Abstract
Trypanosomatids are protozoan parasites that cause devastating vector-borne human diseases. Gene expression regulation of these organisms depends on post-transcriptional control in responding to diverse environments while going through multiple developmental stages of their complex life cycles. In this scenario, non-coding RNAs (ncRNAs) are excellent candidates for a very efficient, quick, and economic strategy to regulate gene expression. The advent of high throughput RNA sequencing technologies show the presence and deregulation of small RNA fragments derived from canonical ncRNAs. This review seeks to depict the ncRNA landscape in trypanosomatids, focusing on the small RNA fragments derived from functional RNA molecules observed in RNA sequencing studies. Small RNA fragments derived from canonical ncRNAs (tsRNAs, snsRNAs, sdRNAs, and sdrRNAs) were identified in trypanosomatids. Some of these RNAs display changes in their levels associated with different environments and developmental stages, demanding further studies to determine their functional characterization and potential roles. Nevertheless, a comprehensive and detailed ncRNA annotation for most trypanosomatid genomes is still needed, allowing better and more extensive comparative and functional studies.
Keywords:
trypanosomatids; tritryp; Trypanosoma; leishmania; non-coding RNA; ncRNA; tRNA; snoRNA; snRNA; siRNA 1. Introduction
Trypanosomatids are flagellated protozoan parasites belonging to the early branching supergroup Discobids, formerly part of the eukaryotic lineage of excavates, which are no longer supported as a monophyletic clade. Thus, these organisms comprise a very early divergent group in the eukaryotic evolutionary tree and show many distinctive features, which are not present in well-studied fungi, plant, or animal cells, providing numerous insights into the early establishment of cell and molecular mechanisms of eukaryotic [1,2]. Trypanosomatids belong to the class Kinetoplastea, named by the peculiar genome architecture of their unique mitochondrion composed of a network of circular DNA molecules called the kinetoplast [3].
Trypanosomes cause devastating vector-borne human diseases, mainly in tropical and sub-tropical regions of the world where their specific transmitting arthropods are endemic. African trypanosomiasis (sleeping sickness) caused by Trypanosoma brucei, American trypanosomiasis (Chagas disease) caused by Trypanosoma cruzi, and leishmaniasis (a set of diverse diseases) caused by various species of the genre Leishmania, are three major parasitoses that impact hundreds of millions of people worldwide [4,5,6]. Trypanosomatids display very complex life cycles with different developmental stages alternating between the invertebrate and the vertebrate hosts, which are usually different mammal species, thus comprising intricate zoonoses [7,8].
Several unusual gene expression mechanisms in trypanosomatids exemplify the divergence across eukaryotic organisms. First and foremost, there is no evidence of canonical core promoter elements for RNA polymerase II to transcribe the protein-coding genes. Instead, a polycistronic transcription starts bi-directionally at divergent strand-switch sites in the genome [9]. The primary polycistronic RNAs are processed into single mature mRNAs by coupled 5′ trans-splicing and 3′ polyadenylation events, leading to the removal of the intergenic regions intervening the genes [10,11]. Since most genes lack introns [12], cis-splicing is a rare event. Due to the absence of transcription initiation control, trypanosomes heavily rely on post-transcriptional events to achieve and maintain differential gene expression [13].
Following the revolution generated by the discovery of microRNAs (miRNAs) in 1993 [14] and the small interfering RNA (siRNA) pathway in 1998 [15], it became clear that non-coding RNAs (ncRNAs) comprise a very efficient, quick, and economic strategy to regulate gene expression, especially for parasite organisms that are constantly exposed to dynamic environmental and host interactions. In addition, the capacity to quantify, characterize, and annotate ncRNAs has increased exponentially with the new generation of massive sequencing technologies, leading to the discovery of new classes of ncRNAs and improving their annotation [16]. Typically, ncRNAs are arbitrarily divided by size into small and long (shorter and longer than 200 nt, respectively) [17]. However, the range of mid-sized ncRNAs (50–400 nt) has been proposed as a distinctive group based on their structures and functional features [18]. Among the small ncRNAs, we can identify: miRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, tRNAs, and vtRNAs. Meanwhile, among the long ncRNAs, we can find: lincRNAs, lncRNAs, rRNAs, and telomerase RNA [16], the acronyms of which can be found in Table 1. The most studied small ncRNAs are the miRNAs and siRNAs, which are processed by Dicer and have functional roles in association with argonaute proteins (AGO). However, more recently, a new category of ncRNAs has bought the attention of the ncRNA field, namely the small RNA fragments derived from tRNAs (the most studied fragments): snoRNAs, snRNAs, and rRNA (Table 1) [19,20,21,22,23,24]. Actually, tRNA fragments and their regulatory functions have been identified across all kingdoms of life [25,26,27], pointing to the need for further studies to expand the knowledge of other ncRNA-derived fragments and their potential roles. The specific cleavage of canonical ncRNAs which produce fragments that functionally mimic miRNAs or directly interact with ribosomes, proteins (such as RNA binding proteins), or other RNAs to modulate gene expression [28,29,30]. Although these fragments were initially considered products of degradation and thus ignored in small RNA-seq analysis, the accumulated evidence supports the generation of fragments by specific processing of ncRNA [31,32,33]. However, many cases lack known processing factors or defined mechanisms of action, leaving an open field for research ahead.
Key and unique ncRNAs were discovered very early in trypanosomes. Pioneering studies regarding editing guide RNAs [34] and splice leader RNA [35] are examples. Most of the ncRNAs classes described later for eukaryotes were found and annotated (rRNAs, tRNAs, snoRNAs, snRNAs, telomerase RNA, and vault RNA) [36,37]. Recently, novel ncRNAs antisense regulators of mRNA translation were added to the catalog [30]. Regarding small RNA fragments derived from canonical ncRNAs, most of the work is focused on tsRNAs (for review see: [38,39]); however, other fragments derived from rRNAs, snoRNAs, and snRNAs were identified [40,41]. The aim of this review is to portray the ncRNA landscape in trypanosomatids, with a special emphasis on the existence and regulatory roles of small RNA fragments derived from functional RNA molecules.

Table 1.
Standardized names for the ncRNAs and derived fragments.
Table 1.
Standardized names for the ncRNAs and derived fragments.
| RNA Biotype (Acronym Used) | Derived Fragments/Small RNA (Acronym Used) | Canonical Process Involved |
|---|---|---|
| Transfer RNA (tRNA) | tRNA-derived RNA fragments (tsRNAs) [42] | Translation |
| Ribosomal RNA (rRNA) | rRNA-derived fragments (sdrRNA) [40] | Translation |
| small nuclear RNAs (snRNAs) | snRNA-derived fragments (snsRNA) [40] | RNA processing |
| Small nucleolar RNAs (snoRNAs) | snoRNA-derived fragments (sdRNAs) [40] | RNA processing |
| Long non-coding RNAs (lncRNAs) | lncRNAs-derived fragments (lncRNAs) [43] | Gene expression |
| Vault RNA (vtRNA) | Vault RNA-derived (svRNAs) [44] | Vault particle |
| Splice leader RNA (SL-RNA) | RNA processing | |
| Editing guide RNAs (gRNAs) | RNA Editing | |
| Small interfering RNAs (siRNAs) | Gene expression | |
| Telomerase template bearing RNA (TR) | Chromosome maintenance |
2. ncRNAs Annotations in the TriTryps Genomes
A hallmark in trypanosomatid biology was set in 2005 with the release of the first full genomic sequence for the three major trypanosomes of public health relevance: T. brucei [45], T. cruzi [46], and L. major [47] that were eventually named as the TriTryps, and the sequenced strains were set as reference (T. brucei TREU 927, T. cruzi CL-Brener, L. major Friedlin). The T. cruzi genome of the hybrid strain CL-Brener was the largest (55 Mb and 12,000 genes) compared with the T. brucei genome of 26 Mb and approximately 9068 genes, and the Leishmania genome of 32.8 Mb with 8311 genes [48]. A comparative analysis of these genomes revealed that a core of protein-coding genes was conserved and displayed a high degree of synteny among the three species, while each organism also displayed an important set of species-specific genes that coded for functions related to the infection process [48]. Since then, a comprehensive database (TriTrypDB) was formed and has been expanded to include genomes of different strains for the TriTryps, as well as many genomic sequences for other trypanosomatid species [49].
Although at first, gene annotations were scarce and not reliable, this has been improved over the years and protein-coding genes now share more comprehensive annotations for most genomes (Figure 1A). A different scenario can be observed for the non-coding RNAs (Figure 1B) when compared with protein-coding genes (Figure 1A). Some genomes have an extensive description of protein-coding genes while lacking non-protein-coding gene annotations. In species where there are numerous strains sequenced (see arrows for T. cruzi and T. brucei in Figure 1A,B), a wide range of a total number of annotated genes can be observed both for protein-coding and non-coding. However, reference strains show less dispersion in the total number of genes (inset plots of Figure 1A,B). This could be explained by the diverse library preparation and sequencing methods utilized that finally has an impact on the genome characterization and annotations. A good example is the application of third-generation sequencing technologies which led to an improvement in the annotation of multicopy gene families that were previously collapsed and underestimated [50]. As can be seen in Figure 1C, there is not a consistent and comprehensive annotation for the ncRNAs of the main 4 biotypes (tRNAs, snoRNAs, snRNAs, rRNAs), reflected by the difference in the total number of features defined in each class for each genome. This is particularly striking for the snoRNA, snRNA, and rRNA classes. Some genomes present annotations for the four ncRNA classes, while others present annotations for only one class. The tRNA class has the most widespread annotations amongst the genomes of different parasites, while the opposite is observed for the snoRNAs class. The heterogeneity of the RNA gene annotation across species together with the scarce annotation of regulatory ncRNAs precludes direct comparisons among species. Overall, the ncRNAs landscape in trypanosomatids requires efforts on the extension and homogenization of the annotations for most genomes to allow comparative and functional studies in this expanding field.
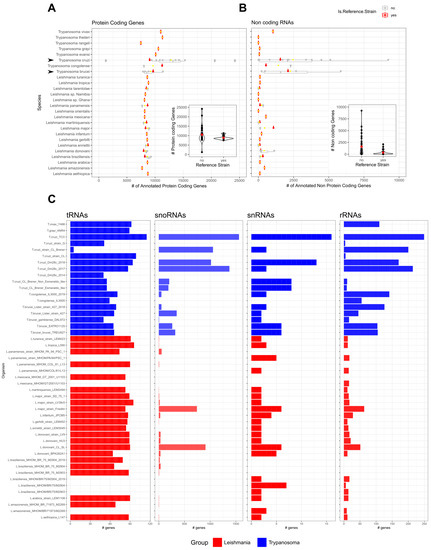
Figure 1.
Outlook of the non-coding genomic annotations available in TriTrypDB. All Leishmania spp. and Trypanosoma spp. genomic data was obtained from TritrypDB (Release 56) for protein-coding genes (A) and non-protein-coding genes (B). The inset plot presents the distribution of the number of genes annotated for all genomes grouped by reference and non-reference strains. For the main ncRNAs biotypes (tRNAs, snoRNAs, snRNAs, and rRNAs) the number of features annotated is presented in bar plots (C) for all genomes with a least 1 ncRNA annotation. Arrows point out species with large numbers of genome strains sequenced.
