LADON, a Natural Antisense Transcript of NODAL, Promotes Tumour Progression and Metastasis in Melanoma
Abstract
:1. Introduction
2. Results
2.1. NODAL Exon 2 Promotes Tumour Growth and Metastasis in A375 Cells
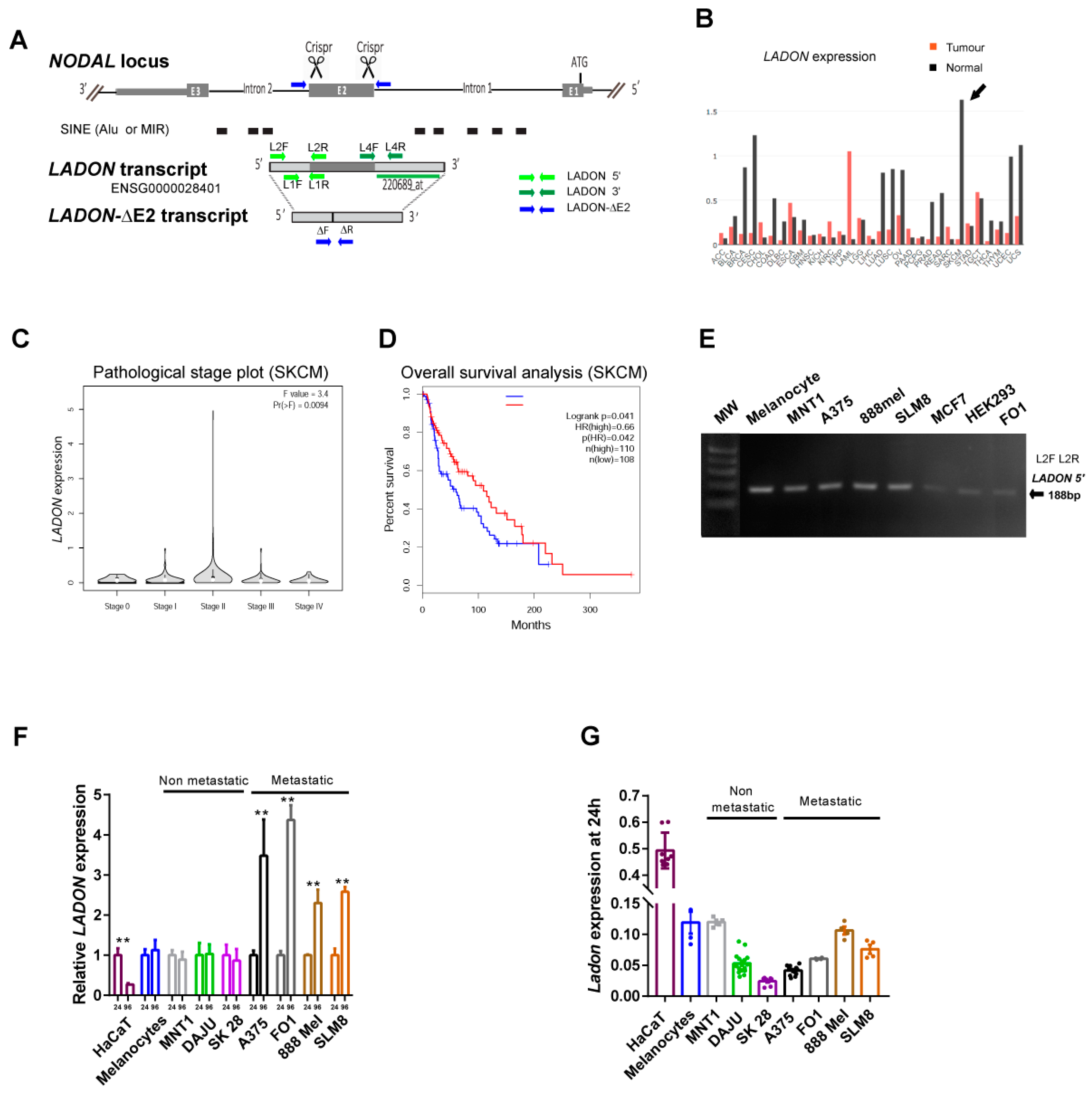
2.2. A Natural Antisense Transcript of NODAL Exon 2 Is Expressed in Melanoma Cells
2.3. Metastatic Melanoma Cell Lines Share the Ability to Upregulate LADON
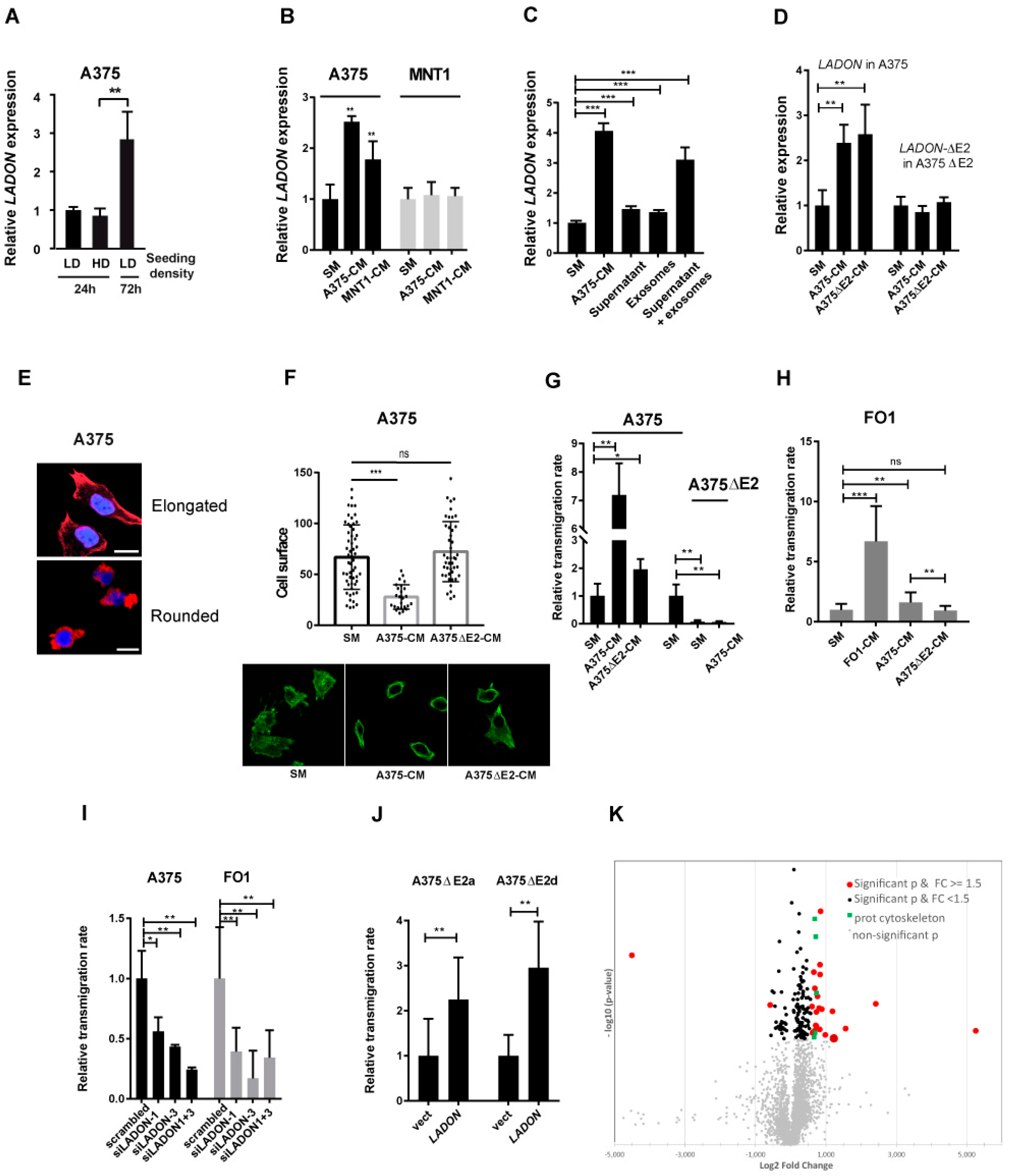
2.4. Melanoma Cells Secrete Factors That Promote LADON Expression
2.5. LADON Expression Promotes Invasion
2.6. LADON Affects the Expression of Oncogenes and Suppressors of Tumour Progression
3. Discussion
4. Materials and Methods
4.1. Plasmids
4.2. Cell Lines and Cell Culture
4.3. Transmigration Assay
4.4. A375 Staining
4.5. Wound Healing/Scratch Assay
4.6. CRISPR/CAS9 Genome Editing
4.7. siRNAs
4.8. RNA Extraction, Reverse Transcription (RT) and Quantitative PCR (Q-PCR)
4.9. CAM Assay
4.10. Histology
4.11. LC-MS/MS Acquisition
4.12. LC-MS/MS Data Processing
4.13. Statistical Analysis
4.14. Data Collection and Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chaffer, C.L.; Weinberg, R.A. A Perspective on Cancer Cell Metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef]
- Quail, D.F.; Siegers, G.M.; Jewer, M.; Postovit, L.-M. Nodal Signalling in Embryogenesis and Tumourigenesis. Int. J. Biochem. Cell Biol. 2013, 45, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Topczewska, J.M.; Postovit, L.-M.; Margaryan, N.V.; Sam, A.; Hess, A.R.; Wheaton, W.W.; Nickoloff, B.J.; Topczewski, J.; Hendrix, M.J.C. Embryonic and Tumorigenic Pathways Converge via Nodal Signaling: Role in Melanoma Aggressiveness. Nat. Med. 2006, 12, 925–932. [Google Scholar] [CrossRef]
- Robertson, E.J. Dose-Dependent Nodal/Smad Signals Pattern the Early Mouse Embryo. Semin. Cell Dev. Biol. 2014, 32, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Bianco, C.; Adkins, H.B.; Wechselberger, C.; Seno, M.; Normanno, N.; De Luca, A.; Sun, Y.; Khan, N.; Kenney, N.; Ebert, A.; et al. Cripto-1 Activates Nodal- and ALK4-Dependent and -Independent Signaling Pathways in Mammary Epithelial Cells. Mol. Cell. Biol. 2002, 22, 2586–2597. [Google Scholar] [CrossRef] [PubMed]
- Papageorgiou, I.; Nicholls, P.K.; Wang, F.; Lackmann, M.; Makanji, Y.; Salamonsen, L.A.; Robertson, D.M.; Harrison, C.A. Expression of Nodal Signalling Components in Cycling Human Endometrium and in Endometrial Cancer. Reprod. Biol. Endocrinol. 2009, 7, 122. [Google Scholar] [CrossRef]
