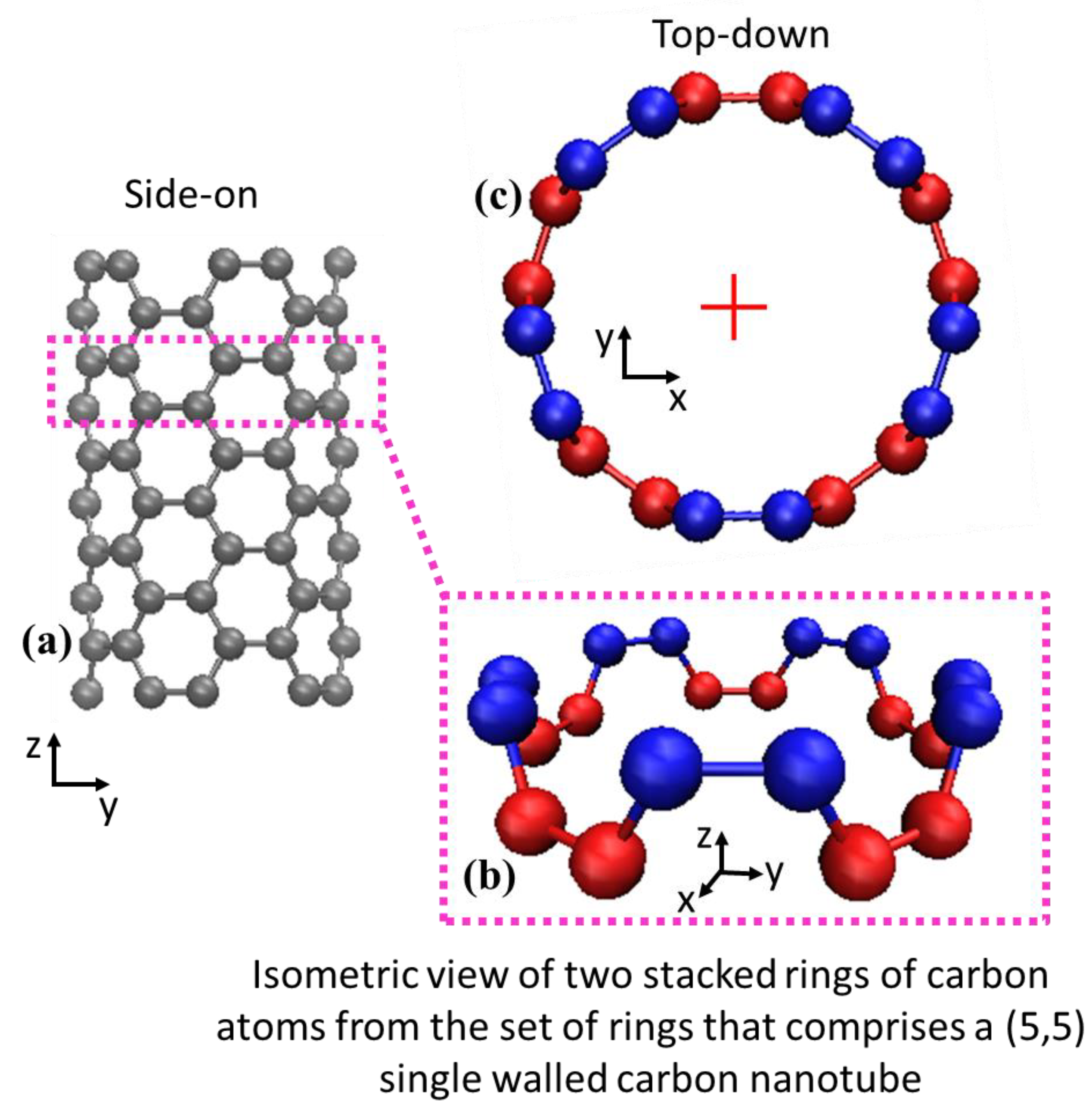
This phase represents a bridge to convert the coarse-grained approximations of nanotubes as filaments into fully atomic carbon nanotubes. The coarse-grained nanotubes function as a central axis upon which carbon atoms are accurately positioned. The process can be envisioned as incrementally building an atomic carbon nanotube as a series of concentric atomic rings stacked atop each other. In the limit of a perfectly straight carbon nanotube (
Figure 1a) the procedure is very simple. Beginning at the base of a bead-spring filament, an initial ring of carbon atoms is placed along the filament’s longitudinal axis, where the carbon atoms are arranged at the known circumferential position for the (5,5) SWNT (e.g., the red atoms in
Figure 2b,c). Another ring is then placed at the known step size
δ along the filament length, rotated by
θ about the axis perpendicular to the plane of the prior ring (e.g., the blue atoms in
Figure 2b,c). Parameters
δ and
θ used in this work are shown in
Table 2 for (5,5) single-walled carbon nanotubes, but adaptation to other arm-chair carbon nanotubes is straightforward. For example, the prevalent (10,10) single-walled carbon nanotube would be built the same way as described here, except for changes to the single carbon ring commensurate with the additional carbon atoms and increased radius, and different
δ and
θ parameters.