3. ncRNA Biotypes
This revision focuses on the regulatory roles of trypanosomatid ncRNAs. The constitutive RNAs involved in mRNA translation (tRNA, rRNA, snoRNA) are presented because of their regulatory potential, most of which is held by the small RNAs derived from them (Figure 2). Other housekeeping RNAs, such as the SL-RNA involved in trans-splicing and the gRNAs implicated in mitochondrial RNA editing are also described due to their prominence in trypanosomatids. Further, ncRNAs are discussed because of their recent involvement in novel regulatory processes (lncRNAs, vtRNA, anti-sense RNAs).
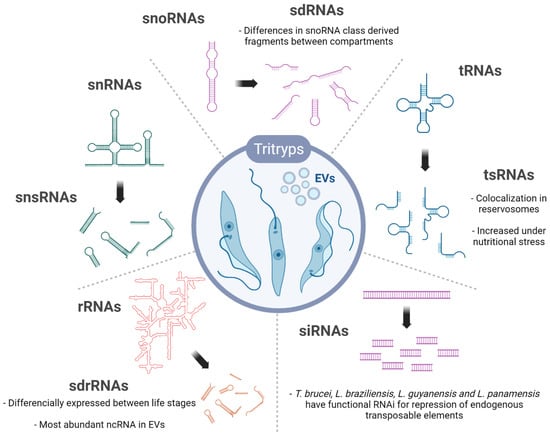
Figure 2.
Overview of the ncRNAs and their derived sncRNAs products identified in trypanosomatids. The sncRNAs represented were found in at least one trypanosomatids, either intracellular and/or in EVs (Extracellular vesicles). snsRNA, sdRNA, tsRNA, sdrRNA are the small RNAs derived from snRNA, snoRNA, tRNA and rRNA, respectively. Specific features or observations are commented on with the text. Created with BioRender.com (accessed on 13 May 2022).
3.1. tRNA and tsRNAs
Transfer RNAs (tRNAs) are one of the most abundant small non-coding RNA in the cell (5–10%), being essential components of the translation machinery delivering the amino acids to the ribosome to form polypeptides [25]. In trypanosomes, the tRNAs are transcribed by RNA polymerase III, and most of them are in clusters of 2–5 repeats in tandems (see review Shikha et al., 2019 for aspects of biogenesis and processing). Currently, the tRNA-derived RNA fragments (tsRNAs) are receiving great attention in RNA biology due to the functional implications that are revealed in all kingdoms of life [25,26,27]. The first report that revealed the presence of fragments derived from tRNA in trypanosomatids (epimastigotes of T. cruzi) was carried out by cloning and sequencing size selected RNAs (22 to 33 nts) [42]. Specifically, 87 clones (~26%) out of 348 correspond to tsRNAs, and ~90% of these fragments were derived from only three tRNAs isoacceptors: tRNA-Asp GUC, tRNA-Glu CUC, and tRNA-Glu UUC. These fragments were mapped to 5′ or 3′ halves of tRNAs (length ranging from 29 to 33 nt), the 5′ halves being the majority. Interestingly, FISH against some tRNA halves showed cytoplasmic granules colocalizing with TcPIWI in reservosomes [51]. Noteworthy is the exposure of late-stationary epimastigotes in increasing levels of extracellular vesicles carrying tsRNAs significantly enhanced by metacyclogenesis in axenic cultures [51]. Supporting the latter, Reifur et al., 2012, with a similar approach, showed that 63% of the cloned sequences correspond to the tsRNA of a median length of 33 nt in T. cruzi metacyclic forms [52]. Strikingly, in a metacyclic stage, most of the fragments (87%) are derived from the 3′ arm of the tRNAs (25% carried the 3′ CCA sequence), in contrast with the arm distribution observed in epimastigotes [42], which could imply a different processing between these two stages [52].
The small non-coding RNA (sncRNA) sequencing by NGS in T. cruzi expanded data of tsRNA in epimastigotes, where most of the tRNAs are precursors of tsRNAs [40], specifically 89% corresponded to 3′ halves with an average length of 38 nt, and 75% of them had the CCA extension. The disagreement with the 3’ tsRNA overrepresentation previously described by Garcia-Silva et al. is likely due to the different methods used. A population of shorter fragments derived from tRNAs (<25 nt) was also identified, but at a much lower frequency [40]. A study of sncRNAs derived from extracellular vesicles (EVs) of epimastigotes and metacyclic trypomastigotes revealed large differences between the proportion of tsRNAs (average length 32 nt) identified in these compartments [53]. Another study also confirmed the presence of tRNAs halves in T. cruzi secreted EVs by microscopy observations using gold-labeled 5′ arm tsRNAs and demonstrated that they could be effectively delivered into mammalian cells, inducing changes in the expression levels of specific genes in mammalian cells susceptible to infection [54]. Later, a comparative analysis of sncRNAs of the intracellular and the extracellular compartments in T. cruzi epimastigotes under nutritional stress confirmed the presence of fragments derived from multiple ncRNAs, including tRNAs [41]. This study found that tsRNAs comprise 45% of all the fragments of EVs, but 31% in their intracellular counterparts. Notably, two major populations of fragments were observed in the intracellular fraction with median lengths of 22 and 34 nts, while a single population with a median of 34 nts was identified in EVs. In a global description of sncRNAs (<30 nt), using small RNA-seq, in T. brucei (slender and procyclic forms), a striking difference in length distribution of sncRNAs derived from each stage of the parasite was described [55]. Key differences were observed in tsRNAs populations, where fragments derived from tRNA-Asp were nearly half of the tsRNA reads in slender form, while in the procyclic form this particular fragment drops down to 6% [55]. Remarkably, the opposite was observed for tsRNAs derived from tRNA-Glu (18% in slender and 74% in procyclic). In the case of leishmanias (L. donovani and L. braziliensis), sncRNA-seq of axenic amastigotes exosomes revealed that the most frequent tsRNAs are derived from tRNA-Asp, tRNA-Gln, tRNA-Glu, and tRNA-Leu. Additionally, there was no correlation with the amino acid usage, as previously observed in T. cruzi. Regardless, the most abundant tsRNAs derived from tsRNA-Asp and tsRNA-Gln were from the 5′ arm [56]. Recently, the study of co-purified sncRNA with ribosomes in the bloodstream and procyclic T. brucei parasites exposed to different environmental conditions revealed many tsRNAs in the ribosome and polysome fraction, most of them 5′ halves of 33 nt [28]. Remarkably, an augmented amount of tsRNAs in stationary phase parasites (maximum in the starved parasites) compared to exponential growth was observed [28]. Additionally, the global amount and the identity of the tsRNAs observed was different among the conditions evaluated. Furthermore, by using an in vitro translation extract of T. brucei, 3′ tRNA-Thr halves were able to stimulate translation by at least 20%, a result that was also confirmed in transfected parasites (Figure 3) [28].
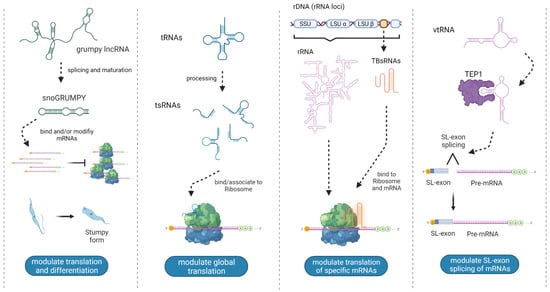
Figure 3.
Regulatory non-canonical ncRNAs with experimentally demonstrated roles reported in trypanosomatids. The illustrations represent the structure, processing and proposed mechanisms of action of the four ncRNAs reported so far [28,30,37,57]. Created with BioRender.com (accessed on 13 May 2022).
Overall, these observations indicate a unique tsRNA expression pattern favoring specific processing, sorting, or stabilization of tsRNAs in diverse conditions, compartments, or life stages of the parasites. Several reports show that different tsRNAs are usually synthesized/processed in normal parasite cell biology while under certain stress, which seems to be a common strategy to respond to the environmental changes. Despite tsRNAs being the most studied sncRNA fragments derived from canonical ncRNAs, further studies are needed to define the molecular mechanism and the precise relation to cell biology, stages, and pathology of the parasites.
3.2. snRNA and snsRNAs
The small nuclear RNAs (snRNAs) are highly conserved U-rich non-coding RNAs located in the nucleus, which are essential in the spliceosome complex, the machinery involved in pre-mRNA processing [58]. The major spliceosome complex which is responsible for the majority of the mRNA splicing events is formed by U1, U2, U4, U6, and U5 snRNAs. Additionally, some organisms possess a second splicing apparatus called the minor spliceosome complex (U11, U12, U4atac, U5 and U6atac snRNAs) which is involved in more specific splicing events [59,60]. The long polycistronically transcribed protein-coding genes (pre-mRNAs) of trypanosomatids are processed by coupling trans-splicing and polyadenylation to individual mRNAs by the spliceosome complex [61]. Particularly, for trans-splicing in trypanosomes, the SL RNA substitutes the function of the U1 snRNA in the complex [62].
Interestingly, small RNA fragments derived from specific locations of the snRNAs were found in eukaryotic model organism [19,63,64], and their possible functional role as a negative regulator of gene expression was hypothesized due to their association with AGO proteins [65,66]. In addition, snsRNAs derived from U2 snRNA were observed to increase in serum from patients with cancer [63,67]. However, more research is needed to determine snRNA-derived fragments’ role and functional mechanism in cell biology.
In trypanosomes, small RNA-seq experiments showed fragments derived from snRNAs with a median of 40 nts. However, 82.1% of the fragments were mapped to snRNAs U4 and U5 [40]. Other studies identify small RNA fragments derived from snRNAs in T. cruzi [41,51,53] and T. brucei [28,36,55], but they no further explore other aspects related to their production, characterization, or possible regulatory function in the studies. In leishmania, the small RNA-seq of exosomal RNA derived from L. donovani and L. braziliensis found snRNA-derived fragments and identified one snsRNA in the top 20 most abundant fragments in the libraries [56].
3.3. snoRNAs and sdRNAs
Small nucleolar RNAs (snoRNAs) are conserved ncRNAs with a length between 60 and 250 nts, encoded in introns of host genes or controlled by independent promoters. Their main function is to serve as a guide for specific base modifications in the ribosomal RNA. Briefly, the snoRNAs are classified into two groups depending on the modification they guide on rRNA: the box C/D snoRNAs serve as a guide for 2′-O-ribose methylation, and the box H/ACA snoRNAs serve as a guide for pseudo-uridylation [68]. While the trypanosomes C/D snoRNAs group has the eukaryotic consensus structure, the H/ACA snoRNAs have only one hairpin instead of two, and carry an AGA sequence box instead of the consensus ACA motif [36]. Trypanosomes snoRNAs carry out peculiar processing and modification of rRNAs (thoroughly reviewed in [69]). Indeed, the trypanosome rRNA holds specific roles in translational regulation and cycling between hosts (see below), which is reflected in the high number of snoRNAs in trypanosomatids genomes [69,70,71]. Recently, a great number of modifications guided by snoRNAs associated with rRNA processing were described and, strikingly, some of them are developmentally regulated [72,73]. Remarkably, the overexpression of some specific snoRNAs that guide modifications in the H69 domain of the trypanosome rRNA was associated with an increasing growth rate of the parasites [30].