- Bodenstine, T.M.; Chandler, G.S.; Seftor, R.E.B.; Seftor, E.A.; Hendrix, M.J.C. Plasticity Underlies Tumor Progression: Role of Nodal Signaling. Cancer Metastasis. Rev. 2016, 35, 21–39. [Google Scholar] [CrossRef]
- Donovan, P.; Dubey, O.A.; Kallioinen, S.; Rogers, K.W.; Muehlethaler, K.; Müller, P.; Rimoldi, D.; Constam, D.B. Paracrine Activin-A Signaling Promotes Melanoma Growth and Metastasis through Immune Evasion. J. Investig. Dermatol. 2017, 137, 2578–2587. [Google Scholar] [CrossRef]
- Hooijkaas, A.I.; Gadiot, J.; van Boven, H.; Blank, C. Expression of the Embryological Morphogen Nodal in Stage III/IV Melanoma. Melanoma Res. 2011, 21, 491–501. [Google Scholar] [CrossRef]
- Strizzi, L.; Hardy, K.M.; Kirschmann, D.A.; Ahrlund-Richter, L.; Hendrix, M.J.C. Nodal Expression and Detection in Cancer: Experience and Challenges. Cancer Res. 2012, 72, 1915–1920. [Google Scholar] [CrossRef]
- Postovit, L.-M.; Margaryan, N.V.; Seftor, E.A.; Kirschmann, D.A.; Lipavsky, A.; Wheaton, W.W.; Abbott, D.E.; Seftor, R.E.B.; Hendrix, M.J.C. Human Embryonic Stem Cell Microenvironment Suppresses the Tumorigenic Phenotype of Aggressive Cancer Cells. Proc. Natl. Acad. Sci. USA 2008, 105, 4329–4334. [Google Scholar] [CrossRef] [PubMed]
- Collignon, J.; Sockanathan, S.; Hacker, A.; Cohen-Tannoudji, M.; Norris, D.; Rastan, S.; Stevanovic, M.; Goodfellow, P.N.; Lovell-Badge, R. A Comparison of the Properties of Sox-3 with Sry and Two Related Genes, Sox-1 and Sox-2. Development 1996, 122, 509–520. [Google Scholar] [CrossRef]
- Ribatti, D. The Chick Embryo Chorioallantoic Membrane (CAM) Assay. Reprod. Toxicol. 2017, 70, 97–101. [Google Scholar] [CrossRef]
- Ota, T.; Suzuki, Y.; Nishikawa, T.; Otsuki, T.; Sugiyama, T.; Irie, R.; Wakamatsu, A.; Hayashi, K.; Sato, H.; Nagai, K.; et al. Complete Sequencing and Characterization of 21,243 Full-Length Human cDNAs. Nat. Genet. 2004, 36, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Kim, D.S.; Lee, M.W.; Lee, T.-H.; Sung, K.W.; Koo, H.H.; Yoo, K.H. Cell Culture Density Affects the Stemness Gene Expression of Adipose Tissue-Derived Mesenchymal Stem Cells. Biomed. Rep. 2017, 6, 300–306. [Google Scholar] [CrossRef]