An example of a straight carbon nanotube is depicted in
Figure 2a and examples of two levels of stacked atomic rings are shown by red and blue layers in
Figure 2b,c.
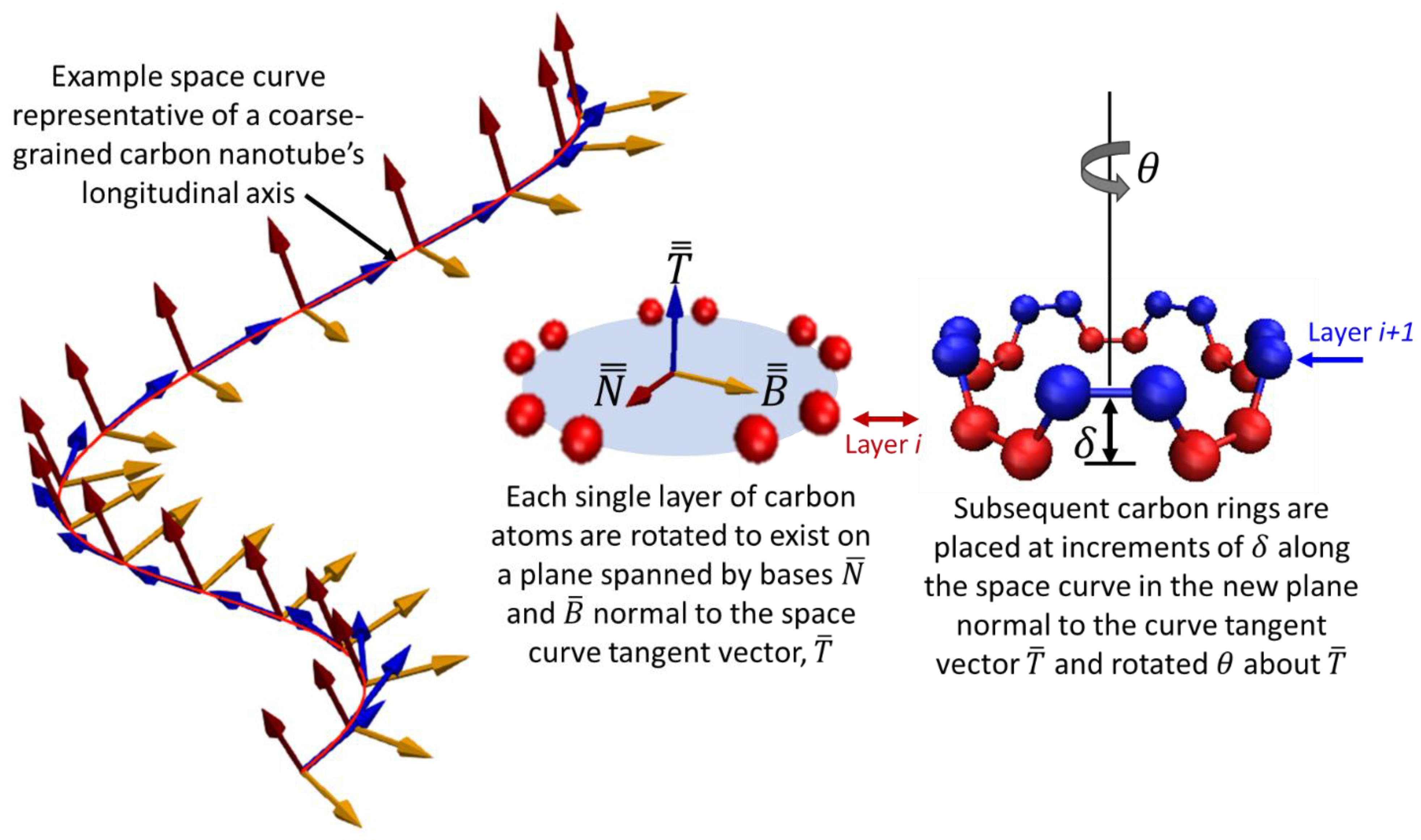
For a straight nanotube, repeated operations of rotating and stacking of atomic rings of carbon would incrementally build a nanotube and are trivial to execute. Real carbon nanotubes are almost never straight however, which complicates the rotation operation of the stacking procedure. The central axis normally exhibits curvature and so an additional rotation must take place for the plane of the atomic ring, but it cannot be arbitrary or else the nanotube lattice will not be maintained. A differential geometry approach to curve framing is used called the Parallel Transport frame [
32,
33]. This approach provides mathematical formalism to create an initial nanotube structure that orients the discrete atomic rings correctly along the axis of the curved bead-spring filament. It is similar in nature to Frenet-Serret frames [
34] that define planes along a space curve, but is more suitable for the present research because the calculations of Frenet-Serret frames requires the second order derivative of the curve which does not exist for straight lines and manifests in abrupt orientation changes when the second derivative approaches and crosses local minima and maxima. Carbon nanotubes in this research exist in both of those cases and so Frenet-Serret frames are not ideal. Janakiev provides a very useful description and implementation examples of parallel transport frames in [
33] that complements the original work by Hanson and Ma [
32] and differentiates it from Frenet-Serret. The concept is illustrated for a generic three-dimensional space curve in
Figure 3 where the red line is a curve defined in three-dimensions, and the parallel transport frames approach is illustrated at finite increments along the curve where local frames are shown to remain consistent.