The discovery of stably accumulating fragments derived from snoRNAs (termed sdRNAs) led to new regulatory functions besides the known canonical main function of this molecule [19,65,74,75,76]. Furthermore, it was evidenced that sdRNAs could act like miRNAs in many model organisms: plants, protozoa, mice, and humans [66,68,74,77,78]. Recently, in addition to the miRNA-like function, it was described that levels of sdRNAs were dependent on stress conditions and may interact with translating ribosomes, promoting protein synthesis suppression in yeast [79]. In T. cruzi, a sncRNA sequencing experiment revealed the presence of sdRNAs with a median length of 35 nts derived from both classes of snoRNAs (C/D box and H/ACA box) [40]. Then, the RNA sequencing of snoRNA-enriched RNPs of T. brucei (implemented for snoRNAs gene annotation) found small RNA fragments of 20–30 nts derived from snoRNAs. Small RNA fragments were identified for 80% of C/D box and 90% of H/ACA box snoRNAs; however, the authors speculate that derived fragments could appear as the result of low-level RNA degradation during preparation and/or manipulation [36]. Posteriorly, several small RNA-seq studies supported the presence of sdRNAs in whole-cell epimastigotes, trypomastigotes, and vesicles in trypanosomatids [28,41,51,53,55]. Intriguingly, Fernandez-Calero et al. remarked that despite the similar proportion of sdRNAs fragments in EVs and their intracellular counterparts, the size distribution of reads was profoundly different between both compartments. EVs sdRNAs presented three main populations with median peaks of 26, 28, and 34 nts, while intracellular sdRNAs showed two populations with median peaks of 33 and 35 nt [41]. However, the major differences between both compartments are associated with the class of snoRNAs from which the fragments derived. The EVs fraction showed 99% of sdRNAs derived from the H/ACA box class, while most of the intracellular sdRNAs derived from the C/D box class. In Leishmania, the small RNA-seq of exosomal RNA derived from L. donovani and L. braziliensis identified a small proportion of sdRNA (~1.5%) [56]. Overall, sdRNAs are identified in trypanosomatids, revealing differences in their levels, sizes, and classes among developmental stages and different subcellular compartments. Despite these observations, the role of the sdRNAs has not been addressed in trypanosomatids yet.
3.4. rRNA and sdrRNA
Ribosomal RNAs (rRNA) are non-coding RNAs transcribed by RNA polymerase I that are indispensable components of a large RNP complex, the ribosome, essential for the translation of all proteins in the cell. Indeed, ribosomal RNA is the most abundant RNA in the cell, reaching at least 80% of the RNA mass of the cell [80]. The processing of the ribosomal RNA in trypanosomes is unique, comprising two large (LSUα and LSUβ) and four small rRNA subunit fragments (srRNA 1,2,4, and 6), with snoRNAs acting as key players in this complex process [69]. The small ribosomal RNA-derived fragments (named both sdrRNA or rRFs) are usually considered by-products of degradation and are discarded or not considered in further small RNA-seq analysis. Nevertheless, several studies have accumulated convincing evidence presenting sdrRNAs as relevant translation regulators, reviewed in [21]. Certainly, the canonical rRNA processing involves different nucleases that might produce small RNA fragments during rRNA maturation that could eventually be functional molecules [19,81]. The systematic study of small RNA sequencing datasets found an extensive presence of sdrRNAs and demonstrated that these are mainly mapped to the rRNA coding regions in the sense orientation (64–70% mapped to large 28S rRNA subunit [82]). Furthermore, the sdrRNAs were found co-immunoprecipitated with Argonaute (AGO) proteins [65,66,82]. Remarkably, the functional role as translation regulators of some sdrRNAs was demonstrated in Neurospora [83] and human models [84]. In fact, Chen et al. demonstrated that RNAi knockout of one of the selected sdrRNAs induces apoptosis and inhibits cell proliferation in H1299 human cells [84]. More recently, meta-analysis of CLASH and AGO-PARCLIP datasets indicates that sdrRNAs are non-randomly generated and confirms their association with AGO proteins [24]. Indeed, this analysis led the authors to identify multiple non-randomly generated pairs of sdrRNAs and cellular transcripts, revealing several motifs of double-stranded binding regions. This observation led the authors to hypothesize a possible mechanism of unwinding mRNAs to regulate their translation [24].
In trypanosomes, a small RNA sequencing experiment of epimastigotes of T. cruzi revealed that 17% of reads derived from rRNA and showed a three-peak length distribution of 20, 33, and 46 nt [40]. Similarly, in a small RNA-seq study on infective metacyclic forms of T. cruzi, 25% of reads mapped to rRNAs [52]. Remarkably, Zheng et al. found sdrRNAs as one of the most differentially expressed sncRNAs (most of them ~20 nt length) in slender versus procyclic forms of T. brucei [55]. Furthermore, one of the studied sdrRNAs, was derived from a hairpin structure at the 3′ end of the 28S rRNA and was present in both stages but significantly increased in the procyclic form (14% of all reads). Furthermore, the authors speculate about a possible functional relevance of this sdrRNA based on the high conservation of its sequence among eight genetically related species [55]. Posteriorly, the small RNA sequencing analysis of RNAs associated with TcPIWI (an AGO/PIWI protein) of T. cruzi epimastigotes revealed that most of the sncRNAs (95%) identified were fragments derived from rRNA [85]. Then, the characterization of the small RNAs of EVs from two stages of T. cruzi (epimastigotes and metacyclic trypomastigotes) determined that the most abundant class in all samples were the sdrRNAs, supporting previous observations [51,53]. Remarkably, Evs secreted by T. cruzi (carrying large amounts of sdrRNAs) provoked changes in the gene expression level of many genes in host HeLa cells upon its incorporation [54]. Indeed, it was observed that T. cruzi trypomastigotes display a 5-fold increase in invasion over cells pre-incubated with trypomastigotes Evs [86]. The comparative analysis of EVs and their intracellular counterpart from T. cruzi epimastigotes under nutritional stress revealed that sdrRNAs were the most abundant class of small RNAs, being 46% and 58%, respectively [41]. However, the length of this sdrRNA was different between both compartments; intracellular distribution showed peaks at 19, 29, and 36 nt, while the 29 nt peak was not observed in EVs. Similar to previous observations, it was determined that sdrRNAs derive from specific locations of the rRNA genes, supporting the idea of site-specific processing rather than random degradation [41]. Additionally, some of these sdrRNAs seem to be differentially enriched between compartments, which could be explained by their specific processing or sorting/packaging [41]. Recently, sdrRNAs were observed in the small RNA interactome of ribosomes in bloodstream and procyclic T. brucei parasites exposed to different environmental conditions [28]. These fragments were the most abundant class of small RNAs in almost all the conditions tested, except for parasites submitted to starvation, where the tsRNA reached its maximum and sdrRNAs reached their minimum. In Leishmania, the small RNA sequencing of exosomes derived from early axenic amastigotes of L. donovani and L. braziliensis which revealed a 31% and 15% of reads mapping to rRNAs, respectively [56]. Remarkably, the peak of the size distribution of these sdrRNAs was different between L. donovani (39 nts) and L. braziliensis (52 nts). In contrast with previous observations, site-specific enrichments could not be determined for the fragments derived from rRNA genes, and 90% of the mappings were distributed in both 28S and 18S rRNA genes [56]. It is worth noting that the small RNA library preparation for this study involves CIP (Calf Intestinal Phosphatase) and TAP (tobacco acid pyrophosphatase) treatment, which would incorporate RNA fragments of different 5′-end nature which could explain the discrepancies mentioned. In addition, new evidence suggests that rRNA genes are also a source of larger (90–500 nt) functional ncRNAs in T. brucei [30]. The later study applied RNA sequencing on the post-ribosomal supernatant revealing the presence of novel developmentally-regulated ncRNAs [30]. These ncRNAs are located and processed from the pre-rRNAs’ internal transcribed spacer (ITS) and external transcribed regions (ETS). Remarkably, by using a UV cross-linking-ligation approach, it was described that these ncRNAs could interact (through base pairing) and move together with ribosomes to the cytoplasm, where they can interact and modulate the translation of a specific subset of mRNAs, evidenced by reporter and western blot assay [30] (Figure 3). Overall, ncRNAs derived from rRNAs can be observed in RNA-seq datasets in several trypanosomatids. Some fragments displayed differential expression over different developmental stages, subcellular compartments, or growth conditions, but these observations were not complemented with a functional approach in trypanosomatids.
3.5. siRNAs
In eukaryotes, RNA interference pathway (RNAi) is broadly conserved, playing key roles in defense against invading viruses, regulation of gene expression, programmed DNA rearrangements, and genome surveillance [87,88]. The RNAi pathway in protist, described for the first time in 1998 in a trypanosomatid [89], is largely dedicated to siRNA. Indeed, since the canonical miRNA biogenesis pathway is not present in Protista, neither precursors and mature miRNAs nor the microprocessor machinery (Drosha, Pasha) exist in trypanosomatids [90].
In the Trypanosoma genus, only T. brucei have functional RNAi, which involves a canonical AGO, two Dicers (one nuclear and one cytoplasmic), and two so-called RNA Interference Factors [91,92,93,94,95]. The main role of this system is the repression of endogenous transposable elements through 24–26 nt siRNAs (TbsiRNAs), most of which are derived from INGI and SLACS elements (~78%) [55,94,96,97]. Additional TbsiRNAs derived from pseudogenes formed between antisense pseudogene and their cognate coding genes (mostly VSG) or pseudogene–pseudogene pairs (mostly RHS), lead to a TbDCL1 (cytoplasmic Dicer) dependent gene repression [98]. Furthermore, TbDCL2 (nuclear Dicer) dependent TbsiRNA (21–27 nt) are known to derive from convergent polycistronic transcription units; yet, no associated gene regulatory activity is currently proven [97,99]. Among Leishmania genus, only the species from the early diverging Viannia subgenera (L. braziliensis, L. guyanensis, L. panamensis) have functional RNAi pathways comprising orthologues of AGO and Dicer genes [100,101]. The 20–25 nt siRNAs of L. braziliensis are mainly derived from SLACS and TATE transposable elements (~75%) [102]. Remarkably, these siRNAs were found in exosomes from L. braziliensis, and thus they might be involved in intercellular communication [56]. Overall, the T. brucei and L. braziliensis RNAi pathways show organism-specific diversifications with implications for the production of dsRNA and siRNA [102]. The size of the siRNA fragments, the modification of the 3´end, and the addition of non-coded Us varies among the groups. The exogenous manipulation of siRNA has quickly become an approach to downregulating gene expression in the RNAi competent trypanosomatids.
Strikingly, the siRNA pathway became non-functional in the remaining trypanosomatids [100,101]. The Leishmania subgenera (L. major, L. donovani, L. mexicata and L. tarentolae) contains only remnant, highly degenerated AGO1 pseudogenes, or lack one or more of the RNAi pathway genes [100,101]. Three evolutionary mechanisms have been proposed as drivers of the RNAi loss: avoidance of dsRNA viral infections strongly relying on the RNAi pathway, loss of mobile genetic elements make the RNAi less necessary, and direct phenotypic selection since this loss affects gene expression [100]. It is worth mentioning that the bilaterian host defense system for transposon silencing, known as the piRNA pathway, is absent in trypanosomatids, and so are the piwi-associated RNAs (piRNAs) [103].