- Xiao, D.; Ohlendorf, J.; Chen, Y.; Taylor, D.D.; Rai, S.N.; Waigel, S.; Zacharias, W.; Hao, H.; McMasters, K.M. Identifying mRNA, MicroRNA and Protein Profiles of Melanoma Exosomes. PLoS ONE 2012, 7, e46874. [Google Scholar] [CrossRef]
- Friedl, P.; Wolf, K. Plasticity of Cell Migration: A Multiscale Tuning Model. J. Cell Biol. 2010, 188, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Moreno, V.; Gadea, G.; Ahn, J.; Paterson, H.; Marra, P.; Pinner, S.; Sahai, E.; Marshall, C.J. Rac Activation and Inactivation Control Plasticity of Tumor Cell Movement. Cell 2008, 135, 510–523. [Google Scholar] [CrossRef]
- Gadea, G.; de Toledo, M.; Anguille, C.; Roux, P. Loss of P53 Promotes RhoA-ROCK-Dependent Cell Migration and Invasion in 3D Matrices. J. Cell Biol. 2007, 178, 23–30. [Google Scholar] [CrossRef]
- Panková, K.; Rösel, D.; Novotný, M.; Brábek, J. The Molecular Mechanisms of Transition between Mesenchymal and Amoeboid Invasiveness in Tumor Cells. Cell. Mol. Life Sci. 2010, 67, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Moreno, V.; Gaggioli, C.; Yeo, M.; Albrengues, J.; Wallberg, F.; Viros, A.; Hooper, S.; Mitter, R.; Féral, C.C.; Cook, M.; et al. ROCK and JAK1 Signaling Cooperate to Control Actomyosin Contractility in Tumor Cells and Stroma. Cancer Cell. 2011, 20, 229–245. [Google Scholar] [CrossRef] [PubMed]
- Sahai, E.; Marshall, C.J. Differing Modes of Tumour Cell Invasion Have Distinct Requirements for Rho/ROCK Signalling and Extracellular Proteolysis. Nat. Cell. Biol. 2003, 5, 711–719. [Google Scholar] [CrossRef]
- Guo, J.; Bian, Y.; Wang, Y.; Chen, L.; Yu, A.; Sun, X. FAM107B Is Regulated by S100A4 and Mediates the Effect of S100A4 on the Proliferation and Migration of MGC803 Gastric Cancer Cells. Cell Biol. Int. 2017, 41, 1103–1109. [Google Scholar] [CrossRef]
- Meeusen, B.; Janssens, V. Tumor Suppressive Protein Phosphatases in Human Cancer: Emerging Targets for Therapeutic Intervention and Tumor Stratification. Int. J. Biochem. Cell Biol. 2018, 96, 98–134. [Google Scholar] [CrossRef]
- Sharma, A.; Mendonca, J.; Ying, J.; Kim, H.S.; Verdone, J.E.; Zarif, J.C.; Carducci, M.; Hammers, H.; Pienta, K.J.; Kachhap, S. The Prostate Metastasis Suppressor Gene NDRG1 Differentially Regulates Cell Motility and Invasion. Mol. Oncol. 2017, 11, 655–669. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhang, D.; Bae, D.-H.; Sahni, S.; Jansson, P.; Zheng, Y.; Zhao, Q.; Yue, F.; Zheng, M.; Kovacevic, Z.; et al. Metastasis Suppressor, NDRG1, Mediates Its Activity through Signaling Pathways and Molecular Motors. Carcinogenesis 2013, 34, 1943–1954. [Google Scholar] [CrossRef]