Consider a discrete curve in space illustrated in
Figure 3, composed of discrete points represented by the matrix
which is an
m × 3 matrix where
m is the number of discrete points that comprise the curve, each point being in three dimensions. The parallel transport of the frames begins with the first derivative computed with respect to the spatial dimensions of the space to obtain the curve’s tangent vectors,
as shown in Equation (7). These are then normalized and are shown in
Figure 3 as the set of blue vectors.
The set of normal vectors to the tangents,
in
Figure 3 are then computed using the normalized cross-product between two successive tangent vectors using Equation (8).
A final basis vector set is then computed from the cross products of sets and .
Each discrete frame spanned by basis vectors
and
from sets
and
shown in
Figure 3 represent the plane orientation of the carbon atoms unit cell, pictured as the red or blue atoms in
Figure 2b,c. The central axis of the coarse-grained nanotube filament is the red curve, with piecewise orientation vectors denoted by the blue vectors in
Figure 3.
At each increment δ along the central axis, the carbon atom ring is rotated θ about the prior increment’s blue vector and then rotated again to coincide with the plane defined by the red and yellow vectors that are each orthogonal to the tangential vector (blue).
Although the methodology presented here was based on parallel transport frames reported in the literature, the application of it to create realistic carbon nanotubes is new.
2.6. Phase III: Atomistic Carbon Nanotube Modeling
Phase I creates individual filaments of realistically curved, looping, and agglomerated carbon nanotube surrogates to serve as a central axis for an atomic representation of a carbon nanotube. Phase II orients the carbon rings along all of the filaments to create atomic carbon nanotubes in the cluster. Phase III then models the carbon atoms individually to allow the whole carbon nanotube cluster to behave as a complex system with atomic resolution.
There are a variety of classical molecular dynamics potentials used for atomistic modeling of carbon structures, but for this work the Tersoff empirical bond order model is chosen [
26] with optimized carbon–carbon model parameters defined by Lindsay and Broido [
25]. Practical experience using other dynamic potential models including ReaxFF and AI-REBO have informed the conclusion that the Tersoff potential provides the best balance of requisite fidelity of carbon nanotube response and computational efficiency as befits the overall problem at hand. Since the Tersoff potential model includes dynamic bonding and bond-breaking commensurate with nanotube deformation, it has enough physical fidelity for the present application while also being stable and efficient for the number of carbon–carbon interactions modeled in this work. The Tersoff potential model is introduced here but the reader is referred to [
26] for greater detail. The general potential energy functional form Equation (2) is defined for the Tersoff model as
in Equation (9) which is the summation of pairwise bond energies
Vij between atoms
i and
j, shown respectively in Equations (9) and (10) which includes both attractive Equation (11) and repulsive Equation (12) potential energy terms, as well as a smoothing function Equation (13) that implements a cut-off for interactions that become vanishingly small with increased separations.
The coefficients
A,
B,
,
,
R, and
D are model parameters defined for the system of interest in [
25], and the
and
terms in Equation (10) are defined in Equation (14) through Equation (18) from [
26] with model coefficients
c, d,
,
α,
β, and
n also defined in [
25].
The Tersoff potential is very widely used for atomic carbon–carbon interactions and the model parameters, and the coefficients used in Equation (10) through Equation (18) do not vary based on a particular application of the methodology. Since the model predicts carbon–carbon interactions, the configuration being carbon nanotubes, graphene, graphite, or amorphous carbon does not matter, just that the Tersoff potential model predicts physical bonding and orientation of individual carbon atoms. The present work uses model coefficients defined using the parameterization for carbon–carbon interactions from [
25], and as implemented in LAMMPS [
24]. The Tersoff potential is used for all atomically resolved motion of the carbon nanotube cluster in Phase III of the presented methodology when relaxing the carbon nanotube cluster to a more energetically favorable configuration.