3.6. vtRNA
Vault RNAs (vtRNAs) are a class of eukaryotic non-coding RNAs of generally 84 to 140 nt, transcribed by the RNA polymerase III, firstly identified for its association with the vault particle [104,105,106]. Although this particle is the largest known cellular ribonucleoprotein complex, its function remains unclear. Intriguingly, as seen in studies in vertebrates, only 5–20% of the total vtRNA transcripts are associated with the vault particle, and therefore, vtRNAs probably have additional independent roles [107,108,109].
The first evidence of the presence of vtRNA in trypanosomatids comes from high-throughput sequencing of small RNAs T. brucei [36]. The vtRNA identified here, TBsRNA-10, is more abundant in the bloodstream than the procyclic form parasites and is mostly localized in a non-nucleolar single focus in the nucleus. It was not until 2019 that TBsRNA-10 was proposed as a vtRNA in T. brucei, based on secondary structure prediction and sequence similarities of short stretches in the 5′ and 3′ regions that form the bulged terminal helix [37]. Furthermore, they demonstrated that TBsRNA-10 is transcribed by pol III and is very efficiently co-immunoprecipitated with a candidate TEP1 protein, the protein that associates with mammalian vtRNAs and is required for the vault particle assembly. The authors also looked for additional examples of vtRNA genes in trypanosomatids genomes and identified candidate vtRNA genes in all searched trypanosomatid species [37]. The expression of the L. braziliensis vtRNA of approximately 190 nt was validated and represents the longest known member of the vtRNA family in eukaryotes. Interestingly, it was demonstrated that the silencing of the vtRNA in the bloodstream form of T. brucei resulted in decreased production of the SL exon-enriched long RNA products (Figure 3). Since the only TROVE containing protein identified in trypanosomatids is TEP1, and no Ro protein or YRNAs has been found in their genomes and transcriptomes, it appears that the vtRNA holds a Y RNA-like function in RNA metabolism [37].
Overall, the recent annotation of vtRNAs opens the opportunity to reanalyze trypanosomatid RNA-seq datasets to study/describe patterns of expression for these ncRNAs. Furthermore, in other organisms, the vtRNAs were identified as precursors of derived sncRNAs (svRNAs) with miRNA-like functions but generated by a different pathway from canonical miRNAs biogenesis [44,110,111,112]. The presence of these svRNAs has not been addressed in trypanosomatids yet.
3.7. Editing gRNAs
RNA editing was first observed in trypanosomes when the insertion of four non-DNA encoded uridine nucleotides was described to restore the coding capacity of the mitochondrial cytochrome C oxidase subunit 2 gene, which presents a frameshift in the genomic sequence [113]. Later, uridine insertions and deletions were found to be frequent and abundant for mitochondrial mRNAs, and this process was confirmed as a post-transcriptional modification on the primary RNAs [114,115,116]. Nowadays, the term RNA editing is used to describe processes in which the sequence information present in an RNA molecule is altered post-transcriptionally, and it can be classified into two general types of events: substitution and insertion/deletion, both affecting all major cellular RNAs (mRNA, rRNA, tRNA) and described widely among eukaryotes [117].
Trypanosomes are distinguished by the presence of a single mitochondrion containing a kinetoplast nucleoid composed of a bipartite mitochondrial genome and histone-like basic proteins. The kinetoplast DNA (kDNA) consists of a densely packed network of interlinked circular DNA molecules of two kinds, referred to as maxicircles and minicircles. Maxicircles are equivalent to the mitochondrial genome in most organisms, thus encoding ribosomal RNAs, two ribosomal proteins, and 16 respiratory-related genes, mostly cryptogenes, meaning they need to be edited in order to restore a protein-coding capacity. This editing is abundant as there can be hundreds of editions for each mitochondrial mRNA. The edition is directed by hundreds of guide-RNAs mostly encoded by the multicopy (102–103) ~1 kb DNA molecules named minicircles [118].
Editing guide RNAs (gRNAs) are small RNAs (50–60 nt) with a 5′ triphosphate and a 3′ oligo-U tail [119]. T3/T7-like RNA polymerases bidirectionally transcribes these RNAs to generate overlapping sense and antisense precursors. Then, these precursors undergo three sequential and well-studied 3′-end processing events involving degradation and uridylation in the mitochondrial 3′ proccesome (MPsome). These events lead to a mature U-tailed sense gRNA that can be incorporated into the editing complex (RESC, RNA-editing substrate-binding complex) and proceed to the editing pathway [120,121]. The model for edition mediated by gRNAs was described based on the perfect sequence complementarity between the gRNA and the mature mRNAs within the edited region [34]. Thus, guide RNAs specify the position and the number of uridines inserted or deleted by hybridizing to pre-mRNA and forming a series of mismatches [122,123]. First, an initial interaction between the gRNA and the pre-edited mRNA is established through a short (10–12 nt) region of complementarity, referred to as the gRNA’s 5′ anchor region. An imperfect duplex is then formed with pre-mRNA and the remaining gRNA sequence that can result in a loop out of single-stranded uridines in mRNA (then a deletion site) or a loop of purine nucleotides in gRNA (forming an insertion site). Later, the first unpaired nucleotide in the mRNA next to the 5′ anchor duplex is cleaved, triggering either a 3′-5′ degradation for the uridines exposed in the mRNA, resulting in a deletion event, or the filling of the single-stranded gap in the duplex, resulting in an insertion event [123].
The nearly complete repertoire of gRNA molecules was determined by RNA sequencing for the procyclic stage of T. brucei [124], describing 642 major sequence classes. Later, the bloodstream from gRNA transcriptome was described using the same protocol [125], and it was noted that RNA editing was developmentally regulated in T. brucei. A different approach was carried out in L. Tarentolae [126], using the PacBio platform to provide unambiguous complete kDNA minicircle sequences. Then, two size-selected mitochondrial-enriched RNA libraries (small under 200 nt, and large over 200 nt) were mapped to the minicircles sequences to verify the transcription of the gRNA genes. This study revealed that both libraries display mappings over both strands of the entire minicircle kDNA, suggesting that transcription is bidirectional and involves almost the full minicircle sequence, which eventually is processed to produce the mature gRNAs molecules. gRNAs comprise a relatively abundant small ncRNA in trypanosomatids that should stay enclosed in the mitochondria. In this context, non-canonical regulatory functions are less likely to emerge and be reported for these molecules.
3.8. Splice Leader RNA
Trans-splicing involves the precise joining of two distinct RNA molecules, and this spliceosomal processing was firstly described in T. brucei when a common 39-nucleotide sequence, named spliced leader (SL), was found at the 5′-end of different variant surface glycoprotein transcripts [35,127]. Later, trans-splicing was involved in the processing all protein-coding genes in trypanosomatids [128], while traditional cis-splicing only takes place in a couple of genes [12,129]. Since trypanosomatid protein-coding genes are transcribed into large polycistronic RNAs, mRNA maturation differs from most eukaryotes [9]. In this scenario, the SL addition coupled with polyadenylation dissects the polycistronic transcripts while providing the unusual cap (namely, cap4) to the mature mRNAs [130]. The leader sequence comes from a small RNA (SL-RNA) of approximately 140 (141 nt for in T. brucei) with a short half-life. The SL-RNA is transcribed from multiple loci in the genome, being this gene is the only one under the control of conventional RNA polymerase II promoter sites in trypanosomes [131,132]. Up to now, there are no extra regulatory functions or modifications for the SL-RNA, despite being one of the more abundantly produced RNAs in trypanosomes.
3.9. Telomerase RNA
Since the initial discovery of telomerase as the solution to the end-replication problem of eukaryotic chromosomes, much progress has been made in the identification of telomerase core components: the catalytic telomerase reverse transcriptase (TERT) and intrinsic template-bearing telomerase RNA (TR) [133]. From the TERT/TR complex, the TERT domain structures are conserved across different species, but the TR molecules vary greatly in sequence, size, and structure (from ~150 nt in ciliates to ~1.800 nt in P. falciparum [134].
Besides common features, T. brucei TR (TbTR) has several unique attributes that suggest a mechanistically different process of telomere maintenance [135]. In T. brucei, the TbTR of 900 nt RNA (longer than vertebrates TR) is encoded by a single-copy gene transcribed by pol II [134,136]. Despite their length, the TbTR presents a unique ciliate-like shorter stem-loop at the 3´ end of the template instead of a typical PK domain [133,134]. In addition, instead of the H/ACA box domain higher eukaryotes TRs, the TbTR presents a consensus sequence of snoRNA C/D box, also present in other kinetoplastids [135,136,137]. This C/D box is generated between the 5´ and 3´ ends of the TbTR and has been shown to bind the C/D snoRNA core protein NOP58 [136]. Recent evidence revealed stage differences in the TbTR structure that influences telomerase activity and consistent population doubling and parasite infection [138]. Since the TR sequence is variable among species, it is interesting to unravel whether TR has non-canonical functions in kinetoplastids and how TR sequence differences affect them. It is worth noting that in model organisms, TR non-canonical functions have been discovered, including a role in developmental myelopoiesis, cell fitness and apoptosis [139,140].
3.10. lncRNAs
Long non-coding RNAs (lncRNAs) are ncRNAs longer than 200 nt that do not code for functional proteins. Several thousand of this RNA biotype are transcribed by pol II in eukaryotic genomes; as an example, the human genome may contain more than 100.000 lncRNA genes [141]. Even though there is debate about the biological function of most lncRNA, the relevance of several of these molecules is well established in the literature (see [142] for a recent review). In trypanosomatids, these molecules were initially described by Kolev et al. in one of the first reports of using RNA-seq in T. brucei [143]. However, evidence of a functional role in these organisms has only recently emerged. Studies estimate several hundred lncRNAs [57,144], mostly present in previously unannotated regions of the T. brucei genome. Only a few have been experimentally characterized with proposed roles in translation regulation [30] parasite development and present a variable expression in culture conditions compared to wild type cells [57,144]. Interestingly, the work by Guegan et al. shows that a lncRNA in T. brucei plays a key role in promoting differentiation to the stumpy form. This effect is mediated by the processing of the lncRNA into a snoRNA, snoGRUMPY, which regulates translation of its mRNA targets [57] (Figure 3). This work points to the significance of this biotype in trypanosomatid biology, a subject that has been mostly unexplored in this group of parasites.
4. Conclusions and Perspectives
The complex trypanosomatid ncRNA landscape displays the presence of both eukaryotic canonical ncRNA and some abundant family-specific ncRNA (SL-RNA and editing gRNA) (Table 1). However, as was shown in Figure 1, a comprehensive and detailed ncRNA annotation for most trypanosomatid genomes is still needed, which will allow for better and more extensive comparative and functional studies. The advent of high throughput RNA-sequencing technologies reveals the presence of small RNA fragments derived from canonical ncRNA and the discovery of new developmentally regulated ncRNAs. Nevertheless, the different RNA-seq studies show some discordant observations that could rely on actual biological differences or be associated with the different library and sequencing approaches used (size-selection, RNA 5′/3′-end modifications, and sequencing technologies) (Table 2). These experimental designs naturally impact the RNA subsetting included in the finally sequenced library. Much effort is currently leading to developing new strategies to reduce biases associated with library and sequencing approaches, and to capture a broader range of ncRNAs present in the cell.

Table 2.