- Zelivianski, S.; Glowacki, R.; Lin, M.F. Transcriptional Activation of the Human Prostatic Acid Phosphatase Gene by NF-κB via a Novel Hexanucleotide-Binding Site. Nucleic Acids Res. 2004, 32, 3566–3580. [Google Scholar] [CrossRef]
- Hastie, C.J.; Carnegie, G.K.; Morrice, N.; Cohen, P.T. A Novel 50 kDa Protein Forms Complexes with Protein Phosphatase 4 and Is Located at Centrosomal Microtubule Organizing Centres. Biochem. J. 2000, 347, 845–855. [Google Scholar] [CrossRef]
- Li, X.; Liang, L.; Huang, L.; Ma, X.; Li, D.; Cai, S. High Expression of Protein Phosphatase 4 Is Associated with the Aggressive Malignant Behavior of Colorectal Carcinoma. Mol. Cancer 2015, 14, 95. [Google Scholar] [CrossRef]
- Findlay, S.D.; Postovit, L.-M. Comprehensive Characterization of Transcript Diversity at the Human NODAL Locus. bioRxiv 2018. [Google Scholar] [CrossRef]
- Hardy, K.M.; Kirschmann, D.A.; Seftor, E.A.; Margaryan, N.V.; Postovit, L.-M.; Strizzi, L.; Hendrix, M.J.C. Regulation of the Embryonic Morphogen Nodal by Notch4 Facilitates Manifestation of the Aggressive Melanoma Phenotype. Cancer Res. 2010, 70, 10340–10350. [Google Scholar] [CrossRef] [PubMed]
- Carreira, S.; Goodall, J.; Denat, L.; Rodriguez, M.; Nuciforo, P.; Hoek, K.S.; Testori, A.; Larue, L.; Goding, C.R. Mitf Regulation of Dia1 Controls Melanoma Proliferation and Invasiveness. Genes Dev. 2006, 20, 3426–3439. [Google Scholar] [CrossRef]
- Hoek, K.S.; Schlegel, N.C.; Brafford, P.; Sucker, A.; Ugurel, S.; Kumar, R.; Weber, B.L.; Nathanson, K.L.; Phillips, D.J.; Herlyn, M.; et al. Metastatic Potential of Melanomas Defined by Specific Gene Expression Profiles with No BRAF Signature. Pigment Cell Res. 2006, 19, 290–302. [Google Scholar] [CrossRef]
- Hoek, K.S.; Eichhoff, O.M.; Schlegel, N.C.; Dobbeling, U.; Kobert, N.; Schaerer, L.; Hemmi, S.; Dummer, R. In Vivo Switching of Human Melanoma Cells between Proliferative and Invasive States. Cancer Res. 2008, 68, 650–656. [Google Scholar] [CrossRef]
- Huarte, M. The Emerging Role of lncRNAs in Cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The Landscape of Long Noncoding RNAs in the Human Transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016, 29, 452–463. [Google Scholar] [CrossRef]