Small RNA transcriptomic studies in Trypanosomes. RNA-sequencing studies that quantify endogenous small RNA fragments (produced without fragmentation steps in the library protocol) are included. RNA biotypes identified in the manuscript are listed and the mainly focused ones are highlighted in bold letters. The 5´/3´-RNA end modifications described in the library preparation are presented in parenthesis. PNK—T4 Polynucleotide Kinase, CIP—Calf Intestinal Phosphatase, TAP—tobacco acid pyrophosphatase.
Small RNA fragments derived from several canonical ncRNAs (tsRNAs, snsRNAs, sdRNAs, and sdrRNAs) were identified as the most commonly studied trypanosomatids, displaying changes in their levels among developmental stages and diverse growing conditions, as well as their distribution over subcellular compartments. These observations demand further studies for characterization and functional analysis with special emphasis on defining possible mechanisms of action. The latter could expose new layers of post-transcriptional gene expression regulation, a crucial level of control in the biology of these parasites. Furthermore, considering that in trypanosomatids the miRNA pathway is absent, while the siRNA pathway is present only in some lineages of the phylogenetic tree, functional characterization may be challenging, but could offer interesting new insights into the mechanism of action.
Funding
This research was funded by the Comisión Sectorial de Investigación Científica (CSIC-UDELAR, Uruguay) (Research Grants M.A.D. I+D 2020 # 566) and the Programa para el Desarrollo de las Ciencias Básicas (PEDECIBA-MEC, Uruguay, Annual Aliquots and Equipment Aid Programs). Postgraduate student fellowships for C.O.-R., S.C., and J.M.T. were provided by the Facultad de Medicina and the Facultad de Ciencias of the University of the Republic (UDELAR), the Comision Academica de Posgrado (CAP-CSIC-UDELAR) and the Agencia Nacional de Investigación e Innovación (ANII).
Acknowledgments
The authors would like to acknowledge the continued and strong support of Beatriz Garat and her expertise in molecular biology of Chagas disease throughout years of research in Uruguay.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Adl, S.M.; Bass, D.; Lane, C.E.; Lukeš, J.; Schoch, C.L.; Smirnov, A.; Agatha, S.; Berney, C.; Brown, M.W.; Burki, F.; et al. Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes. J. Eukaryot. Microbiol. 2019, 66, 4. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J.; Burki, F. Progress towards the Tree of Eukaryotes. Curr. Biol. 2019, 29, R808–R817. [Google Scholar] [CrossRef] [PubMed]
- Cavalcanti, D.P.; de Souza, W. The Kinetoplast of Trypanosomatids: From Early Studies of Electron Microscopy to Recent Advances in Atomic Force Microscopy. Scanning 2018, 2018, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Büscher, P.; Cecchi, G.; Jamonneau, V.; Priotto, G. Human African Trypanosomiasis. Lancet 2017, 390, 2397–2409. [Google Scholar] [CrossRef]
- Pérez-Molina, J.A.; Molina, I. Chagas Disease. Lancet 2018, 391, 82–94. [Google Scholar] [CrossRef]
- Burza, S.; Croft, S.L.; Boelaert, M. Leishmaniasis. Lancet 2018, 392, 951–970. [Google Scholar] [CrossRef]
- Barrett, M.P.; Burchmore, R.J.S.; Stich, A.; Lazzari, J.O.; Frasch, A.C.; Cazzulo, J.J.; Krishna, S. The Trypanosomiases. Lancet 2003, 362, 1469–1480. [Google Scholar] [CrossRef]
- Giordani, F.; Morrison, L.J.; Rowan, T.G.; De Koning, H.P.; Barrett, M.P. The Animal Trypanosomiases and Their Chemotherapy: A Review. Parasitology 2016, 143, 1862–1889. [Google Scholar] [CrossRef]
- Martínez-Calvillo, S.; Vizuet-de-Rueda, J.C.; Florencio-Martínez, L.E.; Manning-Cela, R.G.; Figueroa-Angulo, E.E. Gene Expression in Trypanosomatid Parasites. J. Biomed. Biotechnol. 2010, 2010, 525241. [Google Scholar] [CrossRef]
- LeBowitz, J.H.; Smith, H.Q.; Rusche, L.; Beverley, S.M. Coupling of Poly(A) Site Selection and Trans-Splicing in Leishmania. Genes Dev. 1993, 7, 996–1007. [Google Scholar] [CrossRef]
- Berberof, M.; Vanhamme, L.; Pays, E. Trypanosoma Brucei: A Preferential Splicing at the Inverted Polyadenylation Site of the VSG MRNA Provides Further Evidence for Coupling between Trans-Splicing and Polyadenylation. Exp. Parasitol. 1995, 80, 563–567. [Google Scholar] [CrossRef] [PubMed]
- Mair, G.; Shi, H.; Li, H.; Djikeng, A.; Aviles, H.O.; Bishop, J.R.; Falcone, F.H.; Gavrilescu, C.; Montgomery, J.L.; Santori, M.I.; et al. A New Twist in Trypanosome RNA Metabolism: Cis-Splicing of Pre-MRNA. RNA 2000, 6, 163. [Google Scholar] [CrossRef] [PubMed][Green Version]
- De Gaudenzi, J.G.; Noé, G.; Campo, V.A.; Frasch, A.C.; Cassola, A. Gene Expression Regulation in Trypanosomatids. Essays Biochem. 2011, 51, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis Elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Vickers, K.C.; Roteta, L.A.; Hucheson-Dilks, H.; Han, L.; Guo, Y. Mining Diverse Small RNA Species in the Deep Transcriptome. Trends Biochem. Sci. 2015, 40, 4–7. [Google Scholar] [CrossRef]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA Maps Reveal New RNA Classes and a Possible Function for Pervasive Transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef]
- Boivin, V.; Faucher-Giguère, L.; Scott, M.; Abou-Elela, S. The Cellular Landscape of Mid-Size Noncoding RNA. Wiley Interdiscip. Rev. RNA 2019, 10, e1530. [Google Scholar] [CrossRef]
- Chen, C.J.; Heard, E. Small RNAs Derived from Structural Non-Coding RNAs. Methods 2013, 63, 76–84. [Google Scholar] [CrossRef]
- Wajahat, M.; Bracken, C.P.; Orang, A. Emerging Functions for SnoRNAs and SnoRNA-Derived Fragments. Int. J. Mol. Sci. 2021, 22, 10193. [Google Scholar] [CrossRef]
- Lambert, M.; Benmoussa, A.; Provost, P. Small Non-Coding RNAs Derived From Eukaryotic Ribosomal RNA. Non-Coding RNA 2019, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Martens-Uzunova, E.S.; Olvedy, M.; Jenster, G. Beyond MicroRNA—Novel RNAs Derived from Small Non-Coding RNA and Their Implication in Cancer. Cancer Lett. 2013, 340, 201–211. [Google Scholar] [CrossRef]
- Pan, Q.; Han, T.; Li, G. Novel Insights into the Roles of TRNA-Derived Small RNAs. RNA Biol. 2021, 18, 2157–2167. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Grigoriev, A. Computational Meta-Analysis of Ribosomal RNA Fragments: Potential Targets and Interaction Mechanisms. Nucleic Acids Res. 2021, 49, 4085–4103. [Google Scholar] [CrossRef]
- Li, S.; Xu, Z.; Sheng, J. TRNA-Derived Small RNA: A Novel Regulatory Small Non-Coding RNA. Genes 2018, 9, 246. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G. TRNA-Derived Small RNAs: New Players in Genome Protection against Retrotransposons. RNA Biol. 2018, 15, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Fuchs, G.; Wang, S.; Wei, W.; Zhang, Y.; Park, H.; Roy-Chaudhuri, B.; Li, P.; Xu, J.; Chu, K.; et al. A Transfer-RNA-Derived Small RNA Regulates Ribosome Biogenesis. Nature 2017, 552, 57–62. [Google Scholar] [CrossRef]
- Fricker, R.; Brogli, R.; Luidalepp, H.; Wyss, L.; Fasnacht, M.; Joss, O.; Zywicki, M.; Helm, M.; Schneider, A.; Cristodero, M.; et al. A TRNA Half Modulates Translation as Stress Response in Trypanosoma Brucei. Nat. Commun. 2019, 10, 118. [Google Scholar] [CrossRef]
- Pircher, A.; Gebetsberger, J.; Polacek, N. Ribosome-Associated NcRNAs: An Emerging Class of Translation Regulators. RNA Biol. 2014, 11, 1335–1339. [Google Scholar] [CrossRef]
- Rajan, K.S.; Doniger, T.; Cohen-Chalamish, S.; Rengaraj, P.; Galili, B.; Aryal, S.; Unger, R.; Tschudi, C.; Michaeli, S. Developmentally Regulated Novel Non-Coding Anti-Sense Regulators of MRNA Translation in Trypanosoma Brucei. iScience 2020, 23, 101780. [Google Scholar] [CrossRef]
- Pagès, A.; Dotu, I.; Pallarès-Albanell, J.; Martí, E.; Guigó, R.; Eyras, E. The Discovery Potential of RNA Processing Profiles. Nucleic Acids Res. 2018, 46, e15. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhou, T.; Chen, Q. Exploring the Expanding Universe of Small RNAs. Nat. Cell Biol. 2022, 24, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ender, C.; Meister, G.; Moore, P.S.; Chang, Y.; John, B. Extensive Terminal and Asymmetric Processing of Small RNAs from RRNAs, SnoRNAs, SnRNAs, and TRNAs. Nucleic Acids Res. 2012, 40, 6787–6799. [Google Scholar] [CrossRef]
- Blum, B.; Bakalara, N.; Simpson, L. A Model for RNA Editing in Kinetoplastid Mitochondria: “Guide” RNA Molecules Transcribed from Maxicircle DNA Provide the Edited Information. Cell 1990, 60, 189–198. [Google Scholar] [CrossRef]
- Boothroyd, J.C.; Cross, G.A.M. Transcripts Coding for Variant Surface Glycoproteins of Trypanosoma Brucei Have a Short, Identical Exon at Their 5’ End. Gene 1982, 20, 281–289. [Google Scholar] [CrossRef]
- Michaeli, S.; Doniger, T.; Gupta, S.K.; Wurtzel, O.; Romano, M.; Visnovezky, D.; Sorek, R.; Unger, R.; Ullu, E. RNA-Seq Analysis of Small RNPs in Trypanosoma Brucei Reveals a Rich Repertoire of Non-Coding RNAs. Nucleic Acids Res. 2012, 40, 1282–1298. [Google Scholar] [CrossRef]
- Kolev, N.G.; Rajan, K.S.; Tycowski, K.T.; Toh, J.Y.; Shi, H.; Lei, Y.; Michaeli, S.; Tschudi, C. The Vault RNA of Trypanosoma Brucei Plays a Role in the Production of Trans-Spliced MRNA. J. Biol. Chem. 2019, 294, 15559–15574. [Google Scholar] [CrossRef]
- Shikha, S.; Brogli, R.; Schneider, A.; Polacek, N. TRNA Biology in Trypanosomes. Chimia 2019, 73, 395–405. [Google Scholar] [CrossRef]
- Peng, R.; Santos, H.J.; Nozaki, T. Transfer RNA-Derived Small RNAs in the Pathogenesis of Parasitic Protozoa. Genes 2022, 13, 286. [Google Scholar] [CrossRef]
- Franzén, O.; Arner, E.; Ferella, M.; Nilsson, D.; Respuela, P.; Carninci, P.; Hayashizaki, Y.; Åslund, L.; Andersson, B.; Daub, C.O. The Short Non-Coding Transcriptome of the Protozoan Parasite Trypanosoma Cruzi. PLoS Negl. Trop. Dis. 2011, 5, e1283. [Google Scholar] [CrossRef]
- Fernandez-Calero, T.; Garcia-Silva, R.; Pena, A.; Robello, C.; Persson, H.; Rovira, C.; Naya, H.; Cayota, A. Profiling of Small RNA Cargo of Extracellular Vesicles Shed by Trypanosoma Cruzi Reveals a Specific Extracellular Signature. Mol. Biochem. Parasitol. 2015, 199, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Silva, M.R.; Frugier, M.; Tosar, J.P.; Correa-Dominguez, A.; Ronalte-Alves, L.; Parodi-Talice, A.; Rovira, C.; Robello, C.; Goldenberg, S.; Cayota, A. A Population of TRNA-Derived Small RNAs Is Actively Produced in Trypanosoma Cruzi and Recruited to Specific Cytoplasmic Granules. Mol. Biochem. Parasitol. 2010, 171, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Jalali, S.; Jayaraj, G.G.; Scaria, V. Integrative Transcriptome Analysis Suggest Processing of a Subset of Long Non-Coding RNAs to Small RNAs. Biol. Direct 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Persson, H.; Kvist, A.; Vallon-Christersson, J.; Medstrand, P.; Borg, A.; Rovira, C. The Non-Coding RNA of the Multidrug Resistance-Linked Vault Particle Encodes Multiple Regulatory Small RNAs. Nat. Cell Biol. 2009, 11, 1268–1271. [Google Scholar] [CrossRef]
- Berriman, M.; Ghedin, E.; Hertz-Fowler, C.; Blandin, G.; Renauld, H.; Bartholomeu, D.C.; Lennard, N.J.; Caler, E.; Hamlin, N.E.; Haas, B.; et al. The Genome of the African Trypanosome Trypanosoma Brucei. Science 2005, 309, 416–422. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, N.M.; Myler, P.J.; Bartholomeu, D.C.; Nilsson, D.; Aggarwal, G.; Tran, A.-N.N.; Ghedin, E.; Worthey, E.A.; Delcher, A.L.; Blandin, G.; et al. The Genome Sequence of Trypanosoma Cruzi, Etiologic Agent of Chagas Disease. Science 2005, 309, 409–415. [Google Scholar] [CrossRef]
- Ivens, A.C.; Peacock, C.S.; Worthey, E.A.; Murphy, L.; Aggarwal, G.; Berriman, M.; Sisk, E.; Rajandream, M.-A.; Adlem, E.; Aert, R.; et al. The Genome of the Kinetoplastid Parasite, Leishmania Major. Science 2005, 309, 436–442. [Google Scholar] [CrossRef]
- El-Sayed, N.M.; Myler, P.J.; Blandin, G.; Berriman, M.; Crabtree, J.; Aggarwal, G.; Caler, E.; Renauld, H.; Worthey, E.A.; Hertz-Fowler, C.; et al. Comparative Genomics of Trypanosomatid Parasitic Protozoa. Science 2005, 309, 404–409. [Google Scholar] [CrossRef]
- Aslett, M.; Aurrecoechea, C.; Berriman, M.; Brestelli, J.; Brunk, B.P.; Carrington, M.; Depledge, D.P.; Fischer, S.; Gajria, B.; Gao, X.; et al. TriTrypDB: A Functional Genomic Resource for the Trypanosomatidae. Nucleic Acids Res. 2010, 38, D457–D462. [Google Scholar] [CrossRef]
- Berná, L.; Rodriguez, M.; Chiribao, M.L.; Parodi-Talice, A.; Pita, S.; Rijo, G.; Alvarez-Valin, F.; Robello, C. Expanding an Expanded Genome: Long-Read Sequencing of Trypanosoma Cruzi. Microb. Genom. 2018, 4, e0001779. [Google Scholar] [CrossRef]
- Garcia-Silva, M.R.; Cura Das Neves, R.F.; Cabrera-Cabrera, F.; Sanguinetti, J.; Medeiros, L.C.; Robello, C.; Naya, H.; Fernandez-Calero, T.; Souto-Padron, T.; De Souza, W.; et al. Extracellular Vesicles Shed by Trypanosoma Cruzi Are Linked to Small RNA Pathways, Life Cycle Regulation, and Susceptibility to Infection of Mammalian Cells. Parasitol. Res. 2014, 113, 285–304. [Google Scholar] [CrossRef] [PubMed]
- Reifur, L.; Garcia-Silva, M.R.; Poubel, S.B.; Alves, L.R.; Arauco, P.; Buiar, D.K.; Goldenberg, S.; Cayota, A.; Dallagiovanna, B. Distinct Subcellular Localization of TRNA-Derived Fragments in the Infective Metacyclic Forms of Trypanosoma Cruzi. Mem. Inst. Oswaldo Cruz 2012, 107, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Bayer-Santos, E.; Lima, F.M.; Ruiz, J.C.; Almeida, I.C.; Da Silveira, J.F. Characterization of the Small RNA Content of Trypanosoma Cruzi Extracellular Vesicles. Mol. Biochem. Parasitol. 2014, 193, 71–74. [Google Scholar] [CrossRef]
- Garcia-Silva, M.R.; Cabrera-Cabrera, F.; Cura das Neves, R.F.; Souto-Padrón, T.; de Souza, W.; Cayota, A. Gene Expression Changes Induced by Trypanosoma Cruzi Shed Microvesicles in Mammalian Host Cells: Relevance of TRNA-Derived Halves. Biomed Res. Int. 2014, 2014, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.L.; Wen, Y.Z.; Yang, J.H.; Liao, J.Y.; Shao, P.; Xu, H.; Zhou, H.; Wen, J.Z.; Lun, Z.R.; Ayala, F.J.; et al. Comparative Transcriptome Analysis of Small Noncoding RNAs in Different Stages of Trypanosoma Brucei. RNA 2013, 19, 863–875. [Google Scholar] [CrossRef]
- Lambertz, U.; Oviedo Ovando, M.E.; Vasconcelos, E.J.R.; Unrau, P.J.; Myler, P.J.; Reiner, N.E. Small RNAs Derived from TRNAs and RRNAs Are Highly Enriched in Exosomes from Both Old and New World Leishmania Providing Evidence for Conserved Exosomal RNA Packaging. BMC Genomics 2015, 16. [Google Scholar] [CrossRef]
- Guegan, F.; Rajan, K.S.; Bento, F.; Pinto-Neves, D.; Sequeira, M.; Gumińska, N.; Mroczek, S.; Dziembowski, A.; Cohen-Chalamish, S.; Doniger, T.; et al. A Long Noncoding RNA Promotes Parasite Differentiation in African Trypanosomes. Sci. Adv. 2022, 8, eabn2706. [Google Scholar] [CrossRef]
- Hoskins, A.A.; Moore, M.J. The Spliceosome: A Flexible, Reversible Macromolecular Machine. Trends Biochem. Sci. 2012, 37, 179–188. [Google Scholar] [CrossRef]
- Turunen, J.J.; Niemelä, E.H.; Verma, B.; Frilander, M.J. The Significant Other: Splicing by the Minor Spliceosome. Wiley Interdiscip. Rev. RNA 2013, 4, 61–76. [Google Scholar] [CrossRef]
- Morais, P.; Adachi, H.; Yu, Y.-T. Spliceosomal SnRNA Epitranscriptomics. Front. Genet. 2021, 12, 652129. [Google Scholar] [CrossRef]
- Ambrósio, D.L.; Badjatia, N.; Günzl, A. The Spliceosomal PRP19 Complex of Trypanosomes. Mol. Microbiol. 2015, 95, 885–901. [Google Scholar] [CrossRef] [PubMed]
- Preußer, C.; Jaé, N.; Bindereif, A. MRNA Splicing in Trypanosomes. Int. J. Med. Microbiol. 2012, 302, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Kuhlmann, J.D.; Wimberger, P.; Wilsch, K.; Fluck, M.; Suter, L.; Brunner, G. Increased Level of Circulating U2 Small Nuclear RNA Fragments Indicates Metastasis in Melanoma Patients. Clin. Chem. Lab. Med. 2015, 53, 605–611. [Google Scholar] [CrossRef]
- Mesquita-Ribeiro, R.; Fort, R.S.; Rathbone, A.; Farias, J.; Lucci, C.; James, V.; Sotelo-Silveira, J.; Duhagon, M.A.; Dajas-Bailador, F. Distinct Small Non-Coding RNA Landscape in the Axons and Released Extracellular Vesicles of Developing Primary Cortical Neurons and the Axoplasm of Adult Nerves. RNA Biol. 2021, 18, 832–855. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, A.M.; Ando, Y.; de Hoon, M.J.L.; Tomaru, Y.; Suzuki, H.; Hayashizaki, Y.; Daub, C.O. Deep-Sequencing of Human Argonaute-Associated Small RNAs Provides Insight into MiRNA Sorting and Reveals Argonaute Association with RNA Fragments of Diverse Origin. RNA Biol. 2011, 8, 158–177. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.W.; Pillman, K.A.; Anderson, M.L.; Lawrence, D.M.; Toubia, J.; Goodall, G.J.; Bracken, C.P. Assessing the Gene Regulatory Properties of Argonaute-Bound Small RNAs of Diverse Genomic Origin. Nucleic Acids Res. 2015, 43, 470–481. [Google Scholar] [CrossRef]
- Baraniskin, A.; Nöpel-Dünnebacke, S.; Ahrens, M.; Jensen, S.G.; Zöllner, H.; Maghnouj, A.; Wos, A.; Mayerle, J.; Munding, J.; Kost, D.; et al. Circulating U2 Small Nuclear RNA Fragments as a Novel Diagnostic Biomarker for Pancreatic and Colorectal Adenocarcinoma. Int. J. Cancer 2013, 132. [Google Scholar] [CrossRef]
- Scott, M.S.; Ono, M. From SnoRNA to MiRNA: Dual Function Regulatory Non-Coding RNAs. Biochimie 2011, 93, 1987–1992. [Google Scholar] [CrossRef]
- Rajan, K.S.; Chikne, V.; Decker, K.; Waldman Ben-Asher, H.; Michaeli, S. Unique Aspects of RRNA Biogenesis in Trypanosomatids. Trends Parasitol. 2019, 35, 778–794. [Google Scholar] [CrossRef]
- Rajan, K.S.; Doniger, T.; Cohen-Chalamish, S.; Chen, D.; Semo, O.; Aryal, S.; Saar, E.G.; Chikne, V.; Gerber, D.; Unger, R.; et al. Pseudouridines on Trypanosoma Brucei Spliceosomal Small Nuclear RNAs and Their Implication for RNA and Protein Interactions. Nucleic Acids Res. 2019, 47, 7633–7647. [Google Scholar] [CrossRef]