- Slaby, O.; Laga, R.; Sedlacek, O. Therapeutic Targeting of Non-Coding RNAs in Cancer. Biochem. J. 2017, 474, 4219–4251. [Google Scholar] [CrossRef]
- Wahlestedt, C. Targeting Long Non-Coding RNA to Therapeutically Upregulate Gene Expression. Nat. Rev. Drug Discov. 2013, 12, 433–446. [Google Scholar] [CrossRef]
- Denzler, R.; Agarwal, V.; Stefano, J.; Bartel, D.P.; Stoffel, M. Assessing the ceRNA Hypothesis with Quantitative Measurements of miRNA and Target Abundance. Mol. Cell 2014, 54, 766–776. [Google Scholar] [CrossRef]
- Denzler, R.; McGeary, S.E.; Title, A.C.; Agarwal, V.; Bartel, D.P.; Stoffel, M. Impact of MicroRNA Levels, Target-Site Complementarity, and Cooperativity on Competing Endogenous RNA-Regulated Gene Expression. Mol. Cell 2016, 64, 565–579. [Google Scholar] [CrossRef]
- Jachowicz, J.W.; Strehle, M.; Banerjee, A.K.; Blanco, M.R.; Thai, J.; Guttman, M. Xist Spatially Amplifies SHARP/SPEN Recruitment to Balance Chromosome-Wide Silencing and Specificity to the X Chromosome. Nat. Struct Mol. Biol. 2022, 29, 239–249. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long Non-Coding RNAs: Definitions, Functions, Challenges and Recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Maquat, L.E. LncRNAs Transactivate STAU1-Mediated mRNA Decay by Duplexing with 39 UTRs via Alu Eleme. Nature 2011, 470, 284–290. [Google Scholar] [CrossRef]
- Holdt, L.M.; Hoffmann, S.; Sass, K.; Langenberger, D.; Scholz, M.; Krohn, K.; Finstermeier, K.; Stahringer, A.; Wilfert, W.; Beutner, F.; et al. Alu Elements in ANRIL Non-Coding RNA at Chromosome 9p21 Modulate Atherogenic Cell Functions through Trans-Regulation of Gene Networks. PLoS Genet. 2013, 9, e1003588. [Google Scholar] [CrossRef]
- Hu, S.; Wang, X.; Shan, G. Insertion of an Alu Element in a lncRNA Leads to Primate-Specific Modulation of Alternative Splicing. Nat. Struct. Mol. Biol. 2016, 23, 1011–1019. [Google Scholar] [CrossRef]
- Carrieri, C.; Cimatti, L.; Biagioli, M.; Beugnet, A.; Zucchelli, S.; Fedele, S.; Pesce, E.; Ferrer, I.; Collavin, L.; Santoro, C.; et al. Long Non-Coding Antisense RNA Controls Uchl1 Translation through an Embedded SINEB2 Repeat. Nature 2012, 491, 454–457. [Google Scholar] [CrossRef]
- Johnson, R.; Guigó, R. The RIDL Hypothesis: Transposable Elements as Functional Domains of Long Noncoding RNAs. RNA 2014, 20, 959–976. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Kim, H.K.; Min, S.; Song, M.; Jung, S.; Choi, J.W.; Kim, Y.; Lee, S.; Yoon, S.; Kim, H. Deep learning improves prediction of CRISPR–Cpf1 guide RNA activity. Nat. Biotechnol. 2018, 36, 239–241. [Google Scholar] [CrossRef]