- Chikne, V.; Rajan, K.S.; Shalev-Benami, M.; Decker, K.; Cohen-Chalamish, S.; Madmoni, H.; Biswas, V.K.; Gupta, S.K.; Doniger, T.; Unger, R.; et al. Small Nucleolar RNAs Controlling RRNA Processing in Trypanosoma Brucei. Nucleic Acids Res. 2019, 47, 2609–2629. [Google Scholar] [CrossRef] [PubMed]
- Rajan, K.S.; Zhu, Y.; Adler, K.; Doniger, T.; Cohen-Chalamish, S.; Srivastava, A.; Shalev-Benami, M.; Matzov, D.; Unger, R.; Tschudi, C.; et al. The Large Repertoire of 2’-O-Methylation Guided by C/D SnoRNAs on Trypanosoma Brucei RRNA. RNA Biol. 2020, 17, 1018–1039. [Google Scholar] [CrossRef]
- Barth, S.; Shalem, B.; Hury, A.; Tkacz, I.D.; Liang, X.H.; Uliel, S.; Myslyuk, I.; Doniger, T.; Salmon-Divon, M.; Unger, R.; et al. Elucidating the Role of C/D SnoRNA in RRNA Processing and Modification in Trypanosoma Brucei. Eukaryot. Cell 2008, 7, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Ender, C.; Krek, A.; Friedländer, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A Human SnoRNA with MicroRNA-like Functions. Mol. Cell 2008, 32, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Taft, R.J.; Glazov, E.A.; Lassmann, T.; Hayashizaki, Y.; Carninci, P.; Mattick, J.S. Small RNAs Derived from SnoRNAs. RNA 2009, 15, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Kawaji, H.; Nakamura, M.; Takahashi, Y.; Sandelin, A.; Katayama, S.; Fukuda, S.; Daub, C.O.; Kai, C.; Kawai, J.; Yasuda, J.; et al. Hidden Layers of Human Small RNAs. BMC Genomics 2008, 9, 157. [Google Scholar] [CrossRef]
- Brameier, M.; Herwig, A.; Reinhardt, R.; Walter, L.; Gruber, J. Human Box C/D SnoRNAs with MiRNA like Functions: Expanding the Range of Regulatory RNAs. Nucleic Acids Res. 2011, 39, 675–686. [Google Scholar] [CrossRef]
- Saraiya, A.A.; Wang, C.C. SnoRNA, a Novel Precursor of MicroRNA in Giardia Lamblia. PLoS Pathog. 2008, 4, e1000224. [Google Scholar] [CrossRef]
- Mleczko, A.M.; Machtel, P.; Walkowiak, M.; Wasilewska, A.; Pietras, P.J.; Bąkowska-Żywicka, K. Levels of SdRNAs in Cytoplasm and Their Association with Ribosomes Are Dependent upon Stress Conditions but Independent from SnoRNA Expression. Sci. Rep. 2019, 9, 18397. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Lee, E.S. Non-Coding RNA: What Is Functional and What Is Junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef]
- Zywicki, M.; Bakowska-Zywicka, K.; Polacek, N. Revealing Stable Processing Products from Ribosome-Associated Small RNAs by Deep-Sequencing Data Analysis. Nucleic Acids Res. 2012, 40, 4013–4024. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Zhou, B.; Zhang, F.; Tu, Y.; Hu, Y.; Zhang, B.; Zhai, Q. Profiling and Identification of Small RDNA-Derived RNAs and Their Potential Biological Functions. PLoS ONE 2013, 8, e56842. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-C.; Chang, S.-S.; Choudhary, S.; Aalto, A.P.; Maiti, M.; Bamford, D.H.; Liu, Y. QiRNA Is a New Type of Small Interfering RNA Induced by DNA Damage. Nature 2009, 459, 274–277. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Sun, Y.; Yang, X.; Wu, Z.; Guo, K.; Niu, X.; Wang, Q.; Ruan, J.; Bu, W.; Gao, S. Two Featured Series of RRNA-Derived RNA Fragments (RRFs) Constitute a Novel Class of Small RNAs. PLoS ONE 2017, 12, e0176458. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Silva, M.R.; Sanguinetti, J.; Cabrera-Cabrera, F.; Franzén, O.; Cayota, A. A Particular Set of Small Non-Coding RNAs Is Bound to the Distinctive Argonaute Protein of Trypanosoma Cruzi: Insights from RNA-Interference Deficient Organisms. Gene 2014, 538, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Trocoli Torrecilhas, A.C.; Tonelli, R.R.; Pavanelli, W.R.; da Silva, J.S.; Schumacher, R.I.; de Souza, W.; E Silva, N.C.; de Almeida Abrahamsohn, I.; Colli, W.; Manso Alves, M.J. Trypanosoma Cruzi: Parasite Shed Vesicles Increase Heart Parasitism and Generate an Intense Inflammatory Response. Microbes Infect. 2009, 11, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Shabalina, S.A.; Koonin, E.V. Origins and Evolution of Eukaryotic RNA Interference. Trends Ecol. Evol. 2008, 23, 578. [Google Scholar] [CrossRef]
- Cerutti, H.; Casas-Mollano, J.A. On the Origin and Functions of RNA-Mediated Silencing: From Protists to Man. Curr. Genet. 2006, 50, 81–99. [Google Scholar] [CrossRef]
- Ngô, H.; Tschudi, C.; Gull, K.; Ullu, E. Double-Stranded RNA Induces MRNA Degradation in Trypanosoma Brucei. Proc. Natl. Acad. Sci. USA 1998, 95, 14687–14692. [Google Scholar] [CrossRef]
- Bråte, J.; Neumann, R.S.; Fromm, B.; Haraldsen, A.A.B.; Tarver, J.E.; Suga, H.; Donoghue, P.C.J.; Peterson, K.J.; Ruiz-Trillo, I.; Grini, P.E.; et al. Unicellular Origin of the Animal MicroRNA Machinery. Curr. Biol. 2018, 28, 3288–3295.e5. [Google Scholar] [CrossRef]
- Barnes, R.L.; Shi, H.; Kolev, N.G.; Tschudi, C.; Ullu, E. Comparative Genomics Reveals Two Novel RNAi Factors in Trypanosoma Brucei and Provides Insight into the Core Machinery. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef]
- Durand-Dubief, M.; Bastin, P. TbAGO1, an Argonaute Protein Required for RNA Interference, Is Involved in Mitosis and Chromosome Segregation in Trypanosoma Brucei. BMC Biol. 2003, 1, 2. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Chamond, N.; Tschudi, C.; Ullu, E. Selection and Characterization of RNA Interference-Deficient Trypanosomes Impaired in Target MRNA Degradation. Eukaryot. Cell 2004, 3, 1445–1453. [Google Scholar] [CrossRef][Green Version]
- Patrick, K.L.; Shi, H.; Kolev, N.G.; Ersfeld, K.; Tschudi, C.; Ullu, E. Distinct and Overlapping Roles for Two Dicer-like Proteins in the RNA Interference Pathways of the Ancient Eukaryote Trypanosoma Brucei. Proc. Natl. Acad. Sci. USA 2009, 106, 17933–17938. [Google Scholar] [CrossRef]
- Shi, H.; Djikeng, A.; Tschudi, C.; Ullu, E. Argonaute Protein in the Early Divergent Eukaryote Trypanosoma Brucei: Control of Small Interfering RNA Accumulation and Retroposon Transcript Abundance. Mol. Cell. Biol. 2004, 24, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Djikeng, A.; Shi, H.; Tschudi, C.; Ullu, E. RNA Interference in Trypanosoma Brucei: Cloning of Small Interfering RNAs Provides Evidence for Retroposon-Derived 24-26-Nucleotide RNAs. RNA 2001, 7, 1522–1530. [Google Scholar]
- Tschudi, C.; Shi, H.; Franklin, J.B.; Ullu, E. Small Interfering RNA-Producing Loci in the Ancient Parasitic Eukaryote Trypanosoma Brucei. BMC Genom. 2012, 13, 427. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.-Z.; Zheng, L.-L.; Liao, J.-Y.; Wang, M.-H.; Wei, Y.; Guo, X.-M.; Qu, L.-H.; Ayala, F.J.; Lun, Z.-R. Pseudogene-Derived Small Interference RNAs Regulate Gene Expression in African Trypanosoma Brucei. Proc. Natl. Acad. Sci. USA 2011, 108, 8345–8350. [Google Scholar] [CrossRef]
- Reynolds, D.; Hofmeister, B.T.; Cliffe, L.; Alabady, M.; Siegel, T.N.; Schmitz, R.J.; Sabatini, R. Histone H3 Variant Regulates RNA Polymerase II Transcription Termination and Dual Strand Transcription of SiRNA Loci in Trypanosoma Brucei. PLOS Genet. 2016, 12, e1005758. [Google Scholar] [CrossRef]
- Lye, L.-F.; Owens, K.; Shi, H.; Murta, S.M.F.; Vieira, A.C.; Turco, S.J.; Tschudi, C.; Ullu, E.; Beverley, S.M. Retention and Loss of RNA Interference Pathways in Trypanosomatid Protozoans. PLoS Pathog. 2010, 6, e1001161. [Google Scholar] [CrossRef]
- Matveyev, A.V.; Alves, J.M.P.; Serrano, M.G.; Lee, V.; Lara, A.M.; Barton, W.A.; Costa-Martins, A.G.; Beverley, S.M.; Camargo, E.P.; Teixeira, M.M.G.; et al. The Evolutionary Loss of RNAi Key Determinants in Kinetoplastids as a Multiple Sporadic Phenomenon. J. Mol. Evol. 2017, 84, 104. [Google Scholar] [CrossRef] [PubMed]
- Atayde, V.D.; Shi, H.; Franklin, J.B.; Carriero, N.; Notton, T.; Lye, L.F.; Owens, K.; Beverley, S.M.; Tschudi, C.; Ullu, E. The Structure and Repertoire of Small Interfering RNAs in Leishmania (Viannia) Braziliensis Reveal Diversification in the Trypanosomatid RNAi Pathway. Mol. Microbiol. 2013, 87, 580–593. [Google Scholar] [CrossRef]
- Grimson, A.; Srivastava, M.; Fahey, B.; Woodcroft, B.J.; Chiang, H.R.; King, N.; Degnan, B.M.; Rokhsar, D.S.; Bartel, D.P. Early Origins and Evolution of MicroRNAs and Piwi-Interacting RNAs in Animals. Nature 2008, 455, 1193–1197. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.L.; Rome, L.H. Isolation and Characterization of a Novel Ribonucleoprotein Particle: Large Structures Contain a Single Species of Small RNA. J. Cell Biol. 1986, 103, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Vilalta, A.; Kickhoefer, V.A.; Rome, L.H.; Johnson, D.L. The Rat Vault RNA Gene Contains a Unique RNA Polymerase III Promoter Composed of Both External and Internal Elements That Function Synergistically. J. Biol. Chem. 1994, 269, 29752–29759. [Google Scholar] [CrossRef]
- Kedersha, N.L.; Heuser, J.E.; Chugani, D.C.; Rome, L.H. Vaults. III. Vault Ribonucleoprotein Particles Open into Flower-like Structures with Octagonal Symmetry. J. Cell Biol. 1991, 112, 225–235. [Google Scholar] [CrossRef]
- Kickhoefer, V.A.; Rajavel, K.S.; Scheffer, G.L.; Dalton, W.S.; Scheper, R.J.; Rome, L.H. Vaults Are Up-Regulated in Multidrug-Resistant Cancer Cell Lines. J. Biol. Chem. 1998, 273, 8971–8974. [Google Scholar] [CrossRef] [PubMed]