| Protein | Ratio Changes A375ΔE2 vs. A375 | (p) ANOVA | Filamentous Actin | Stress Fiber | Lamellipodium | Accession |
|---|---|---|---|---|---|---|
| Dynactin subunit 4 (DCTN4) | 1.664529632 | 0.01351308 | ■ | Q9UJW0 | ||
| 1phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 (PLCG1) | 1.637021061 | 0.00271601 | ■ | 19174 | ||
| Fermitin family homolog 2 (FERMT2) | 1.618822307 | 0.0415036 | ■ | ■ | ■ | Q96AC1 |
| PDZ and LIM domain protein (PDLIM4) | 1.606767329 | 0.0016411 | ■ | ■ | ■ | P50479 |
| Nck-associated protein 1 (NCKAP1) | 1.580463985 | 0.047219 | ■ | ■ | Q9Y2A7 |
| Protein | Ratio Changes A375ΔE2 vs. A375 | (p) ANOVA | Fonction | Accession |
|---|---|---|---|---|
| Prostatic acid phosphatase (ACPP) | 38.17912767 | 0.03938661 | tumor suppressor, regulated by p65 | P15309 |
| Transcription factor p65 (RELA) | 5.3358974 | 0.01828842 | transcription factor, cell proliferation, apoptosis, and oncogenesis | Q04206 |
| Gamma-aminobutyric acid receptor-associated protein-like2 (GABARAPL2) | 2.947432444 | 0.03702373 | Ubiquitin-like modifier involved in intra-Golgi traffic | P60520 |
| Protein FAM107B (FAM107B) | 2.274639991 | 0.02268322 | tumor suppressor | Q9H098 |
| Protein NDRG1 (NDRG1) | 2.33790023 | 0.04885314 | metastasis suppressor | Q92597 |
| Serine/threonine-protein phosphatase 4 regulatory subunit 2 (PPP4R2) | −22.78074964 | 0.03702373 | overexpression in cancer promotes cell growth and invasion | P60520 |
| Chemicals, Peptides, and Recombinant Proteins | Source | Identifier |
|---|---|---|
| D-MEM/F-12 (1:1) (1X), liquid plus Glutamax, | LifeTechnologies Carlsbad, CA, USA | 31331028 |
| FBS Foetal Bovine Serum, Origin: E.U. Approved (South American) | Life Technologies | 10270106 |
| Penicillin/Streptomycin | Life Technologies | 15140-148 |
| Trypsin 0.5% EDTA | Thermo Fisher Waltham, MA, USA, États-Unis | 25300054 |
| DAPI (4′, 6-diamidino-2-phenylindole) | Invitrogen Molecular Probes (Thermo Fisher) | D1306 |
| ACTIVIN | CELL guidance systems | GFM29 |
| SB431542 | Millipore, Guyancourt, France | 616461 |
| Dharmafect 1 | Dharmacon, Lafayette, CO, USA | T-2001 |
| NODAL | Biotechne, Minneapolis, MN USA | 1315-ND-025 |
| Experimental Models: Cell Lines | ||