- Nandy, C.; Mrázek, J.; Stoiber, H.; Grässer, F.A.; Hüttenhofer, A.; Polacek, N. Epstein-Barr Virus-Induced Expression of a Novel Human Vault RNA. J. Mol. Biol. 2009, 388, 776–784. [Google Scholar] [CrossRef]
- van Zon, A.; Mossink, M.H.; Schoester, M.; Scheffer, G.L.; Scheper, R.J.; Sonneveld, P.; Wiemer, E.A. Multiple Human Vault RNAs. Expression and Association with the Vault Complex. J. Biol. Chem. 2001, 276, 37715–37721. [Google Scholar] [CrossRef]
- Sajini, A.A.; Choudhury, N.R.; Wagner, R.E.; Bornelöv, S.; Selmi, T.; Spanos, C.; Dietmann, S.; Rappsilber, J.; Michlewski, G.; Frye, M. Loss of 5-Methylcytosine Alters the Biogenesis of Vault-Derived Small RNAs to Coordinate Epidermal Differentiation. Nat. Commun. 2019, 10, 2550. [Google Scholar] [CrossRef]
- Miñones-Moyano, E.; Friedländer, M.R.; Pallares, J.; Kagerbauer, B.; Porta, S.; Escaramís, G.; Ferrer, I.; Estivill, X.; Martí, E. Upregulation of a Small Vault RNA (SvtRNA2-1a) Is an Early Event in Parkinson Disease and Induces Neuronal Dysfunction. RNA Biol. 2013, 10, 1093–1106. [Google Scholar] [CrossRef] [PubMed]
- Fort, R.S.; Garat, B.; Sotelo-Silveira, J.R.; Duhagon, M.A. VtRNA2-1/Nc886 Produces a Small RNA That Contributes to Its Tumor Suppression Action through the MicroRNA Pathway in Prostate Cancer. Non-Coding RNA 2020, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Benne, R.; Van Den Burg, J.; Brakenhoff, J.P.J.; Sloof, P.; Van Boom, J.H.; Tromp, M.C. Major Transcript of the Frameshifted CoxII Gene from Trypanosome Mitochondria Contains Four Nucleotides That Are Not Encoded in the DNA. Cell 1986, 46, 819–826. [Google Scholar] [CrossRef]
- Shaw, J.M.; Feagin, J.E.; Stuart, K.; Simpson, L. Editing of Kinetoplastid Mitochondrial MRNAs by Uridine Addition and Deletion Generates Conserved Amino Acid Sequences and AUG Initiation Codons. Cell 1988, 53, 401–411. [Google Scholar] [CrossRef]
- van der Spek, H.; van den Burg, J.; Croiset, A.; van den Broek, M.; Sloof, P.; Benne, R. Transcripts from the Frameshifted MURF3 Gene from Crithidia Fasciculata Are Edited by U Insertion at Multiple Sites. EMBO J. 1988, 7, 2509–2514. [Google Scholar] [CrossRef]
- Harris, M.E.; Moore, D.R.; Hajduk, S.L. Addition of Uridines to Edited RNAs in Trypanosome Mitochondria Occurs Independently of Transcription. J. Biol. Chem. 1990, 265, 11368–11376. [Google Scholar] [CrossRef]
- Gray, M.W. Evolutionary Origin of RNA Editing. Biochemistry 2012, 51, 5235–5242. [Google Scholar] [CrossRef]
- Shlomai, J. The Structure and Replication of Kinetoplast DNA. Curr. Mol. Med. 2004, 4, 623–647. [Google Scholar] [CrossRef]
- Blum, B.; Simpson, L. Guide RNAs in Kinetoplastid Mitochondria Have a Nonencoded 3’ Oligo(U) Tail Involved in Recognition of the Preedited Region. Cell 1990, 62, 391–397. [Google Scholar] [CrossRef]
- Madina, B.R.; Kumar, V.; Metz, R.; Mooers, B.H.M.; Bundschuh, R.; Cruz-Reyes, J. Native Mitochondrial RNA-Binding Complexes in Kinetoplastid RNA Editing Differ in Guide RNA Composition. RNA 2014, 20, 1142–1152. [Google Scholar] [CrossRef][Green Version]
- Aphasizhev, R.; Sbicego, S.; Peris, M.; Jang, S.H.; Aphasizheva, I.; Simpson, A.M.; Rivlin, A.; Simpson, L. Trypanosome Mitochondrial 3′ Terminal Uridylyl Transferase (TUTase): The Key Enzyme in U-Insertion/Deletion RNA Editing. Cell 2002, 108, 637–648. [Google Scholar] [CrossRef][Green Version]
- Seiwert, S.D.; Heidmann, S.; Stuart, K. Direct Visualization of Uridylate Deletion In Vitro Suggests a Mechanism for Kinetoplastid RNA Editing. Cell 1996, 84, 831–841. [Google Scholar] [CrossRef][Green Version]
- Aphasizhev, R.; Aphasizheva, I. Mitochondrial RNA Editing in Trypanosomes: Small RNAs in Control. Biochimie 2014, 100, 125. [Google Scholar] [CrossRef] [PubMed]
- Koslowsky, D.; Sun, Y.; Hindenach, J.; Theisen, T.; Lucas, J. The Insect-Phase GRNA Transcriptome in Trypanosoma Brucei. Nucleic Acids Res. 2014, 42, 1873–1886. [Google Scholar] [CrossRef] [PubMed]
- Kirby, L.E.; Sun, Y.; Judah, D.; Nowak, S.; Koslowsky, D. Analysis of the Trypanosoma Brucei EATRO 164 Bloodstream Guide RNA Transcriptome. PLoS Negl. Trop. Dis. 2016, 10, e0004793. [Google Scholar] [CrossRef] [PubMed]
- Simpson, L.; Douglass, S.M.; Lake, J.A.; Pellegrini, M.; Li, F. Comparison of the Mitochondrial Genomes and Steady State Transcriptomes of Two Strains of the Trypanosomatid Parasite, Leishmania Tarentolae. PLoS Negl. Trop. Dis. 2015, 9, e0003841. [Google Scholar] [CrossRef]
- van der Ploeg, L.H.T.; Liu, A.Y.C.; Michels, P.A.M.; de Lange, T.; Borst, P.; Majumder, H.K.; Weber, H.; Veeneman, G.H.; Boom, J. Van RNA Splicing Is Required to Make the Messenger RNA for a Variant Surface Antigen in Trypanosomes. Nucleic Acids Res. 1982, 10, 3591–3604. [Google Scholar] [CrossRef]
- Agabian, N. Trans Splicing of Nuclear Pre-MRNAs. Cell 1990, 61, 1157–1160. [Google Scholar] [CrossRef]
- Tan, T.H.P.; Pach, R.; Crausaz, A.; Ivens, A.; Schneider, A. TRNAs in Trypanosoma Brucei: Genomic Organization, Expression, and Mitochondrial Import. Mol. Cell. Biol. 2002, 22, 3707–3717. [Google Scholar] [CrossRef]
- Perry, K.L.; Watkins, K.P.; Agabian, N. Trypanosome MRNAs Have Unusual “Cap 4” Structures Acquired by Addition of a Spliced Leader. Proc. Natl. Acad. Sci. USA 1987, 84, 8190–8194. [Google Scholar] [CrossRef]
- Gilinger, G.; Bellofatto, V. Trypanosome Spliced Leader RNA Genes Contain the First Identified RNA Polymerase II Gene Promoter in These Organisms. Nucleic Acids Res. 2001, 29, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Milhausen, M.; Nelson, R.G.; Sather, S.; Selkirk, M.; Agabian, N. Identification of a Small Rna Containing the Trypanosome Spliced Leader: A Donor of Shared 5′ Sequences of Trypanosomatid MRNAs? Cell 1984, 38, 721–729. [Google Scholar] [CrossRef]
- Podlevsky, J.D.; Chen, J.J.L. Evolutionary Perspectives of Telomerase RNA Structure and Function. RNA Biol. 2016, 13, 720–732. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, R.; Sanford, S.; Basu, S.; Park, M.; Pandya, U.M.; Li, B.; Chakrabarti, K. A Trans-Spliced Telomerase RNA Dictates Telomere Synthesis in Trypanosoma Brucei. Cell Res. 2013, 23, 537–551. [Google Scholar] [CrossRef]
- Davis, J.A.; Chakrabarti, K. Telomerase Ribonucleoprotein and Genome Integrity—An Emerging Connection in Protozoan Parasites. Wiley Interdiscip. Rev. RNA 2021, 1–20. [Google Scholar] [CrossRef]
- Gupta, S.K.; Kolet, L.; Doniger, T.; Biswas, V.K.; Unger, R.; Tzfati, Y.; Michaeli, S. The Trypanosoma Brucei Telomerase RNA (TER) Homologue Binds Core Proteins of the C/D SnoRNA Family. FEBS Lett. 2013, 587, 1399–1404. [Google Scholar] [CrossRef]
- Logeswaran, D.; Li, Y.; Podlevsky, J.D.; Chen, J.J.-L. Monophyletic Origin and Divergent Evolution of Animal Telomerase RNA. Mol. Biol. Evol. 2021, 38, 215–228. [Google Scholar] [CrossRef]
- Dey, A.; Chakrabarti, K. Current Perspectives of Telomerase Structure and Function in Eukaryotes with Emerging Views on Telomerase in Human Parasites. Int. J. Mol. Sci. 2018, 19, 333. [Google Scholar] [CrossRef]
- Alcaraz-Pérez, F.; García-Castillo, J.; García-Moreno, D.; López-Muñoz, A.; Anchelin, M.; Angosto, D.; Zon, L.I.; Mulero, V.; Cayuela, M.L. A Non-Canonical Function of Telomerase RNA in the Regulation of Developmental Myelopoiesis in Zebrafish. Nat. Commun. 2014, 5, 3228. [Google Scholar] [CrossRef]
- Raghunandan, M.; Decottignies, A. The Multifaceted HTR Telomerase RNA from a Structural Perspective: Distinct Domains of HTR Differentially Interact with Protein Partners to Orchestrate Its Telomerase-Independent Functions. BioEssays 2021, 43, 2100099. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, J.; Li, Y.; Song, T.; Wu, Y.; Fang, S.; Bu, D.; Li, H.; Sun, L.; Pei, D.; et al. NONCODEV6: An Updated Database Dedicated to Long Non-Coding RNA Annotation in Both Animals and Plants. Nucleic Acids Res. 2021, 49, D165–D171. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Kolev, N.G.; Franklin, J.B.; Carmi, S.; Shi, H.; Michaeli, S.; Tschudi, C. The Transcriptome of the Human Pathogen Trypanosoma Brucei at Single-Nucleotide Resolution. PLOS Pathog. 2010, 6, e1001090. [Google Scholar] [CrossRef]
- Kim, H.C.; Jolly, E.R. LncRNAs Are Differentially Expressed between Wildtype and Cell Line Strains of African Trypanosomes. Non-Coding RNA 2022, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Madej, M.J.; Alfonzo, J.D.; Hüttenhofer, A. Small NcRNA Transcriptome Analysis from Kinetoplast Mitochondria of Leishmania Tarentolae. Nucleic Acids Res. 2007, 35, 1544–1554. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Silva, M.R.; Tosar, J.P.; Frugier, M.; Pantano, S.; Bonilla, B.; Esteban, L.; Serra, E.; Rovira, C.; Robello, C.; Cayota, A. Cloning, Characterization and Subcellular Localization of a Trypanosoma Cruzi Argonaute Protein Defining a New Subfamily Distinctive of Trypanosomatids. Gene 2010, 466, 26–35. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).