| A375M | American Type Culture Collection, Manassas, VA, USA | cat# ATCC CRL-1619 |
| Melanocyte | From Nathalie Andrieu | N/A |
| MNT1, FO1 | From Lionel Larue | N/A |
| SLM8 | From Manuelle Viguier | N/A |
| 888 Mel | From Alain Mauviel | N/A |
| Oligonucleotides | ||
| Primers | See Table S2 | N/A |
| ON-TARGETplus Non-targeting Control siRNAs | Dharmacon | D-001810-01-05 |
| ON-TARGETplus LADON siRNAs 1,2 and 3 | Dharmacon | SO-2770618G |
| Recombinant DNA/Plasmids | ||
| pcDNA3.1 | Thermo Fisher Scientific | V79520 |
| pcDNA3.1 GFP | Thermo Fisher Scientific | |
| pcDDA3.1 LADON full-length | This paper | N/A |
| Critical Commercial Assays | ||
| SUPERSCRIPT III VILO (50 réactions) | Thermo Fisher Scientific | 11755050 |
| LightCycler FastStart DNA Master SYBR Green I | Roche | 12239264001 |
| RiboCellin | EUROBIO, Les Ulis France | RC1000 |
| Software and Algorithms | ||
| Graph Prism | GraphPad Software | Graphpad Prism 10.0.3 |
| Adobe Illustrator | Adobe | Adobe illustrator CS3 |
| Image J | [50] | ImageJ |
| UCSC | [51] | UCSC |
| Excel | Microsoft | Microsoft 2019 |
| CFX Maestro | Bio-Rad | CFX Maestro |
| GPP sgRNA Design | [52] | portals.broadinstitute.org/gppx/crispick/public |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dutriaux, A.; Diazzi, S.; Bresesti, C.; Hardouin, S.; Deshayes, F.; Collignon, J.; Flagiello, D. LADON, a Natural Antisense Transcript of NODAL, Promotes Tumour Progression and Metastasis in Melanoma. Non-Coding RNA 2023, 9, 71. https://doi.org/10.3390/ncrna9060071
Dutriaux A, Diazzi S, Bresesti C, Hardouin S, Deshayes F, Collignon J, Flagiello D. LADON, a Natural Antisense Transcript of NODAL, Promotes Tumour Progression and Metastasis in Melanoma. Non-Coding RNA. 2023; 9(6):71. https://doi.org/10.3390/ncrna9060071
Chicago/Turabian StyleDutriaux, Annie, Serena Diazzi, Chiara Bresesti, Sylvie Hardouin, Frédérique Deshayes, Jérôme Collignon, and Domenico Flagiello. 2023. "LADON, a Natural Antisense Transcript of NODAL, Promotes Tumour Progression and Metastasis in Melanoma" Non-Coding RNA 9, no. 6: 71. https://doi.org/10.3390/ncrna9060071
APA StyleDutriaux, A., Diazzi, S., Bresesti, C., Hardouin, S., Deshayes, F., Collignon, J., & Flagiello, D. (2023). LADON, a Natural Antisense Transcript of NODAL, Promotes Tumour Progression and Metastasis in Melanoma. Non-Coding RNA, 9(6), 71. https://doi.org/10.3390/ncrna9060071






